Abstract
The Polycomb-repressive complex 2 (PRC2) is important for maintenance of stem cell pluripotency and suppression of cell differentiation by promoting histone H3 lysine 27 trimethylation (H3K27me3) and transcriptional repression of differentiation genes. Here we show that the tumour-suppressor protein BRCA1 interacts with the Polycomb protein EZH2 in mouse embryonic stem (ES) and human breast cancer cells. The BRCA1-binding region in EZH2 overlaps with the noncoding RNA (ncRNA)-binding domain, and BRCA1 expression inhibits the binding of EZH2 to the HOTAIR ncRNA. Decreased expression of BRCA1 causes genome-wide EZH2 re-targeting and elevates H3K27me3 levels at PRC2 target loci in both mouse ES and human breast cancer cells. BRCA1 deficiency blocks ES cell differentiation and enhances breast cancer migration and invasion in an EZH2-dependent manner. These results reveal that BRCA1 is a key negative modulator of PRC2 and that loss of BRCA1 inhibits ES cell differentiation and enhances an aggressive breast cancer phenotype by affecting PRC2 function.
Keywords: BRCA1, breast cancer, embryonic stem cell, epigenetic gene silencing, PRC2
Introduction
The evolutionally conserved Polycomb group (PcG) proteins play pivotal roles in transcription repression by forming chromatin-modifying complexes termed Polycomb-repressive complexes (PRCs) such as PRC1 and PRC2 (Kennison, 1995; Schwartz and Pirrotta, 2007; Simon and Kingston, 2009; Margueron and Reinberg, 2011). PRC2 contains four core subunits of EZH2, SUZ12, EED and RbAp46/48 in humans or E(z), Su(z)12, esc and Nurf55 in Drosophila. EZH2 is an enzymatic subunit of PRC2 that contains a SET domain catalysing histone H3 lysine 27 trimethylation (H3K27me3; Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Muller et al, 2002). This chromatin mark is commonly associated with silencing of differentiation genes and has key roles in developmental patterning in organisms ranging from plants and flies to humans. PRC2 is important for embryonic stem (ES) cell self-renewal, cell-fate decisions and generation of inducible pluripotent stem (iPS) cells (Boyer et al, 2006; Lee et al, 2006; Ezhkova et al, 2009; Onder et al, 2012). EZH2 is often overexpressed or mutated in human solid and hematologic tumours and has been implicated in cancer progression (Varambally et al, 2002; Bracken et al, 2003; Kleer et al, 2003; Morin et al, 2010).
Mounting evidence indicates that the potent function of EZH2 is tightly regulated in diverse biological contexts. EZH2 expression is regulated by RB/p130-E2F pathways, microRNA 101 and sex hormones (Bracken et al, 2003; Varambally et al, 2008; Bohrer et al, 2010). Noncoding RNAs (ncRNAs) such as HOTAIR and XIST bind to and facilitate PRC2 occupancy on chromatin (Rinn et al, 2007; Zhao et al, 2008; Gupta et al, 2010). The protein kinase AKT phosphorylates EZH2 at serine 21, which inhibits PRC2-mediated H3K27me3 and gene silencing but activates Polycomb-independent oncogenic functions of EZH2 (Cha et al, 2005; Xu et al, 2012). Cyclin-dependent kinase 1 (CDK1) and CDK2 phosphorylate EZH2 at threonine 350 (T350) and 487 (T487) residues and regulate PRC2 recruitment to its target loci (Chen et al, 2010; Kaneko et al, 2010; Wei et al, 2011). T350 phosphorylation also enhances EZH2 binding to HOTAIR and XIST ncRNAs and accelerates turnover of phosphorylated EZH2 (Kaneko et al, 2010; Wu and Zhang, 2011). Moreover, the Jumonji C-containing protein Jarid2 has been shown to interact with and regulate PRC2 enzymatic activity and target gene occupancy in ES cells (Peng et al, 2009; Shen et al, 2009; Landeira et al, 2010; Li et al, 2010; Pasini et al, 2010).
Breast cancer susceptibility gene 1 (BRCA1) was identified as a hereditary cancer susceptibility gene (Miki et al, 1994). Heterozygous germline mutations of BRCA1 predispose women to breast and ovarian cancer with a lifetime risk up to 85% by age 70 years (King et al, 2003; Wooster and Weber, 2003). Strikingly, the majority of breast cancers arising in BRCA1 mutation carriers are of the basal-like phenotype with unique characteristics such as lack of estrogen receptor (ER) but expression of basal or myoepithelial cell markers cytokeratins (CKs) CK5/6, CK14 and CK17 (Foulkes et al, 2003; Sorlie et al, 2003; Foulkes, 2004; Lakhani et al, 2005). It has therefore been suggested that BRCA1 tumours are originated from basal-like stem cells (Foulkes, 2004; Vassilopoulos et al, 2008). To date, a large number of biochemical activities have been linked to BRCA1 function, which include DNA damage response and repair (Scully et al, 1997; Cortez et al, 1999), transcription regulation (Chapman and Verma, 1996; Harkin et al, 1999), chromatin remodelling (Bochar et al, 2000), heterochromatin maintenance (Zhu et al, 2011), among others. Moreover, the NH2-terminal RING domain and the COOH-terminal BRCT domain have been identified as two major functional domains of BRCA1 (Huen et al, 2010). However, how BRCA1 regulates cell differentiation and how BRCA1 deregulation contributes to development of aggressive phenotypes of basal-like breast tumours remain elusive. In the present study we report that BRCA1 binds to EZH2 and modulates its functions in regulation of transcription repression, ES cell differentiation and breast cancer cell migration and invasion.
Results
BRCA1 associates with the PRC2 complex in ES and breast cancer cells
It has been shown previously that expression of PRC2 target genes are downregulated in ER-negative, basal-like breast cancers in comparison to other subtypes of breast cancer (Ben-Porath et al, 2008). Given that the vast majority of breast cancers arising from BRCA1 mutation carriers are of basal-like phenotype (Foulkes et al, 2003; Sorlie et al, 2003), we hypothesized that BRCA1 functions as a negative regulator of PRC2 and that loss of BRCA1 enhances PRC2 function. To test this hypothesis, we assessed whether BRCA1 protein interacts with PRC2. The 293T cells were transfected with myc-tagged EZH2 and cell lysates were subjected to co-immunoprecipitation (co-IP). As expected, the PRC2 core components SUZ12 and EED were detected in the anti-myc immunoprecipitants (Figure 1A). BRCA1 protein was also immunoprecipitated by anti-myc antibody, but not nonspecific IgG (Figure 1A). In contrast, BRCA1-associated RING domain protein 1 (BARD1) was not found in this complex (Figure 1A), suggesting a specific interaction between EZH2 and BRCA1. Neither BARD1 overexpression nor T350 phosphorylation on EZH2 affected the interaction (Supplementary Figure S1A and B). Next, we examined if endogenous BRCA1 interacts with PRC2. Ethidium bromide was added in co-IP assays to exclude DNA/chromatin as a potential mediator of protein interaction as reported previously (van der Vlag and Otte, 1999; Pasini et al, 2008). Endogenous BRCA1 protein along with endogenous PRC2 proteins Suz12 and Eed were immunoprecipitated by anti-Ezh2 from R1 mouse ES cells (Figure 1B). Reciprocally, endogenous PRC2 proteins Ezh2 and Suz12 were immunoprecipitated by anti-Brca1 antibody from AB2.2 mouse ES cells (Figure 1C). Parallel interactions were detected in human breast cancer cell line MCF7 (Figure 1D). Together, both endogenous and ectopically expressed BRCA1 and EZH2 interact with each other in various cell types.
Figure 1.
BRCA1 interacts with the PRC2 complex in ES and breast cancer cells. (A) Co-IP of myc–EZH2 with BRCA1 and PRC2 components SUZ12 and EED in 293T cells. The 293T cells transfected with myc-tagged EZH2 were subjected to Co-IP with anti-myc antibody. #Nonspecific proteins reacted with the antibodies used. The proportion (%) of each protein involved in the interaction is determined by dividing the immunoblotting band density of immunoprecipitant with the band density of input (1%), and shown in the right side of each blot. (B) Co-IP of endogenous Brca1 and PRC2 proteins by anti-Ezh2 antibody in R1 mouse ES cells. #Nonspecific proteins reacted with anti-BRCA1 antibody. (C) Co-IP of endogenous Ezh2, Suz12 and Brca1 proteins by anti-Brca1 antibody in AB2.2 mouse ES cells. (D) Co-IP of endogenous EZH2, SUZ12 and BRCA1 proteins by anti-BRCA1 antibody in MCF7 cells. (E) Effect of RNase A treatment on BRCA1–EZH2 interaction. The 293T cells transfected with HA–BRCA1 and myc–EZH2 and cell lysates were treated with or without RNase A prior to co-IP with anti-myc antibody (left) and RT–PCR for the presence of GAPDH mRNA.
Source data for this figure is available on the online supplementary information page.
Identification of region(s) in BRCA1 and EZH2 responsible for their interaction
We performed glutathione S-transferase (GST) pull-down assays to determine if BRCA1 directly interacts with EZH2 and which region(s) in BRCA1 and EZH2 proteins mediates their interaction. EZH2, SUZ12, EED and Ezh1 were produced individually by in vitro transcription and translation and incubated separately with bacteria-purified five GST-BRCA1 fusion proteins (Figure 2A). Only EZH2, but not SUZ12, EED and Ezh1 interacted with BRCA1, although similar amounts of proteins were used (Figure 2B). There were two regions in BRCA1 (BR2, amino acids 252–551 and BR3, amino acids 501–1021) that interacted with EZH2 (Figure 2B). When the recombinant PRC2 complex containing Ezh2, SUZ12, EED and RbAp48 copurified from insect Sf9 cells was employed in GST pull-down assay, we found that the most NH2-terminal region in BRCA1 (BR1, amino acids 1–304) also interacted with EZH2 and EED in the PRC2 complex (Figure 2C), indicating that other subunits of PRC2 contribute to the interaction of BRCA1 with EZH2. The interaction between BRCA1 NH2-terminus and EZH2 was confirmed by co-IP and mass spectrometry analyses (Supplementary Figure S1C–F). Reciprocally, recombinant BRCA1 proteins purified from Sf9 cells were incubated with GST-EZH2 fusion proteins. As shown in Figure 2D, BRCA1 interacted with a region in EZH2 containing amino acids 341–559, which overlaps with the ncRNA binding domain 1 (ncRBD1, amino acids 342–370) (Kaneko et al, 2010). Because the EZH2 interaction region in BRCA1 contains a RING domain and a similar domain in other proteins enables to bind to RNA (Lai et al, 1998), we sought to determine if the BRCA1–EZH2 interaction is RNA mediated. Cell lysates were treated with RNase A prior to co-IP. The effectiveness of RNase A treatment was evident by complete depletion of GAPDH mRNA from the cell lysates (Figure 1E, right). However, RNase A treatment had little or no effect on the BRCA1–EZH2 interaction (Figure 1E, left), suggesting that their interaction is not RNA mediated.
Figure 2.
Analysis of the regions in BRCA1 and EZH2 responsible for their interaction. (A) Schematic diagram of the five GST-BRCA1 fusion proteins (BR1 to BR5). NLS, nuclear localization signal. (B) GST pull-down assay detecting the regions of BRCA1 that bind to PRC2 proteins. Top, western blot analysis of in vitro transcribed and translated Polycomb proteins EZH2, SUZ12, EED and Ezh1 pulled down by GST or GST-BRCA1 fusion proteins BR1-BR5. #Nonspecific proteins reacted with anti-EZH2 antibody. Bottom, Coomassie blue staining of GST and GST-BRCA1 fusion proteins BR1–BR5. Asterisks indicate proteins with correct molecular masses. (C) Top, western blot analysis of Ezh2 and EED in baculovirally expressed PRC2 complex pulled down by GST or GST-BRCA1 fusion proteins. Bottom, Coomassie blue staining of GST and GST-BRCA1 fusion proteins BR1–BR5. Asterisks indicate proteins with correct molecular masses. (D) GST pull-down assay detecting the regions of EZH2 that bind to BRCA1. Top, schematic diagram of the four GST-EZH2 fusion proteins designated as EZ1 to EZ4. EBD, EED binding domain; ncRBD1, ncRNA binding domain 1. Middle, western blot analysis of baculovirally expressed BRCA1 pulled down by GST or GST-EZH2 fusion proteins EZ1-EZ4. #Nonspecific proteins reacted with anti-BRCA1 antibody. Bottom, Coomassie blue staining of GST and GST-EZH2 fusion proteins EZ1–EZ4. Asterisks indicate proteins with correct molecular mass.
Source data for this figure is available on the online supplementary information page.
BRCA1 regulates genome-wide occupancy of EZH2 on chromatin
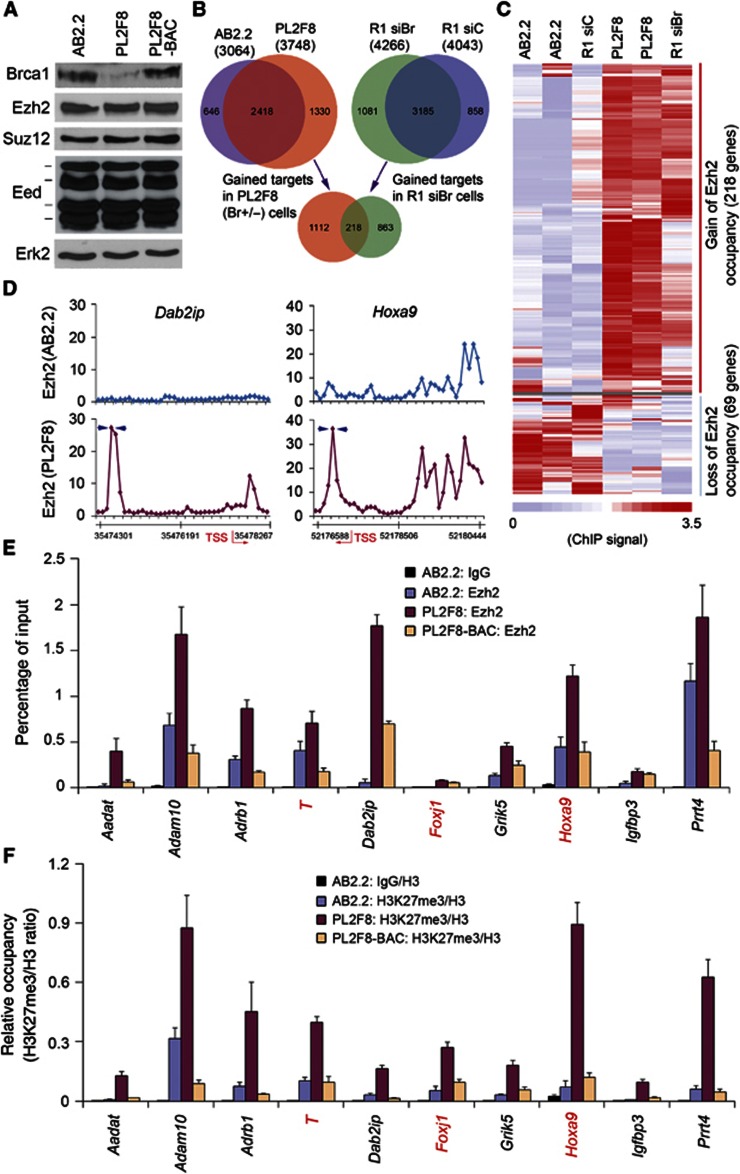
The interaction between BRCA1 and EZH2 prompted us to determine if BRCA1 affects EZH2 occupancy on its target genes. We employed a chromatin immunoprecipitation coupled with DNA microarray (ChIP-chip) approach to interrogate genome-wide chromatin occupancy of EZH2 in two independent mouse ES cell lines. Consistent with the previous report (Chang et al, 2009), Brca1 protein levels were significantly reduced in Brca1-heterozygous PL2F8 cells in comparison to wild-type AB2.2 ES cells, but was restored in PL2F8-BAC cells rescued with human BRCA1 BAC DNA (Figure 3A and Supplementary Figure S2A). Brca1-heterozygous deletion resulted in gain of Ezh2 occupancy on 1330 de novo targets (Figure 3B and Supplementary Table S1) and the result was reproducible (Supplementary Figure S2B). Brca1-heterozygous deletion also caused loss of Ezh2 occupancy on 646 pre-existing targets (Figure 3B). These results were unlikely caused by the effect of BRCA1 on cell proliferation since little or no difference in cell cycle distribution was observed between AB2.2 and PL2F8 cells (Supplementary Figure S2C). Similarly, Brca1 knockdown by small interference RNA (siRNA) in R1 mouse ES cells (Supplementary Figure S2D) induced gain of Ezh2 occupancy on 1081 de novo targets but loss of occupancy on 858 targets (net gain of 223 targets; Figure 3B and Supplementary Table S1). The overlap (218 genes) of the gained Ezh2 targets between PL2F8 and R1 siBr cells was relatively small, but reproducible and statistically significant (Figure 3B and C and Supplementary Figure S2E). This is probably due to a large degree of genetic diversity among 129 substrains (Simpson et al, 1997; Threadgill et al, 1997) from which AB2.2 and R1 ESC lines were established and the overt differences in Brca1 protein level in these two ESC lines both before and after Brca1 knockdown or knockout (Supplementary Figure S2F). It is worth noting that overexpression HOTAIR ncRNA also resulted in a biphasic effect on EZH2 targeting in breast cancer cells (Gupta et al, 2010). Gene ontology (GO) analysis of the 218 de novo Ezh2 targets showed that these genes are enriched for diverse functions including development, cell growth and adhesion (Supplementary Figure S3A). Additionally, Brca1-heterozygous deletion in PL2F8 cells enhanced Ezh2 occupancy on 1089 pre-existing PRC2 targets including Hoxa9 (Figure 3D and Supplementary Table S1). Similarly, silencing of Brca1 in R1 cells also increased Ezh2 occupancy on a large number of pre-existing PRC2 targets (Supplementary Table S1). Brca1 deficiency-enhanced chromatin enrichment of Ezh2 in PL2F8 ES cells was confirmed by ChIP–qPCR at the promoters of 7 de novo and 3 pre-existing target genes including development regulators and tumour growth and metastasis suppressor genes Brachyury (T), Dab2ip, Igfbp3, Adrb1 and Adam10 (Peng et al, 2009; Xie et al, 2010) (Figure 3E). Restored BRCA1 expression in PL2F8-BAC cells reversed Ezh2 occupancy at these loci (Figure 3A and E). Collectively, these data indicate that reduced expression of BRCA1 broadens genome-wide occupancy of EZH2 on chromatin in murine ES cells.
Figure 3.
Effect of Brca1 expression on genome-wide Ezh2 targeting in mouse ES cells. (A) Western blot analysis of expression of indicated proteins in AB2.2, PL2F8 and PL2F8-BAC mouse ES clones. Expression of extracellular signal-regulated kinase 2 (Erk2) was included as a loading control. Data shown are from one representative experiment out of two independent experiments that gave similar results. (B) Venn diagram showing de novo Ezh2 targeting genes shared between PL2F8 cells with Brca1 heterozygous deletion and R1 mouse ES cells with Brca1 knockdown. The overlap is statistically significant with P=8.59E–31 (hypergeometric test). (C) Heat map showing genes with gain or loss of Ezh2 occupancy due to decreased expression of Brca1 in three Brca1-proficient replicates (two AB2.2 plus one R1 siC) and three Brca1-deficient replicates (two PL2F8 plus one R1 siBr). ChIP signals are depicted using the colour scale at the bottom. (D) Examples of Ezh2 ChIP-chip results for the indicated loci in AB2.2 and PL2F8 ES cells. TSS, transcription start site. Arrows in blue indicate the locations of primers for ChIP–qPCR. (E) ChIP–qPCR analysis of Ezh2 occupancy on target loci in AB2.2, PL2F8 and PL2F8-BAC ES cells. Data are shown as means±s.d. from experiments with three replicates (n=3). Genes labelled in red are known EZH2 targets in mouse ES cells. (F) ChIP–qPCR analysis of H3K27me3 in PRC2 target loci identified by ChIP-chip in AB2.2, PL2F8 and PL2F8-BAC ES cells. Data are means±s.d. from three individual experiments (n=3). H3K27me3 and total H3 ChIP results were first normalized to input DNA and the H3K27me3/H3 ratio was determined by their normalized values. Genes labelled in red are known EZH2 targets.
Source data for this figure is available on the online supplementary information page.
BRCA1 regulates H3K27me3 levels at EZH2 target loci
Next, we sought to determine if BRCA1 affects H3K27me3 levels at EZH2 target loci. Brca1-heterozygous deletion had no effect on the level of PRC2 proteins Ezh2, Suz12 and Eed in PL2F8 cells compared to AB2.2 cells (Figure 3A), while it resulted in a substantial increase in H3K27me3 in the promoters of de novo and pre-existing PRC2 targets examined (Figure 3F). This effect was completely reversed by restored expression of BRCA1 in PL2F8-BAC ES cells (Figure 3F). To determine whether BRCA1 regulates PRC2 function in human breast cancer cells, endogenous BRCA1 was knocked down by two independent BRCA1-specific siRNAs in MCF7 cells (Figure 4A). Silencing of BRCA1 resulted in a significant increase in EZH2 occupancy and H3K27me3 levels in the promoter of HOXA9 (Figure 4B, left). Accordingly, overexpression of BRCA1 decreased the binding of EZH2 to the HOXA9 promoter and increased HOXA9 mRNA expression (Figure 4C and D). Consistent with the findings that Dab2ip is not a PRC2 target in wild-type mouse ES cells (Ku et al, 2008), and that it is highly bound by Ezh2 due to heterozygous deletion of Brca1 in PL2F8 ES cells (Figure 3D–F), EZH2 binding and H3K27me3 levels significantly increased at the DAB2IP locus in BRCA1-knockdown MCF7 cells (Figure 4B, right). BRCA1 knockdown also increased EZH2 occupancy and H3K27me3 levels in the promoter of HOXA9 in MCF10A cells and other four EZH2 target loci in MCF7 cells (Figure 4E and Supplementary Figure S3B). Consistent with these observations, BRCA1 knockdown decreased the expression of HOXA9, DAB2IP and FOXJ1 proteins in MCF7 cells (Figure 4A). Thus, BRCA1 negatively regulates PRC2 function and loss of BRCA1 increases H3K27me3 levels at PRC2 target loci in both mouse ES and human breast cancer cells.
Figure 4.
BRCA1 negatively regulates EZH2 recruitment and H3K27me3 levels at PRC2 target loci in breast cancer cells. (A) Western blot analysis of expression of BRCA1 and PRC2 target proteins HOXA9, DAB2IP and FOXJ1 in MCF7 cells transfected with control (siC) or two independent BRCA1-specific siRNAs. ERK2, loading control. (B) MCF7 cells were transfected with control and BRCA1-specific siRNAs as in (A) and subjected to ChIP analysis with anti-EZH2, anti-H3K27me3 antibodies or control IgG and qPCR analysis (means± s.d., n=3). *P<0.05 comparing EZH2 enrichment in siBR#1 and #2 transfected cells to that in siC transfected cells; **P<0.01 comparing H3K27me3 enrichment in siBR#1 and #2 transfected cells to that in siC transfected cells. (C, D) Effects of BRCA1 overexpression on EZH2 recruitment to the HOXA9 gene promoter and HOXA9 mRNA expression in MCF7 cells. At 24 h after transfection, cells were subjected to western blot, ChIP–qPCR (means± s.d., n=3) (C) and RT–qPCR (means± s.d., n=3) (D). CV, control vector; *P<0.05 and **P<0.01. (E) ChIP–qPCR analysis of EZH2 and H3K27me3 enrichment on HOXA9 gene promoter in MCF10A cells (means± s.d., n=3). **P<0.01 comparing EZH2 enrichment in siBR#1 and #2 transfected cells to that in siC transfected cells; *P<0.05 comparing H3K27me3 enrichment in siBR#2 transfected cells to that in siC transfected cells.
Source data for this figure is available on the online supplementary information page.
BRCA1 inhibits HOTAIR-enhanced EZH2 recruitment to its target loci
Since BRCA1 inhibits EZH2 occupancy on chromatin (Figure 3E and 4B) without affecting PRC2 component expression (Figure 3A), we sought to determine if BRCA1 influences PRC2 recruitment to its target loci. Several transcription regulators (e.g., YY1) and long ncRNAs (such as HOTAIR and XIST) have been shown to interact with and facilitate PRC2 targeting in mammalian cells (Caretti et al, 2004; Rinn et al, 2007; Zhao et al, 2008). Because the binding region of BRCA1 in EZH2 (amino acids 341–559) overlaps with the ncRBD1 (amino acids 342–370) (Kaneko et al, 2010), we examined if BRCA1 interferes EZH2 binding to ncRNAs. RNA immunoprecipitation (RIP) assay showed that expression of BRCA1 inhibited and BRCA1 knockdown increased EZH2 binding to HOTAIR in MCF7 cells (Figure 5A and B). Consistent with the finding that HOTAIR is important for PRC2 targeting (Rinn et al, 2007; Kaneko et al, 2010), expression of HOTAIR increased the binding of EZH2 to the HOXA9 promoter in MDA-MB-231 cells, but this effect was blocked by forced expression of BRCA1 (Figure 5C, left). BRCA1 also inhibited HOTAIR-enhanced EZH2 occupancy on HOXA9 promoter in MCF7 cells and BJ human fibroblasts (Figure 5D). However, BRCA1 expression failed to inhibit EZH2 occupancy on EZH2 target loci in HOTAIR-knockdown MCF7 cells (Supplementary Figure S4A–C), suggesting that BRCA1-mediated inhibition of EZH2 occupancy on chromatin is mainly mediated through HOTAIR in MCF7 cells. Moreover, the HOXA9 mRNA level was repressed by HOTAIR expression in MDA-MB-231 cells and this was completely reversed by forced expression of BRCA1 (Figure 5C, right). Thus, BRCA1 inhibits the binding of HOTAIR to EZH2 and abolishes HOTAIR-enhanced recruitment of PRC2 to its target gene HOXA9 in human breast cancer cells and fibroblasts.
Figure 5.
BRCA1 regulation of ncRNA-mediated recruitment of EZH2 on its targeting gene loci. (A) Effect of BRCA1 overexpression on the binding of HOTAIR ncRNA to EZH2. Transfected MCF7 cells were harvested for western blots and RIP with anti-myc or nonspecific IgG. Lysates from AB2.2 and Brca1-heterozygous PL2F8 mouse ESC lines were used as positive/negative controls to define the BRCA1 band on western blots. Retrieved HOTAIR ncRNA was analysed by RT–qPCR (n=3). *P<0.01. (B) Effect of BRCA1 knockdown on the binding of HOTAIR ncRNA to EZH2. MCF7 cells were transfected with control (siC) and BRCA1-specific siRNAs (siBR) and subjected to western blots and RIP with anti-EZH2 or nonspecific IgG. Retrieved HOTAIR ncRNA was analysed by RT–qPCR (n=3). *P<0.01. (C) Effect of BRCA1 on HOTAIR-enhanced binding of EZH2 to the HOXA9 promoter (left) and HOXA9 mRNA expression (right). MDA-MB-231 cells were transfected with plasmids as indicated and subjected to western blots. The binding of EZH2 to the HOXA9 promoter was analysed by ChIP using anti-EZH2 antibody (ChIP–qPCR, n=3, left) and RT–qPCR (n=3, right). * P<0.01. (D) MCF7 cells and BJ fibroblasts were transfected with the indicated plasmids and subjected to ChIP assay with anti-EZH2 antibody and qPCR analysis (n=3). *P<0.01. (E, F) R1 mouse ES cells were transfected with pools of control (siC) or Brca1-specific siRNAs (siBr) and subjected to western blot analysis (E). #Nonspecific protein reacted with anti-Brca1 antibody. The other set of cells were harvested for ChIP assay (F). Extracted nuclei were pretreated with or without RNase A and subjected to ChIP analysis with anti-Ezh2 antibody. Enrichment of Ezh2 on target loci were analysed by ChIP–qPCR (n=3). Data are shown as means±s.d. from three individual experiments. *P<0.05.
Source data for this figure is available on the online supplementary information page.
BRCA1 is involved in G2/M cell cycle checkpoint control. As expected, expression of wild-type BRCA1 in MDA-MB-231 cells increased cell cycle distribution at G2/M (Supplementary Figure S5A and B). In contrast, expression of BRCA1-N (amino acids 1–1100), a COOH-terminal truncation mutant of BRCA1, had no effect on G2/M distribution (Supplementary Figure S5A and B). This result is consistent with a previous report that stable expression of a COOH-terminal deletion mutant of BRCA1 had little or no effect on cell growth (Harkin et al, 1999). In agreement with the finding that BRCA1-N interacts with EZH2 in vitro and in cells (Figure 2C and Supplementary Figure S1C), ectopic expression of BRCA1-N not only inhibited EZH2 occupancy on chromatin at its target loci, but also increased its target gene expression (Supplementary Figure S5C and D). Importantly, these effects of BRCA1-N were similar to those mediated by wild-type BRCA1 (Supplementary Figure S5C and D). Together, these data rule out the possibility that the effects of BRCA1 on EZH2 chromatin occupancy are due to cell cycle alterations.
We also investigated how Brca1 affects PRC2 targeting in mouse ES cells. While a recent study demonstrates that the cognate mouse Hotair ncRNA is poorly conserved in sequence (Schorderet and Duboule, 2011), genome-wide RIP-seq analysis reveals that there are more than 200 long ncRNAs bound to PRC2 in mouse ES cells (Zhao et al, 2010). It is worth noting that pretreatment of nuclear extracts with RNases invariably abolished Ezh2 binding with all the ncRNAs examined (Zhao et al, 2010), whereas RNase treatment of nuclear extracts does not generally affect chromatin and nuclei ChIP efficiency (Yap et al, 2010). To determine if ncRNAs also play a role in Brca1 regulation of Ezh2 targeting in mouse ES cells, endogenous Brca1 was knocked down by gene-specific siRNAs in R1 ES cells and nuclear extracts were mock treated or treated with RNase A prior to nuclei ChIP assay. Similar to the results obtained in MCF7 cells (Figure 4A), Brca1 knockdown decreased Hoxa9 and Foxj1 protein expression in R1 ES cells (Figure 5E), indicating that Brca1 knockdown affects PRC2 function in these cells. Brca1 knockdown substantially increased Ezh2 binding to Brachyury, Foxj1 and Hoxa9 promoters (Figure 5F). Importantly, this effect was abolished by RNase A treatment (Figure 5F). These data suggest that Brca1 may regulate PRC2 targeting in mouse ES cells via a mechanism similar to that mediated by HOTAIR in human breast cancer cells.
Brca1 affects mouse ES cell differentiation via regulation of Ezh2
PRC2 plays a pivotal role in H3K27me3-mediated silencing of developmental regulators that control human and mouse ES cell differentiation (Boyer et al, 2006; Lee et al, 2006). We sought to examine whether BRCA1 affects ES cell differentiation via EZH2. R1 mouse ES cells were transfected with control siRNAs or Brca1 and/or Ezh2 siRNA in combination with or without HA-tagged human BRCA1. Cells were then cultured in suspension in the absence of leukemia inhibitory factor (LIF). Cells transfected with control siRNAs underwent differentiation by forming embryoid bodies (EBs) in a time-dependent manner (Figure 6A and B, column 1). Western blot analysis showed that Brca1 was effectively silenced by siRNAs (Figure 6A). No apparent phenotypic difference was detected between Brca1-knockdown and control cells at the early time points (2 days) of culture (Figure 6B, column 2). However, the prolonged (4–8 days) culture severely compromised EB formation in Brca1-knockdown cells (Figure 6B, column 2). This effect was rescued by ectopic expression of human BRCA1, which is resistant to mouse Brca1 siRNAs (Figure 6A and B, column 5). Thus, reduced expression of Brca1 impairs mouse ES cell differentiation. The inhibitory effect of Brca1 was largely diminished by concomitant Ezh2 knockdown (Figure 6A and B, column 3). Ezh2 knockdown alone had no drastic effect on EB formation (Figure 6B, column 4). Consistent with the morphology data, expression of stem cell markers Dax1 and Eras (Takahashi et al, 2003; Pasini et al, 2010) was much higher in Brca1-knockdown cells compared to differentiation-unaffected cells in the other four groups (Figure 6C). Together, these data suggest that Ezh2 plays a key role in Brca1 deficiency-induced blockage of mouse ES cell differentiation.
Figure 6.
Effect of Brca1 expression on murine ES cell differentiation. (A) R1 ES cells were transfected as indicated and subjected to western blot analysis at 48 h after transfection. (B) R1 cells were transfected as in (A). Cells were cultured on low-attachment plates in the absence of LIF. Images were taken 2, 4, 6 and 8 days after plating. Scale bar, 200 μm. (C) R1 cells were transfected as in (A). At 6 days after cultured in suspension, cells were harvested for RT–qPCR analysis of expression of stem cell markers Dax1 and Eras (means±s.d., n=3). *P<0.05 comparing Dax1 and Eras expression in the siBr-transfected group to every other group.
Source data for this figure is available on the online supplementary information page.
BRCA1 regulates breast cancer migration and invasion via EZH2
Given that expression of the PRC2 target proteins HOXA9 and DAB2IP is modulated by BRCA1 in MCF7 breast cancer cells (Figure 4A) and that both proteins play important roles in inhibition of cell migration and invasion (Chen et al, 2010; Gilbert et al, 2010; Xie et al, 2010), we sought to determine whether BRCA1 regulates breast cancer cell migration and invasion via EZH2. Treatment of MDA-MB-231 breast cancer cells by a pool of BRCA1-specific siRNAs completely depleted endogenous BRCA1 (Figure 7A). Like in MCF7 cells, BRCA1 silencing decreased DAB2IP protein levels in MDA-MB-231 cells (Figure 7A). Notably, BRCA1 knockdown increased breast cancer cell migration (Figure 7B) and invasion (Figure 7D). The quantified data are shown in Figures 7C and E, respectively. The effect of BRCA1 silencing on DAB2IP expression (Figure 7A), migration (Figure 7B and C) and invasion (Figure 7D and E) was partially reversed by concomitant EZH2 knockdown, suggesting that the effect of BRCA1 on cell migration and invasion is mediated, at least in part, through EZH2. As a control, knockdown of EZH2 alone resulted in a slight increase in DAB2IP expression (Figure 7A), a modest but statistically significant decrease in cell migration (Figure 7B and C) and a marked decrease in invasion (Figure 7D and E). Together, BRCA1 inhibits EZH2-mediated repression of tumour metastasis-inhibitory genes DAB2IP and HOXA9 (Gilbert et al, 2010; Xie et al, 2010) and blocks EZH2-enhanced migration and invasion of breast cancer cells.
Figure 7.
BRCA1 regulates human breast cancer cell migration and invasion via EZH2. (A) MDA-MB-231 cells were transfected with HOTAIR and siRNAs as indicated. At 48 h after transfection, one set of cells were harvested and subjected to western blot analysis. #Nonspecific protein reacted with anti-BRCA1 antibody. The density of DAB2IP was determined by normalizing to ERK2 first and then to the normalized value in siC-transfected cells. (B, C) At 48 h after transfection as in (A), artificial wounds were created on cells grown in confluence. Images were taken at 0, 24 and 48 h after wound (B). Scale bar, 200 μm. The wound widths were measured and quantified in (C) (means±s.d., n=5). *P<0.05. (D, E) After transfection as in (A), cells were used for transwell invasion assays. Images for invasive cells were taken at 48 h after transfection (D). Scale bar, 50 μm. Cell invasion was quantified by measuring the number of invasive cells per high-power field (HPF, × 400) (means±s.d., n=5) from five random fields (E). *P<0.01.
Source data for this figure is available on the online supplementary information page.
Discussion
The striking bias observed in BRCA1 mutation carriers towards developing aggressive basal-like breast cancers has driven significant interest in understanding how BRCA1 might regulate gene expression and cell fate. Mediating global gene silencing, the PRC2 chromatin-modifying complex has been implicated in stem cell differentiation (Boyer et al, 2006; Lee et al, 2006; Pasini et al, 2007; Ezhkova et al, 2009) and cancer progression (Varambally et al, 2002; Bracken et al, 2003; Kleer et al, 2003). In this report we provide evidence that BRCA1 interacts with EZH2 in vitro and in cultured cells. Reduction of BRCA1 levels in ES cells leads to genome-wide re-targeting of EZH2 and concomitant increase in H3K27me3 levels in PRC2 target loci. BRCA1 deficiency also blocks ES cell differentiation and enhances breast cancer cell migration and invasion, and these effects are mediated, at least in part, through EZH2. Thus, we have identified modulation of EZH2 functions as a novel mechanism of BRCA1 in regulation of gene expression, stem cell differentiation and cancer aggressive phenotypes.
BRCA1 is detected in many protein complexes with diverse functions including DNA damage response and repair, transcription regulation and chromatin remodelling (Huen et al, 2010). Through the RING domain at the NH2-terminus, BRCA1 forms a heterodimer with BARD1, a well-studied BRCA1 partner protein that has been implicated in regulation of genomic stability, DNA repair and transcription (Meza et al, 1999; Huen et al, 2010). Our in vitro protein binding and co-IP assays demonstrated that BRCA1 binds to the PRC2 complex through its NH2-terminal (amino acids 1–1021), but not the COOH-terminal portion (amino acids 1022–1863). However, we found that neither BARD1 was detected in the BRCA1–PRC2 complex nor BARD1 overexpression affected BRCA1–EZH2 interaction. Thus, our data reveal that BRCA1 associates with PRC2 in a protein complex different from the BRCA1–BARD1 complex, suggesting that BRCA1–PRC2 interaction works independent of the BRCA1–BARD1 heterodimer and thereby may not interfere with the heterodimer function of BRCA1–BARD1 in DNA damage. Moreover, there is an emerging link between PcG and the DNA damage response in mammalian cells (Liu et al, 2009; Chou et al, 2010; Lukas et al, 2011; Vissers et al, 2012). We found that the binding of EZH2 by BRCA1 was substantially reduced following UV irradiation in 293T cells (Supplementary Figure S6). These data implicate that BRCA1-mediated inhibition of EZH2 can be attenuated in response to DNA damage, which is consistent with the notion that activated EZH2 might enhance DNA repair by repressing gene transcription at sites of damage (Chou et al, 2010; Vissers et al, 2012). How DNA damage affects the BRCA1–EZH2 interaction warrants further investigation.
BRCA1 has been implicated in mammary luminal epithelial differentiation and basal-like breast cancer development (Furuta et al, 2005; Liu et al, 2008; Lim et al, 2009; Molyneux et al, 2010; Proia et al, 2011). However, which biochemical activity of BRCA1 mediates such a function is not fully understood. Using mouse ES cells as a working model, we demonstrate that Brca1 knockdown blocks mouse ES cell differentiation in a manner dependent on Ezh2. We provide evidence that decreased expression of Brca1 broadens genome-wide occupancy of Ezh2 on chromatin and represses Ezh2 target gene expression in mouse ES cell lines. We further show that loss of BRCA1 induces aggressive (migratory/invasive) phenotypes of breast cancer and that these effects are mediated through EZH2. Based upon these results, we envisage a model that reduced BRCA1 expression augments function of PRC2, thereby preventing committed cell lineage differentiation and/or favouring undesired reprogramming of differentiated BRCA1-mutated luminal cells into basal-like, aggressive breast adenocarcinoma cells. Our hypothesis is supported by previous findings that the function of PRC2 is crucial for the maintenance of stem cell properties and reprogramming of somatic cells into iPS cells (Boyer et al, 2006; Lee et al, 2006; Pasini et al, 2007; Ezhkova et al, 2009; Pereira et al, 2010; Onder et al, 2012). Moreover, this notion is consistent with the report that the synthetic lethality of BRCA1-mutant breast cancers is attributed to EZH2 inhibition (Puppe et al, 2009). It is hence warranted to explore if EZH2 plays a causal role in Brca1 deletion-induced formation of basal-like breast tumours in animals (Liu et al, 2007).
Findings from mouse and human ES cell studies suggest that PRC2 plays a key role in maintenance of ES cell pluripotency and H3K27me3-mediated transcriptional repression of cell lineage-specific differentiation genes (Boyer et al, 2006; Lee et al, 2006). Further studies show that PRC2 targets are bivalent genes that often contain the transcription repression mark H3K27me3 and the activation mark H3K4me3 (Bernstein et al, 2006), suggesting that PRC2 target genes are poised for activation during differentiation. Consistent with this notion, it has been shown that Suz12-null mouse ES cells fail to undergo retinoic acid-induced neuronal differentiation (Pasini et al, 2007) and Eed-null ES cells exhibit severe defect in mesoendoderm (ME) differentiation (Shen et al, 2008). Thus, it has been speculated that PRC2 complexes are required for both suppression and activation of differentiation genes in ES cells (Ezhkova et al, 2009). In contrast to the findings in Suz12- and Eed-null ES cells, we found that EZH2 knockdown did not impair ES cell differentiation. Similar to our observation, it has been shown previously that Ezh2-null ES cells have less severe defect in ME differentiation than Eed-null cells (Shen et al, 2008). One plausible explanation is that Ezh1, a homologue of Ezh2, has complementary but nonredundant roles in mediating H3K27me3 and ES cell function (Margueron et al, 2008; Shen et al, 2008). Additionally, different from Ezh2-null ES cells, the residual Ezh2 proteins in Ezh2-knockdown ES cells (Figure 6A) may enable to fine-tune the balance between suppression and activation of differentiation genes.
Although it is not entirely clear how PRC2 targets specific chromatin regions in mammalian cells (Kaneko et al, 2010), long ncRNAs HOTAIR and XIST have been shown to bind to and recruit the PRC2 complex in cis or in trans (Rinn et al, 2007; Zhao et al, 2008). A functional motif, designated as ncRBD1, is critical for EZH2 binding to HOTAIR and XIST (Kaneko et al, 2010). Consistent with the observation that the BRCA1 binding region in EZH2 overlaps with ncRBD1, we show that BRCA1 overexpression inhibits, but BRCA1 knockdown enhances, EZH2 binding to HOTAIR. BRCA1 overexpression also abolishes HOTAIR-augmented binding of EZH2 on the promoter of PRC2 target gene HOXA9 in human breast cancer cells and fibroblasts. Despite the fact that the cognate mouse Hotair ncRNA is poorly conserved in sequence (Schorderet and Duboule, 2011), we provide evidence that Brca1 knockdown-induced Ezh2 recruitment at PRC2 target loci was abolished by RNase treatment in mouse ES cells. Thus, our data suggest that inhibition of EZH2 binding to ncRNAs could be one of the potential mechanisms that mediate BRCA1 interference of PRC2 targeting in both ES and breast cancer cells.
In closing, BRCA1 has been implicated in transcription regulation and mammary luminal epithelial differentiation, and loss of BRCA1 was shown to lead to basal-like breast cancers that are migratory and invasive; however, the underlying mechanism is largely unclear. We uncovered a previously uncharacterized interaction between the tumour-suppressor BRCA1 and the oncoprotein EZH2. Reduction of BRCA1 levels broadens genome-wide EZH2 occupancy on chromatin and elevates H3K27me3 levels at PRC2 target loci. We further showed that decreased BRCA1 expression blocks ES cell differentiation and augments breast cancer migration and invasion in a manner dependent on EZH2. Thus, our study has identified the BRCA1–EZH2 interaction as a key mechanism mediating BRCA1 regulation of gene expression, stem cell differentiation and cancer aggressive phenotypes. Our findings suggest that the PRC2 complex may serve as a viable target for the treatment of human basal-like breast cancers carrying a mutated or methylated BRCA1 gene.
Materials and methods
Plasmids
Myc-tagged EZH2 plasmid was kindly provided by Mien-Chie Hung (the University of Texas M. D. Anderson Cancer Center, Houston, TX, USA). Baculovirus expression vectors for mouse Ezh2, human SUZ12, EED and RbAp48 were kindly provided by Yi Zhang (the University of North Carolina, Chapel Hill, NC, USA). Mouse Ezh1 was kindly provided by Xiaohua Shen (Tsinghua University, Beijing, China). The mammalian expression vector for HA-tagged BRCA1 was kindly provided by Fergus Couch (Mayo Clinic College of Medicine, Rochester, MN, USA). V5-tagged expression vectors for wild-type BRCA1, BRCA1-N (amino acids 1–1100), were constructed by PCR amplifying and subcloning the corresponding fragments of BRCA1 into pcDNA3.1D vector (Invitrogen). A series of BRCA1 fragments (1–304, 252–551, 501–1021, 1022–1500 and 1051–1863) and EZH2 (1–173, 174–340, 341–559 and 560–751) were amplified by PCR and subcloned into pGEX-4T-1 vector (GE Healthcare) for construction of recombinant GST-BRCA1 and GST-EZH2 fusion proteins, respectively. The FLAG-BARD1 expression vector was kindly provided by Zhenkun Lou (Mayo Clinic, Rochester, MN, USA). The retrovirus-based HOTAIR expression vector LZRS-HOTAIR constructed by the laboratory of Howard Chang (Stanford University School of Medicine, Stanford, CA, USA) (Gupta et al, 2010) was obtained from Addgene and subcloned into the backbone vector pcDNA3.1 (Invitrogen).
Antibodies and reagents
Antibodies used are as follows: rabbit polyclonal anti-BRCA1 antibody was described previously (Yu et al, 2003); anti-EZH2 (XP) (Cell Signaling Technology); anti-EED, anti-SUZ12 (3C1.2) (Millipore); anti-HOXA9, anti-DAB2IP, anti-H3 (total), anti-H3K27me3 (Abcam); anti-SUZ12 (D-10), anti-myc, anti-ERK2 (Santa Cruz Biotechnology); anti-FOXJ1, anti-HA (Covance); and anti-FLAG (Sigma Aldrich). Antibodies against total and phosphorylated CHK1 were kindly provided by Zhenkun Lou (Mayo Clinic).
Cell culture and transfection
MCF7, MDA-MB-231, 293T and BJ cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Hyclone). MCF10A cells were cultured in DMEM/F12 supplemented with 5% horse serum (Invitrogen), 1% penicillin/streptomycin (Invitrogen), 0.5 μg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), 10 μg/ml insulin (Sigma) and 20 ng/ml recombinant human EGF (Peprotech). Mouse wild-type (AB2.2), Brca1-heterozygous mutant (PL2F8) and human BRCA1-rescued PL2F8 (PL2F8-BAC) ES cell lines were kindly provided by Shyam K Sharan (Chang et al, 2009). R1 mouse ES cell line was purchased from ATCC. Mouse ES cells were cultured in KNOCKOUTTM DMEM medium (Invitrogen) supplemented with 15% ES cell-screened FBS (Hyclone), 0.1 mM 2-mercaptoethanol, 1 mM L-glutamine (Invitrogen) and 103 unit/ml LIF (Millipore) on 0.1% gelatin-coated dishes. Cells were cultured at 37°C supplied with 5% CO2. Transfection of cells was performed by electroporation using an Electro Square Porator ECM 830 (BTX) as we described previously (Huang et al, 2006) or using Lipofectamine 2000 (Invitrogen). Approximately 75–90% transfection efficiencies were routinely achieved (Huang et al, 2006).
RNA interference
siRNAs for human BRCA1 (siBR #1, 5′-GGGAUACCAUGCAACAUAA-3′ and siBR #2, 5′-CUAGAAAUCUGUUGCUAUG-3′), smart pools of siRNAs against human BRCA1 and EZH2, mouse Brca1 and Ezh2 and nonspecific control siRNAs were purchased from Dharmacon. siRNA for human HOTAIR (siHOTAIR GAACGGGAGUACAGAGAGAUU) was synthesized by Dharmacon. siRNA transfection of cells was performed following the manufacturer’s instruction.
RT–qPCR
Total RNA was isolated from cells and cDNA was synthesized using the Super-Script kit from Invitrogen. Two-step real-time PCR was performed using the SYBR Green Mix (Bio-Rad) and an iCycler iQTM system (Bio-Rad) as we described previously (Huang et al, 2006). Both forward and reverse primers were used at a final concentration of 200 nM. The primer sequences used for PCR are described in Supplementary Table S2.
Co-immunoprecipitation and western blotting
Immunoprecipitations were performed as described previously (van der Vlag and Otte, 1999; Huang et al, 2006; Pasini et al, 2008). Briefly, cells were harvested and lysed using glass Dounce homogenizer in cell lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate and 1% protease inhibitor cocktails; Sigma Aldrich). Ethidium bromide was added to a final concentration of 50 μg/ml to disrupt DNA–protein interactions in endogenous protein co-IP experiments. Cell lysis was centrifuged and the supernatant (5∼10 mg) was incubated with indicated antibodies (5–10 μg) and protein-G beads (Invitrogen) at 4°C overnight. The beads were washed five times with cell lysis buffer and the precipitated proteins were subjected to western blot analysis. For RNase A treatment, cell lysates were mock treated or treated with 50 μg/ml RNase A at 37°C for 10 min and then subjected to co-IP experiments as described above. For western blotting, protein samples were prepared by lysing cells in lysis buffer (1% Nonidet P-40 and 0.1% SDS in 1 × PBS solution plus protease inhibitors). Equal amounts of proteins (50–100 μg) from cell lysates were denatured in sample buffer (Invitrogen), subjected to SDS–polyacrylamide gel electrophoresis and were transferred to nitrocellulose membranes (Bio-Rad). The membranes were immunoblotted with specific primary antibodies, horseradish peroxidase-conjugated secondary antibodies and visualized by SuperSignal West Pico Stable Peroxide Solution (Thermo Scientific).
ChIP and RIP assays
ChIP assay was performed as previously described (Boyer et al, 2005). Briefly, cells were crosslinked with 1% formaldehyde for 10 min at room temperature. Crosslinking was quenched by adding 2.5 M glycine to a final concentration of 125 mM. Cells were scraped and washed twice with 1 × PBS. Cell nuclei were extracted by resuspending cells in lysis buffer 1 (50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40 and 0.25% Triton X-100) for 10 min at 4°C and lysis buffer 2 (10 mM Tris–HCl, pH 8.0, 200 mM NaCl and 1 mM EDTA) for 10 min at room temperature. Cell nuclei were lysed with lysis buffer 3 (10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.1% sodium deoxycholate) and sonicated to solubilize and shear crosslinked DNA. Cell lysis was centrifuged and the supernatant was incubated overnight at 4°C with 30 μl protein G beads (Invitrogen) that has been preincubated with 10 μg of indicated antibody. The beads were washed seven times with radioimmune precipitation buffer (RIPA) buffer (50 mM HEPES-KOH, pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40 and 0.7% Na-Deoxycholate) and one time with TE containing 50 mM NaCl. The precipitated DNA was eluted by heating at 65°C and crosslinking was reversed by overnight incubation at 65°C. The ChIP DNA was extracted with a PCR purification kit (Qiagen) and was subjected to real-time PCR amplification using the primers specific for the promoters of genes analysed (Supplementary Table S2). The data for the occupation are expressed as a ratio of the cycle threshold for the immunoprecipitated chromatin DNA versus the cycle threshold for the input (5%) samples and further normalized to the values in control cells. RIP was performed as described previously (Kaneko et al, 2010).
ChIP-chip assay
Fragmented DNA obtained from ChIP assay was repaired by DNA Terminator End Repair Kit (Lucigen Corporation, Middleton, WI) and purified by QIAquick PCR purification kit as described previously (Murphy et al, 2010). Then, two unidirectional linkers were annealed (JW102 5′-GCGGTGACCCGGGAGATCTGAATTC-3′; JW103 5′-GAATTCAGATC-3′) and were ligated to the blunt end DNA at 16 °C overnight. The purified products were amplified to 2 μg by LM–PCR as described previously (Murphy et al, 2010). NimbleGen Mouse ChIP-chip 3 × 720K RefSeq Promoter Arrays (Roche) were used for hybridization. Relative enrichments were calculated by dividing the normalized level of ChIP DNA to that of input DNA at corresponding locus. To identify DNA promoter regions with significant association, the enrichment for each probe on the array was calculated as the ratio of the intensities of the ChIP product (Cy5) to control input chromatin (Cy3). Probes covered 4000, bp for each of 20 404 promoter proximal regions in the mouse genome (MM9).
Nuclear ChIP assay
Prior to ChIP assay, nuclear extraction and RNase A treatment were performed as previously described (Yap et al, 2010). Briefly, R1 mouse ES cells were transfected with Brca1-specific siRNA or control siRNA (Dharmacon). At 48 h post transfection, cell nuclei were extracted using cell membrane lysis buffer (10 mM HEPES, pH 8.0, 1.5 mM MgCl2, 0.5 mM DTT, 0.01% Nonidet P-40 and 1% protease inhibitor cocktails) and treated in TMS buffer (50 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2) with or without 50 μg/ml RNase A (Invitrogen) at room temperature for 30 min. Then, cell nuclei were crosslinked with 1% formaldehyde and subjected to ChIP assay as described above.
Embryoid body formation assay
Embryoid body formation assays were performed as described previously (Li et al, 2010). Briefly, cells were plated at a density of 3 × 105 per 10-cm Ultra-low-Attachment Dishes (Corning) containing 15 ml of ES medium without LIF, and the medium was changed every other day. The pictures of embryoid body formation were taken every other day.
Wound healing assay
MDA-MB-231 cells were transiently transfected with pools of BRCA1-specific siRNA, EZH2-specific siRNA or control siRNA (Dharmacon) as indicated. Artificial wounds were created on the cell monolayer when cells were grown to confluence. Migrated cells and wound healing were visualized at 0, 24 and 48 h. For each group, at least three artificial wounds were photographed immediately and at the time points indicated after the wound formation. Cell migration was evaluated by measuring the difference in wound width.
In vitro invasion assay
In vitro invasion assay was conducted by using BioCoat Matrigel invasion chamber (BD Biosciences) according to the manufacturer’s protocol. MDA-MB-231 cells were transfected with BRCA1-specific siRNA, EZH2-specific siRNA or control siRNA as indicated, cultured in the insert for 48 h and stained with Diff-Quick stain. At least five fields for each group were photographed after staining. Invasion was evaluated by counting the number of the invaded cells.
Statistics
Experiments were carried out with three or more replicates. Statistical analyses were performed using two-tailed Student’s t-test. P<0.05 is considered statistically significant. Hypergeometric test was used to determine the statistical significance of the EZH2 new target genes overlapped between PL2F8 cells with Brca1-heterozygous deletion and Brca1-knockdown R1 cells.
Supplementary Material
Acknowledgments
We thank Mien-Chie Hung, Yi Zhang, Zhenkun Lou, Xiaohua Shen and Fergus Couch for plasmids and reagents; Shyam Sharan for matched Brca1 wild-type, deficient and rescued mouse ES cells; and Jeffrey Simon and Aswathy Rai for providing purified PRC2 complexes. This work was supported in part by grants from the National Institutes of Health (CA134514 and CA130908 to HH and K99/R00CA129565 to JY), the Department of Defense (W81XWH-09-1-622 to HH) and the American Cancer Society (the Research Scholar Award RSG-12-085-01 to JY).
Author contributions: LW (Lan Wang), XZ, SC, LD and JZ (Jian Zhong) performed the experiments described in the manuscript. JCZ, LW (Liguo Wang), AS, JZ (Jizu Zhi) and JY performed ChIP-chip data and statistical analyses. AK performed mass spectrometry and data analysis. YM and JC provide reagents. HH conceived the study. LW (Lan Wang), XZ, SC, YM, JC and HH wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R (2000) BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102: 257–265 [DOI] [PubMed] [Google Scholar]
- Bohrer LR, Chen S, Hallstrom TC, Huang H (2010) Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: a potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology 151: 5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K (2003) EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22: 5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V (2004) The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC (2005) Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310: 306–310 [DOI] [PubMed] [Google Scholar]
- Chang S, Biswas K, Martin BK, Stauffer S, Sharan SK (2009) Expression of human BRCA1 variants in mouse ES cells allows functional analysis of BRCA1 mutations. J Clin Invest 119: 3160–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MS, Verma IM (1996) Transcriptional activation by BRCA1. Nature 382: 678–679 [DOI] [PubMed] [Google Scholar]
- Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, Simon JA, Huang H (2010) Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol 12: 1108–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA 107: 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ (1999) Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD (2004) BRCA1 functions as a breast stem cell regulator. J Med Genet 41: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA (2003) Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 95: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Furuta S, Jiang X, Gu B, Cheng E, Chen PL, Lee WH (2005) Depletion of BRCA1 impairs differentiation but enhances proliferation of mammary epithelial cells. Proc Natl Acad Sci USA 102: 9176–9181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, Benezra M, Licht JD, Boudreau NJ, Tsai KK, Welm AL, Feldman MD, Weber BL, Weaver VM (2010) HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest 120: 1535–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, Haber DA (1999) Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell 97: 575–586 [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Lou Z, Chen J, Tindall DJ (2006) CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314: 294–297 [DOI] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D (2010) Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24: 2615–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA (1995) The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet 29: 289–303 [DOI] [PubMed] [Google Scholar]
- King MC, Marks JH, Mandell JB (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302: 643–646 [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 100: 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Freedman DA, Levine AJ, McLendon GL (1998) Metal and RNA binding properties of the hdm2 RING finger domain. Biochemistry (Mosc) 37: 17005–17015 [PubMed] [Google Scholar]
- Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, Chang-Claude J, Hamann U, Cornelisse CJ, Devilee P, Beckmann MW, Nestle-Kramling C, Daly PA, Haites N, Varley J, Lalloo F et al. (2005) Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res 11: 5175–5180 [DOI] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L et al. (2010) Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12: 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ (2009) Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907–913 [DOI] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, Ahn BH, Kumar NG, Rovira II, Xu XL, van Lohuizen M, Motoyama N, Deng CX, Finkel T (2009) Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, Dontu G, Wicha MS (2008) BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci USA 105: 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, Kerkhoven RM, van Vliet MH, Wessels LF, Peterse JL, Berns A, Jonkers J (2007) Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA 104: 12111–12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J (2011) More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza JE, Brzovic PS, King MC, Klevit RE (1999) Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J Biol Chem 274: 5659–5665 [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W et al. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71 [DOI] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, Reis-Filho JS, Smalley MJ (2010) BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7: 403–417 [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R et al. (2010) Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, Melnick A, Heckert LL, Zarkower D, Bardwell VJ (2010) Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci USA 107: 13360–13365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, Lander ES, Armstrong SA, Daley GQ (2012) Chromatin-modifying enzymes as modulators of reprogramming. Nature 483: 598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev 22: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, Koseki H, Merkenschlager M, Fisher AG (2010) ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6: 547–556 [DOI] [PubMed] [Google Scholar]
- Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, Gilmore H, Tung N, Naber SP, Schnitt S, Lander ES, Kuperwasser C (2011) Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 8: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppe J, Drost R, Liu X, Joosse SA, Evers B, Cornelissen-Steijger P, Nederlof P, Yu Q, Jonkers J, van Lohuizen M, Pietersen AM (2009) BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to Polycomb Repressive Complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res 11: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D (2011) Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet 7: e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM (1997) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435 [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ (1997) Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet 16: 19–27 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Mitsui K, Yamanaka S (2003) Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature 423: 541–545 [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Yee D, Matin A, Nadeau JH, Magnuson T (1997) Genealogy of the 129 inbred strains: 129/SvJ is a contaminated inbred strain. Mamm Genome 8: 390–393 [DOI] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP (1999) Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet 23: 474–478 [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM (2008) Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322: 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629 [DOI] [PubMed] [Google Scholar]
- Vassilopoulos A, Wang RH, Petrovas C, Ambrozak D, Koup R, Deng CX (2008) Identification and characterization of cancer initiating cells from BRCA1 related mammary tumors using markers for normal mammary stem cells. Int J Biol Sci 4: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers JH, van Lohuizen M, Citterio E (2012) The emerging role of Polycomb repressors in the response to DNA damage. J Cell Sci 125: 3939–3948 [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC (2011) CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 13: 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster R, Weber BL (2003) Breast and ovarian cancer. N Engl J Med 348: 2339–2347 [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y (2011) Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem 286: 28511–28519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Gore C, Liu J, Pong RC, Mason R, Hao G, Long M, Kabbani W, Yu L, Zhang H, Chen H, Sun X, Boothman DA, Min W, Hsieh JT (2010) Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc Natl Acad Sci USA 107: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, Xu H, Cato L, Thornton JE, Gregory RI, Morrissey C, Vessella RL, Montironi R, Magi-Galluzzi C, Kantoff PW, Balk SP et al. (2012) EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338: 1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.