Abstract
Tumour-associated oncogenes induce unscheduled proliferation as well as genomic and chromosomal instability. According to current models, therapeutic strategies that block oncogene activity are likely to selectively target tumour cells. However, recent evidences have revealed that oncogenes are only essential for the proliferation of some specific tumour cell types, but not all. Indeed, the latest studies of the interactions between the oncogene and its target cell have shown that oncogenes contribute to cancer development not only by inducing proliferation but also by developmental reprogramming of the epigenome. This provides the first evidence that tumorigenesis can be initiated by stem cell reprogramming, and uncovers a new role for oncogenes in the origin of cancer. Here we analyse these evidences and propose an updated model of oncogene function that can explain the full range of genotype–phenotype associations found in human cancer. Finally, we discuss how this vision opens new avenues for developing novel anti-cancer interventions.
Keywords: cancer stem cell, cancer therapy, mouse model, oncogenes, tumoural reprogramming
Introduction
For decades, contemporary cancer research has been mainly focused on the altered controls of proliferation in tumoural cells. This has been reflected in the therapeutic approaches employed in the clinic to treat the patients: with very few exceptions, anti-cancer treatments are targeted at the mechanisms of abnormal tumoural growth. These problems result in the eventual failure of therapy, that is often accompanied by the development of drug resistance and by metastatic dissemination. For this reason, an urgent goal of cancer research is to understand how to counteract the mechanisms that underlie the ability of normal cells to become cancer cells in the first place. The complexity of the properties of cancer cells was distilled by Hanahan and Weinberg (2011) into ‘nine essential alterations in cell physiology that collectively dictate malignant growth’. Cancer cells are the foundation of the disease: they initiate the tumours and drive cancer progression forward, and they are the ones carrying the oncogenic and tumour suppressor mutations that define cancer as a genetic disease (Hanahan and Weinberg, 2011). However, we still do not understand sufficiently well the underlying mechanisms leading to the origin of these cells, so as to have a sizable impact on cancer mortality (Jemal et al, 2009). As a result, our progress is incremental and largely empirical, leading only to slight improvements in treatments, surgical interventions or radiation regimes. These may provide some benefit, but they seem unable of bringing the disease itself to an end.
Thus, a complete understanding of the cancer process requires a more detailed knowledge of the mechanisms giving rise to neoplastic growth, and is a prerequisite, not only for the understanding of the genesis of human cancer but also for the identification of the molecular events responsible for cancer maintenance. In spite of this, all the aspects related to the alterations of the normal developmental regulatory mechanisms in carcinogenesis have received comparatively little attention during the process leading to the definition of the hallmarks of cancer cells. But in fact, if cellular fate was immovable, cancer would not be possible, since no new lineages could be generated other than the normal, physiological ones. Here is where the mechanisms regulating tumour cellular identity play an essential role in allowing cancers to arise and hopefully, as we will discuss, they might be the key to its eradication. The aim of this review is to discuss the impact that oncogenes have in establishing the identity of the tumour cell, and how a better understanding of this previously unexplored mode of action of the oncogenes should lead us to a deeper knowledge of carcinogenesis, and to the development of new treatments.
Cancer cells and oncogene addiction
During the last four decades, scientific research has clearly demonstrated the relevance of oncogenes in human cancer. Since the discovery that human tumours contain activated oncogenes (Der et al, 1982; Goldfarb et al, 1982; Parada et al, 1982; Pulciani et al, 1982; Santos et al, 1982; Shih and Weinberg, 1982), many efforts have been made to understand their causal role in cancer development. All this work has shown that oncogene expression is not only required for cancer initiation but also for the maintenance of the disease, and has kept oncogenes in the limelight as the central anti-cancer therapeutic targets. When the oncogenic expression is driven by tissue-specific promoters in genetically engineered mouse models, tumours arise at high frequency, but they regress when the inducing stimulus is switched off (Chin et al, 1999; Huettner et al, 2000; Boxer et al, 2004), therefore suggesting that oncogenes are indeed the Achilles’ heel of cancers (Weinstein, 2002). This current model of cancer is in agreement with the fact that, in human cancers, all cancerous cells, with independence of the cellular heterogeneity existing within the tumour, carry the same initiating oncogenic genetic lesions. Overall, these observations seem to point towards an homogenous mode of action for oncogenes within cancer cells, since the brief inactivation of the different single tumour-inducing oncogenes can cause cancer remission in these model systems. Unfortunately however, the therapies based on this cancer model fail to eradicate tumours in humans (see below). These clinical observations suggest that, in human patients, oncogene-induced tumorigenesis might not be reversible through the unique inactivation of the gene defect(s) initiating cancer development. But then, what are the mechanisms of tumour relapse by which tumours evolve to escape oncogene dependence?
These therapeutic failures cannot be explained just by invoking the existence of cancer stem cells (CSCs) or by the known cellular plasticity of tumours. Indeed, both aspects only imply that a tumoural population that is genetically homogeneous may nevertheless appear as phenotypically heterogeneous, due to the presence of cells in different states of differentiation (Hanahan and Weinberg, 2011). However, the aforementioned observations, derived from human targeted-therapy failure, might suggest that oncogenes have a mode of action that is not homogenous throughout the cancer cell population. This would explain the different sensitivity towards anti-oncogene-targeted therapies among the different cancer cellular stages. Recent in vivo genetic evidences have shown that human oncogenes are capable of reprogramming early stem/precursor cells towards specific differentiated tumour cell fates, but they are not required within the malignant cells. These results not only highlight a previously unrecognized role for human oncogenes but also provide evidence for a previously unmodeled process of tumorigenesis, in which the programming of the malignant phenotype has already taken place at the stem cell stage.
Oncogene–target cell interaction
In the last years, a new recognition of the role of aberrant differentiation at the root of cancer has arisen, mainly driven by the coming of age of the ‘cancer stem cell’ theory. From this point of view, a comprehensive knowledge of the developmental mechanisms by which normal target cells acquire their tumour identity is essential to understand cancer development.
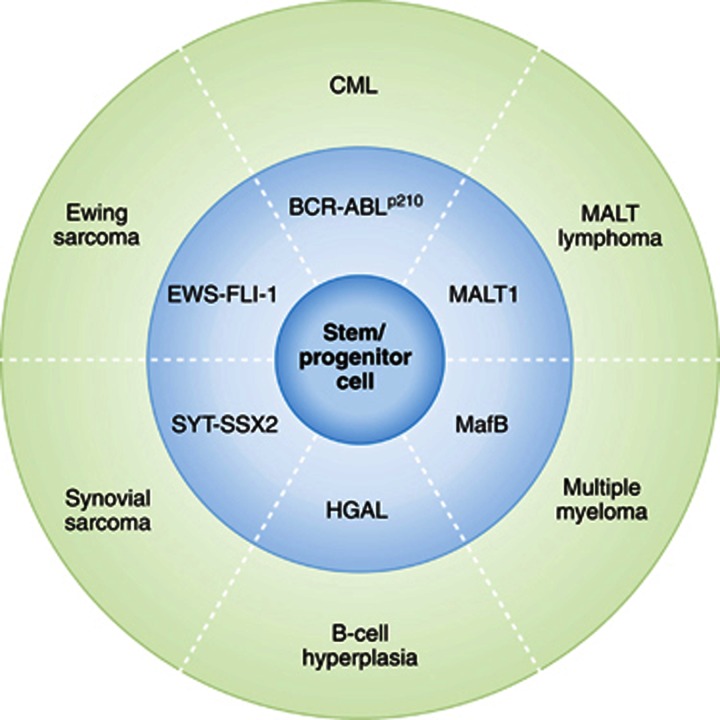
Cytogenetic and molecular genetic analyses have identified that many types of cancer are specifically associated with consistent defined genetic events (Mitelman et al, 2013). The expression of each one of these genetic lesions is associated almost exclusively with a characteristic subgroup of human cancer (Figure 1). Not only are these genetic lesions of clinical importance, as they may serve as unequivocal diagnostic markers, but they also provide important clues to the understanding of the cellular mechanisms behind cancer development. However, these genotype–phenotype correlations established in humans have demanded during the last three decades that we explain the nature of this intimate association between each genetic lesion and the phenotype (particular type of cancer) with which it is associated. Two different hypotheses have been considered to explain this link (Figure 2). In the classical view of the initiation and progression of cancer, the initiating genetic alteration takes place and is required for the immortalization of a committed/differentiated target cell (Figure 2A). Such cell will afterwards acquire additional genetics hits over time. The acquisition of additional hits aggravates the deregulation of the behaviour of the differentiated target cell, therefore leading to the clinically recognized features of cancers. This is the model that has traditionally been assumed in the study of oncogenesis, taking for granted that the phenotype of the tumour cells was a reflection of that of the normal cell that gave rise to the tumour in the first place. Since the beginning, there were already some classical examples in which this was clearly not the case, for example, chronic myelogenous leukaemia (CML), where Fialkow et al (1977) first suggested nearly 40 years ago that the disease arose from rare transformed hematopoietic stem cells (HSCs), since the t(9;22) chromosomal translocation could be found in most types of differentiated haematopoietic cells. But, in most cases, cancerous cells do share similarities with some non-pathological differentiated cell types. Therefore, for every kind of cancer, the cell-of-origin was assumed to be the corresponding normal differentiated cell.
Figure 1.
Examples of cancer types generated directly from mouse stem/progenitor cells by tumour fate reprogramming. Human cancer is associated to specific and consistent genetic events. Each one of these genetic defects is observed exclusively in characteristic subgroups of human cancer. In mouse models, as illustrated, specific genotype alterations associated to human cancer (medium circle) give rise to specific phenotypes (outer circle) when targeted to the stem cell/progenitor compartment (see text for details).
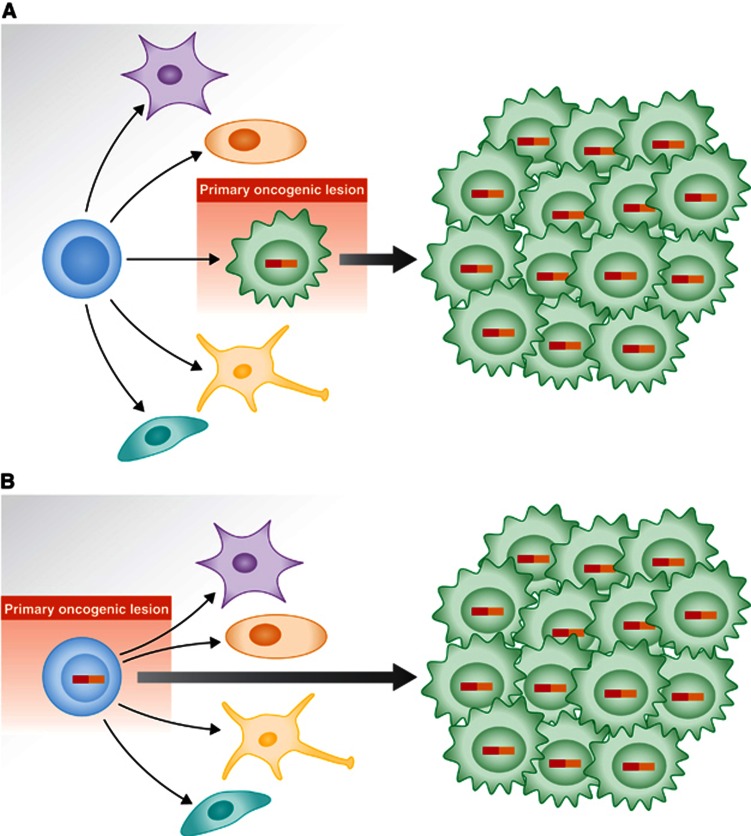
Figure 2.
Proposed model for the role of human cancer gene defects in tumour cell fate specification. (A) Traditionally, the human cancer genetic defects have been thought to act on cells already committed to a differentiation program, in such a way that the tumoural phenotype is derived from that of the initial differentiated target cell. (B) Alternatively, the latest findings support a view in which the oncogenic lesion acts on stem/progenitor cells by imposing a given, oncogene-specific, tumour-differentiated cell fate.
The other way of interpreting the genotype–phenotype correlations observed between genetic lesions and a given tumoural type is to consider the possibility that the oncogene is directly responsible of imposing the specific characteristics of the tumour phenotype (Figure 2B). This is in fact what happens in CML, where the oncogene is expressed at the stem cells but the phenotype manifests as a granulocytic expansion.
Cancer stem cell theory
The CSC hypothesis is in good agreement with the interpretation of oncogenesis presented in Figure 2B (Reya et al, 2001; Sanchez-Garcia et al, 2007). The term tumour/CSC (or cancer-maintaining cell, see the definition of terms in Figure 3) was first coined nearly 40 years ago to explain the observation that only a small subset of multiple myeloma cells were capable of clonogenic growth (Hamburger and Salmon, 1977a; Hamburger and Salmon, 1977b), and it was demonstrated experimentally for the first time for a human cancer by Bonnet and Dick (1997). The CSC theory proposes that tumours are stem cell-based tissues, like any other one in the organism. This implies the existence of a hierarchical structure within the tumour and, most importantly, that not all the cells forming the tumoural mass are equally competent for regenerating the tumour, and that the phenotype of the tumoural cells is, to a large degree, genetically programmed by the oncogene from the tumour stem cell stage. This would imply that, both in the cases of tumour regeneration after transplantation, and of tumour relapse after therapy, only a certain tumoural subpopulation (the cells possessing stem properties) is responsible for the cancer recurrence. The existence of CSCs is the reason why current therapies are incapable, in most cases, of eradicating the disease, as these cells are, in general, resistant to antiproliferative therapies and no other options are available due to toxicities to sensitive normal stem cells. Indeed, in the same way that normal stem cells continue giving rise to normal tissues after chemotherapy (regrowth of lost hair, regeneration of the hematopoietic compartments, reparation of intestinal mucosae), CSCs regenerate the tumour as well. There are a few exceptions to this rule, such as testicular carcinoma, where tumour stem cells are more sensitive to the chemotherapy cocktail than normal stem cells, and then cancer can be cured. There are discrepancies among researchers and, most probably, big differences among different cancer types, in the calculated proportions of CSCs within a tumour, ranging from very few to a large 25% (Quintana et al, 2008; Cobaleda and Sanchez-Garcia, 2009; Vicente-Duenas et al, 2009a; Vicente-Duenas et al, 2009b). Nevertheless, independently of this fact, the most relevant aspect is that not all the cells composing the tumour mass possess the capacity of regenerating it. The question that follows is then: nowadays, it is accepted that the tumour mass comes from the cancer stem cells but, where do they come from themselves? What is their origin?
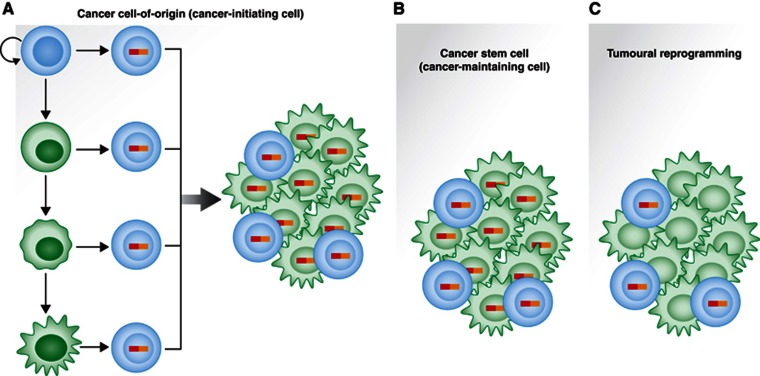
Figure 3.
Developmental biology of cancer cells. (A) Cancer cell-of-origin (or cancer-initiating cell): the cell where the first genetic lesion linked to the development of the tumour takes place. It might be located anywhere within the physiological differentiation pathway. It does not need to have any phenotypic relationship with the final phenotype of the tumour cells (either stem or differentiated). (B) CSC (cancer-maintaining cell): those cells that have the capacity to regenerate all the cellular diversity of the tumour. They retain broad self-renewal potential and differentiation potential. They arise initially from the cancer cell-of-origin, and then they can self-propagate. (C) Tumoural reprogramming: the process by which the initial oncogenic lesion(s) can ‘reset’ the epigenetic and/or transcriptome status of an initially healthy cell (the cancer cell-of-origin), therefore establishing a new, pathological differentiation program ultimately leading to cancer development, where the oncogenic lesion(s) does not need to be present anymore once the initial cancer fate-inducing change has taken place.
The nature of the cancer cell-of-origin
This cancer cell-of-origin (or cancer-initiating cell, not to be confused with the CSCs which, as we have explained, are the cancer-maintaining cells of a tumour that is already developed) is the, initially, healthy cell (it doesn’t necessarily have to be a stem cell) that will be reprogrammed by the oncogenic hit(s) and will finally give rise to a (pre)tumoural cell with stem cell properties (Figure 3A). From a developmental point of view, there are two possibilities to be considered in this context. One option is that the cell-of-origin that suffers the first oncogenic hit(s) is a stem cell: in this case, a new tumoural stem cell will be reprogrammed to generate the new pathological tissue instead of the normal one. In the case of CML, by restricting the expression of the oncogenic alteration to the stem cell/progenitor compartment in genetically modified mice, it has been possible to generate a tumour very similar to the human one, with its cellular variability (Perez-Caro et al, 2009; Vicente-Duenas et al, 2009b). In the case of intestinal cancers, when the oncogenic hit (activation of the Wnt signalling pathway) is directed to the stem cell compartment in mouse models, intestinal adenomas are generated that maintain their internal developmental hierarchy; however, when the targeting of the oncogenic hits is directed to differentiated intestinal epithelial cells, only short-lived, small microadenomas appear (Barker, 2008; Barker et al, 2009; Zhu et al, 2009). These data strongly suggest that the tumour must have its origin in the crypt stem cells. Something similar happens in the context of tumours of the nervous system; when oncogenic lesions associated to astrocytomas are targeted to tissue progenitors (the subventricular zone), tumours originate, while only astrogliosis happens when the targeted cells are differentiated adult parenchymal cells (Alcantara Llaguno et al, 2009). These examples and others (Dirks, 2008; Joseph et al, 2008; Zheng et al, 2008) prove that the initiating event can take place in a normal stem cell, even if the mature tumour is composed by differentiated cells, and that the oncogenic lesions possess reprogramming capacity, leading to the appearance of a mature tumoural differentiated population (Vicente-Duenas et al, 2009b).
There is, however, other option regarding the nature of the cancer cell-of-origin: it might happen that a differentiated cell is the original target of the initiating oncogenic hit(s), and in the subsequent process of oncogene-mediated tumoural reprogramming, it regains stem cell characteristics to become a true CSC. This possibility has, however, two prerequisites: on the cell side, it implies that, even though it is a differentiated cell, it must possess enough plasticity so as to be reprogrammable (at least, by this specific oncogene). On the oncogene side, the alteration must be able of activating the required programs to finally confer stem cell properties (at least, for this specific target cell). It has been demonstrated that some oncogenes can generate CSCs when they are introduced into committed target cells; this is the case for MOZ-TIF2 (Huntly et al, 2004), MLL-AF9 (Krivtsov et al, 2006; Somervaille and Cleary, 2006), MLL-ENL (Cozzio et al, 2003), MLL-GAS (So et al, 2003) or PML-RARa (Guibal et al, 2009; Wojiski et al, 2009). MLL-AF9, for example, can confer the property of self-renewal to committed granulocyte–macrophage progenitors, by activating in them a stem cell-like program (Krivtsov et al, 2006). Similarly, c-Myc can originate epithelial CSCs by inducing an embryonic stem cell-like transcriptional program in differentiated epithelial cells (Wong et al, 2008). However, other oncogenes are unable of conferring self-renewal properties, like in the case of BCR-ABLp190 (Huntly et al, 2004). In these cases, where the oncogene cannot confer stem cell properties, it might however originate a pre-cancerous cell that can afterwards, with the accumulation of additional alterations leading to the acquisition of stem cell properties, give finally rise to a CSC (Chen et al, 2007). For example, it has recently been described in intestinal tumorigenesis that the inflammatory tumour microenvironment can, through elevated NF-κB signalling, lead to an enhanced Wnt activation that, in turn, causes dedifferentiation of non-stem cells and acquisition of tumour-initiating capacity (Schwitalla et al, 2013). In human gliomas, MEF promotes the acquisition of stem cell characteristics by upregulating Sox2 (Bazzoli et al, 2012), and other studies also suggest that gliomas can originate by dedifferentiation of neurons and astrocytes (Friedmann-Morvinski et al, 2012). This reprogramming effect of oncogenes does not have to be restricted to cells of the same lineage; for example, adipocyte-restricted activation of Sonic hedgehog signalling in mice gives rise to aggressive rhabdomyosarcomas, indicating that adipocyte progenitors can be the cell-of-origin of these tumours, that can therefore originate from non-skeletal muscle precursors (Hatley et al, 2012). In the initiation of basal carcinoma, constitutively active expression of an Smoothened mutant (SmoM2) in the adult epidermis leads to the appearance of tumour-initiating cells with the characteristics of embryonic hair follicle progenitors, caused by the reprogramming of adult interfollicular tumour-initiating cells (Youssef et al, 2012). Many of these aspects can be recapitulated, to a certain degree, in vitro, where CSC-like cells can be generated by oncogenic reprogramming of human somatic cells during neoplastic transformation (Scaffidi and Misteli, 2011). Indeed, human differentiated cells can be transformed in vitro by the action of telomerase, an oncogenic H-RasV12 mutant and the concomitant inhibition of p53 and pRB; during in vitro transformation, a subset of the fibroblasts are reprogrammed to a more primitive, multipotent cell type, possessing CSC characteristics and generating hierarchically organized tumours (Scaffidi and Misteli, 2011).
The tumoural stem cell reprogramming hypothesis
Cellular reprogramming refers to ‘the concept of rewiring the epigenetic and transcriptional network of one cell state to that of a different cell type’ (Hanna et al, 2008). When this process is induced by molecularly defined means it is called direct reprogramming. The term ‘reprogramming’ is very frequently used as it was restricted to direct reprogramming to induced pluripotent stem cells (iPSCs) by means of a specific cocktail of transcription factors (Takahashi and Yamanaka 2006; Takahashi et al, 2007). However, as defined above, ‘reprogramming’ also encompasses other processes of cell fate change, not necessarily involving the four Yamanaka transcription factors, and not always implying the existence of iPSCs as an end point (Figure 4). Accordingly, tumoural reprogramming is the process by which an oncogene (or cancer genetic alteration) can ‘reset’ the epigenetic and/or transcriptome status of an initially healthy cell (the cancer cell-of-origin), therefore establishing a new, pathological differentiation program ultimately leading to cancer development (Figures 3C and 4). Therefore, the parallelism with reprogramming to pluripotency is clear, (Krizhanovsky and Lowe, 2009; Perez-Caro et al, 2009). The existence of this process is, however, difficult to demonstrate in human cancers, since the initiating oncogenic lesions that reprogram the fate of the tumour-target cells are genetically preserved in the cancer-maintaining cells, and also throughout the different pathological cellular intermediates, until the final tumour differentiated cells, thus making it difficult to ascertain their molecular role at the different stages of tumour biology.
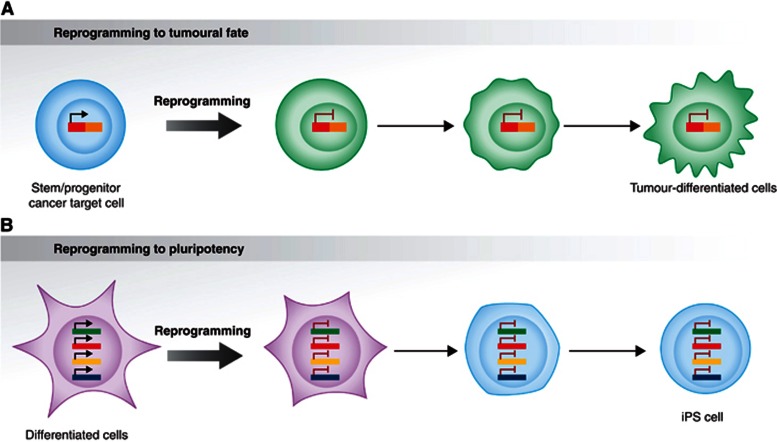
Figure 4.
Tumour stem cell reprogramming versus reprogramming to pluripotency. (A) Recent in vivo genetic evidences have shown that human oncogenes are capable of reprogramming early stem/precursor cells towards specific differentiated tumour cell fates, but they are not required afterwards, within the malignant cells. (B) ‘Hit-and-run’ reprogramming has grounding in other contexts outside of cancer, such as during induced pluripotent stem (iPS) cell formation in vitro. However, unlike the tumour stem cell reprogramming, the iPS process initiates in a differentiated cell (see text for details).
One important conceptual and biological aspect of the molecular mechanisms of reprogramming to pluripotency is the fact that, once the way to pluripotency has been opened and the reprogramming per se has taken place, the pluripotent condition does not require the inducing factors anymore for its maintenance (Papp and Plath, 2013). Therefore, by parallelism, if CSCs are generated by a (tumoural) reprogramming process, then maybe the oncogenes that initiate tumour formation might not be required for tumour progression (Krizhanovsky and Lowe, 2009; Perez-Caro et al, 2009). This would explain the cases in which a pre-cancerous lesion exists stably as the single aberration in an abnormal cell population that will only progress to an open tumour when secondary hits occur. The initiating lesion would be the driving force in the reprogramming process, essential for tumorigenesis. However, once reprogramming has taken place, this initiating hit would only be a passenger mutation within the CSC, either without a significant function anymore, or even performing a different role, unrelated to the reprogramming one, in tumour expansion or proliferation. This mode of action would explain why well-designed targeted therapies fail in eradicating the disease, in spite of their apparent efficacy against the main tumour mass (see below).
In vivo experimental model of tumoural stem cell reprogramming
To be able to demonstrate this lack of homogeneity in the mode of actions of oncogenes throughout the biological history of the tumour, it would be necessary to dissect and isolate the function that the oncogene is playing at the earliest stages of the disease, at the level of the cell-of-origin. Indeed, to prove that the maintenance of the expression of the oncogene is not necessary for tumour progression beyond the initial step of reprogramming, one would need to find a way of restricting the expression of the oncogene to the stem/progenitor compartment. Such a system would also allow us to prove, if this was indeed the case, that the oncogenes that initiate tumour formation might be dispensable for tumour progression and/or maintenance. To shed light on this issue and to elucidate if cancer is a stem cell-driven tissue, we have used the locus control region of the mouse Sca1 (Ly6E.1) gene to restrict the expression of selected, specific human cancer-associated oncogenes to the stem cell compartment in a transgenic mouse setting (Perez-Caro et al, 2009; Vicente-Duenas et al, 2012a; Vicente-Duenas et al, 2012c; Romero-Camarero et al, 2013). We initially focused our studies on BCR-ABLp210+ CML, since this is widely accepted to be a stem cell disorder. CML starts as a prolonged chronic phase, characterized by high leukocyte counts and enlarged spleen and liver. Within a median time of 5 years (in the absence of treatment), the chronic phase develops into a blast crisis that is indistinguishable from an acute leukaemia. The current treatment of choice, the specific BCR-ABL inhibitor STI571 (imatinib) is able to eliminate the BCR-ABL-expressing differentiated cells that constitute the bulk of the tumour, but it cannot eliminate BCR-ABL-expressing CSCs (see below).
In transgenic mice, when the expression of BCR-ABL is restricted to the stem/progenitor cells, the Sca1-BCR-ABLp210 mice develop a CML that very closely recapitulates the main features of human disease. In these Sca1-BCR-ABLp210 mice, according to the experimental design, although tumour initiation takes place at the stem cell/progenitor compartment, all the leukaemic differentiated cells, which form the main mass of the tumour, have already switched off the expression of the oncogene. Therefore, BCR-ABL is not expressed in lineage-positive hematopoietic cells, not even the tumoural ones. However, it is conceivable that BCR-ABL target genes could continue to be expressed in the absence of BCR-ABL, by a ‘hit-and-run’ mode of action in which the oncogene would turn genes on in stem cells but would not be required afterwards for maintaining their expression at later stages of development. However, neither the BCR-ABL protein nor any known downstream signalling were detected in the committed non-progenitor cells of Sca1-BCR-ABLp210 mice, indicating that BCR-ABL downstream targets are effectively switched off after the silencing of BCR-ABL. How is it then possible that CML develops efficiently in these mice, when in human cancers all leukaemic cells (not only the CSC) carry the oncogenic genetic lesions? This can only be explained if BCR-ABL is actually reprogramming, by a ‘hit-and-run’ mechanism, the hematopoietic stem/progenitor cells (HS/P-Cs) for malignancy. In summary, these results represented the first in vivo genetic evidence mechanistically connecting tumorigenesis and reprogramming of early progenitors, and they uncovered the possibility of a reprogramming-like mechanism in cancer development. Other examples of tumoural stem cell reprogramming, in which the fate-changing effect of the oncogene takes place at the stem cell level (in contraposition to reprogramming to pluripotency, that implies starting from a differentiated cell, see Figure 4) have been described for other types of tumours (Figure 1). For example, the EWS-FLI-1 fusion gene, associated with 85–90% of Ewing sarcoma family tumours, induces expression of the embryonic stem cell genes OCT4, SOX2, and NANOG in human pediatric mesenchymal stem cells but not in their adult counterparts, reprogramming them towards Ewing sarcoma CSCs (Riggi et al, 2010). Also, the synovial sarcoma-associated oncogene SYT-SSX2 can reprogram mesenchymal stem cells by dictating their commitment to a pro-neural lineage and most likely this constitutes the primary event in this type of tumour (Garcia et al, 2012).
Reprogramming in malignancies supposed to originate from mature cells
An essential question that remained to be answered in vivo to prove the reprogramming capacity of human oncogenes, is whether normal progenitors can also be reprogrammed to give rise to terminally differentiated tumour cells by those other oncogenes that are specifically associated with malignancies commonly believed to originate from mature, differentiated cells. Recent data provide proof-of-principle evidence of the fact that, at least in the context of multiple myeloma (MM) (Vicente-Duenas et al, 2012c), mucosa-associated lymphoid tissue (MALT) lymphoma (Vicente-Duenas et al, 2012a) and HGAL-associated lymphoma (Romero-Camarero et al, 2013), this is possible. Indeed, genetically engineered mouse models have shown that hematopoietic stem/progenitor cells (HS/P-Cs) can be directly reprogrammed into either tumour plasma-like cells or clonal extranodal B-cell lymphomas, in vivo by an oncogene that in human patients is associated either with MM or with MALT lymphoma, respectively, therefore proving the oncogene capacity of programming and propagating the tumoural cellular identity. This oncogene-mediated reprogramming is, however, permissive in that the reprogrammed Sca1+ population can nevertheless complete a multistage differentiation pathway involving an initial commitment to the B-cell lineage and a subsequent differentiation to plasma cells, and to all the other hematopoietic lineages. In human CLL, it has recently been shown that the propensity to generate malignant B cells is acquired very early, at the HSC stage, long before the cells become B cells (Kikushige et al, 2011). As a consequence of this reprogramming, patient-derived HSCs show an abnormal expression of lymphoid-related genes, presumably reflecting their cell-intrinsic pathological priming into the lymphoid lineage. Thus, these results provide evidence that the oncogenic proteins expressed in stem/progenitor cells can have selective impacts that depend on their intrinsic molecular properties, and in turn provide a rationale for the strikingly consistent associations between different chromosomal translocations, their unique resulting fusion genes and the final cancer phenotypes. Thus, these results support the view of cancer also as a disease of cell differentiation, and not only of multiplication, highlighting a previously unrecognized role for oncogenes in cancer, and also providing evidence for a previously unmodelled process for tumorigenesis in which the programming of the malignant phenotype has already taken place at the stem cell stage. Here one must take into account that ‘hit-and-run’ fate programming is not something happening only in non-physiological conditions, like cancer. For example, during normal hematopoietic differentiation, molecular cues such as IL7 and erythropoietin are required to induce, to instruct, or to allow commitment to a given differentiation program, but are not required thereafter, once the fate has been established.
Tumoural stem cell reprogramming and therapeutic implications
The main therapeutical consequence of these findings is that, if cancer occurs via a reprogramming-like mechanism, oncogenes that initiate tumour formation might be dispensable for tumour-target cell survival and/or tumour progression. In this context, mutations that activate oncogenes would have a driving role in the reprogramming process, but may act as passenger mutations thereafter, or may have a secondary role in evolved tumour cell clones. This is well illustrated by the BCR-ABL kinase inhibitor imatinib/STI571, which can target with high efficiency the differentiated tumour cells of chronic myeloid leukaemia (CML) but cannot kill the BCR-ABL-expressing stem cells (Corbin et al, 2011; Chomel et al, 2011; Chu et al, 2011; Hamilton et al, 2012; Kumari et al, 2012), since it does not seem to interfere with the reprogramming role that the chimeric oncogene is playing in this cellular context (Graham et al, 2002; Barnes and Melo, 2006; Perez-Caro et al, 2009). These findings therefore have important implications for understanding and therapeutically targeting tumour cell pathobiology. Indeed, in the stem cell-driven Sca1-BCR-ABL model, the expression of the oncogene is restricted to the stem/progenitor compartment, but is nevertheless capable of generating a full-blown CML with all its differentiated cellular components, and it is not controllable by the use of imatinib (Perez-Caro et al, 2009). Of course, the fact that CML development can be recapitulated in mice by limiting oncogene expression to Sca1+ cells implies that eliminating oncogene function in human patients is not going to interfere with neither the survival of the CSC nor the formation of differentiated tumour cells, because it suggests that the oncogene is programming a (epi)genetic regulatory state in stem cells, that in some way persists during hematopoietic development, and which imposes a CML-specific tumour phenotype. This observation also applies to other cancer-initiating gene defects, as we have described. Indeed, when the oncogene expressed under the control of Sca1 regulatory sequences is MafB, associated with human multiple myeloma, the tumoural differentiation is directed to this pathology (Vicente-Duenas et al, 2012c). The same holds true for the MALT-lymphoma-associated protein MALT1 (Vicente-Duenas et al, 2012a), or for the lymphoma-associated protein HGAL (Romero-Camarero et al, 2013; Figure 1). Therefore, one same pattern of ectopic oncogene expression leads to different tumoural phenotypes, clearly driven in each case by the reprogramming capacities of each individual oncogene when expressed at the stem cell level. This series of results represent the most striking in vivo demonstration of the genotype–phenotype correlations seen for human oncogenes in human cancer (Perez-Caro et al, 2009; Sanchez-Garcia, 2009; Alizadeh and Majeti, 2011; Ben-David et al, 2011; Moran-Crusio et al, 2011; Vicente-Duenas et al, 2012a; Vicente-Duenas et al, 2012c; Romero-Camarero et al, 2013). In the specific case of CML, for which an oncogene-targeted therapy already exists in the clinic, these data show that CML stem cell survival is BCR-ABL kinase-independent and suggest that curative approaches in CML must focus on kinase-independent mechanisms of resistance (Perez-Caro et al, 2009). The results on the failure of imatinib to eradicate CML in Sca1-BCR-ABLp210 mice have been later confirmed in human patients (Corbin et al, 2011; Chomel et al, 2011; Chu et al, 2011; Hamilton et al, 2012; Kumari et al, 2012), this being the first time a preclinical model anticipates human CSC-therapeutic response. Therefore, these combined studies of the effect of BCR-ABL in cancer development show that leukaemia stem cells are not oncogene addicted, and are most likely extensible to many other pathologies (multiple myeloma, MALT lymphoma, CLL, etc.). These findings challenge the currently accepted models for the origin of human hematopoietic cancers and for the role of oncogenes.
Tumour suppressors can act as barriers for tumoural stem cell reprogramming
In analogy to what happens in reprogramming to pluripotency (Zhao et al, 2008; Banito et al, 2009; Hong et al, 2009; Kawamura et al, 2009; Krizhanovsky and Lowe, 2009; Li et al, 2009; Marion et al, 2009; Utikal et al, 2009), the efficiency of the oncogene-induced tumoural reprogramming of normal HS/P-Cs to terminally differentiated malignant cells is enhanced by p53 deficiency, at least in the cases of BCR-ABL (CML) and MafB (MM) (Velasco-Hernandez et al, 2012; Vicente-Duenas et al, 2012b). These results suggest that the absence of the tumour suppressor does not have an instructive role in the genesis of tumour cells, but just a permissive one, preventing cells with damage from being successfully terminally reprogrammed. In addition, this further supports the interpretation that the driving force of the tumoural reprogramming process is the oncogene itself, and that it is just the need of maintaining genetic integrity what prevents the reprogrammed cells with any kind of damage to progress along the newly programmed malignant pathway (just like in iPSCs generation). Along these lines, transient restoration of p53 slows down CML disease progression and significantly extends the survival of leukemic animals (Velasco-Hernandez et al, 2012). The mechanism responsible for this effect is the p53-mediated apoptotic death of primitive leukemia cells, therefore suggesting that reestablishing p53 function might be a therapeutic strategy for the eradication of leukemic stem cells and for the prevention of disease progression.
Fidelity of tumour lineage conversion by oncogenes: epigenetic memory
During cancer development, a new tumour cell identity is established and maintained in a manner that is specific for each different oncogene. This raises another interesting question: what are the similarities and differences between the different oncogenic ways of resetting the genome? There is still much to learn about the mechanisms that underlie tumour reprogramming, and, by analogy to the iPSCs generation process, it is conceivable that tumour reprogramming may take place in distinct stages.
The term ‘epigenetic memory’ in a reprogramming process refers to the retention of transcriptional properties or chromatin features of the starting cell type that remain after reprogramming (Feinberg et al, 2006; Iacobuzio-Donahue, 2009). The persistence of epigenetic memory is a critical issue in the reprogramming field because it has the potential of modifying the behavior of the reprogrammed cells, and could have important consequences for cancer disease-modelling studies, and for any future clinical application. We have demonstrated that there is epigenetic memory in tumour plasma cells produced as a result of the reprogramming of stem cells by the MafB oncogene (Vicente-Duenas et al, 2012c). This suggests that an epigenetic mark of transcriptional activity persisted through the reprogramming process, setting a pattern inherited throughout B-cell development, similar to the DNA methylation changes taking place during somatic erythroid cell differentiation (Shearstone et al, 2011), and showing that HS/P-Cs can be epigenetically reprogrammed to terminally differentiated tumour cells by defined oncogenes associated to human neoplasias. Similar to iPSCs generation, this work clearly showed that tumoural generation involves, in the first place, epigenetic rather than genetic modifications to generate the myriad of tumour-differentiated cells that emerge from a tumour stem cell.
A new approach to complement cancer treatment: reprogramming the cancer epigenome to an alternative lineage cell fate
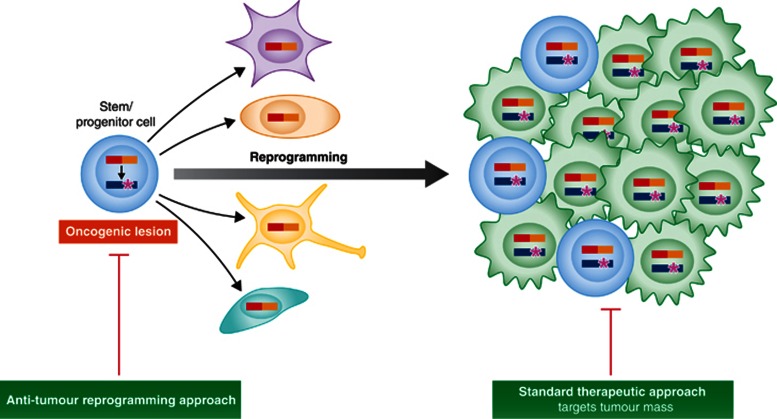
Tumour stem cell reprogramming as a cancer driver opens a clear hope for cancer treatment, since epigenetic modifications, unlike genetic changes, can be erased, manipulated, and reinitiated (Figure 5). Indeed, it has already been proven that incorporation of targeted epigenetic agents to the standard chemotherapy is a promising approach to the treatment of relapsed pediatric ALL (Bhatla et al, 2012). In different experimental settings, the results suggest that cancer cells can be reprogrammed to a non-tumoural fate, losing their malignancy. For example, it is possible, using nuclear transplantation, to reprogram melanoma cells (Hochedlinger et al, 2004), embryonal carcinomas (Blelloch et al, 2004) and even to clone mouse embryos from brain tumours (Li et al, 2003). All these findings indicate that it can be completely viable to reprogram tumour cells. However, like for any other therapeutic approach, a precise knowledge of the epigenetic rewiring is necessary before we can attempt a successful intervention. Indeed, it has recently been shown that malignant glioblastoma neural stem cells can be reprogrammed to iPSCs (Stricker et al, 2013). However, these iPSCs can differentiate into mesodermal lineages losing their malignancy, but not into neural lineages, where they maintain their malignant nature (Stricker et al, 2013). In any case, it is clear that reprogramming the cancer epigenome to an alternative lineage cell fate can suppress malignancy (Figure 5).
Figure 5.
Opportunities for therapeutic intervention using tumour stem cell reprogramming as a target. Tumour stem cell reprogramming largely relies on epigenetic modifications. These, unlike genetic changes, can be erased, manipulated, and reinitiated, therefore implying that anti-tumour reprogramming strategies can provide a new window of opportunity to interfere with the cancer fate-inducing change.
This tumoural epigenetic reprogramming is conceptually different from the ‘epigenetic progenitor’ model of cancer, proposing that cancer formation involves a general, non-targeted epigenetic disruption at the level of progenitor cells, followed by an initiating mutation and then by genetic and epigenetic plasticity. However, in both contexts, epigenetic alterations in cancer serve as potent surrogates for genetic mutations and are driving forces of the initial tumoural development (Feinberg et al, 2006; Iacobuzio-Donahue, 2009).
What lies ahead?
The enormous increase in our understanding of the biology of tumour cells in the last four decades did not have a proportional impact in our capacity of curing the disease, since the treatment of most cancers has not changed significantly during this time, and the decreasing mortality has been mostly the result of early detection and prevention, rather than the consequence of effective therapeutics (Etzioni et al, 2003; Jemal et al, 2009). Therefore, the cells and the genetic lesions responsible for maintaining the disease remain an essential topic of research, since they are the ones to blame for the tumour’s resistance to conventional therapies, recurrence, and metastasis. We have seen the evidences showing that oncogenes contribute to cancer development not just by inducing proliferation, but rather because of their capacity to developmentally reprogram the epigenome of the mutated target cell. Stem cell reprogramming (where the maintenance of oncogene expression is not critical for the generation of differentiated tumour cells) seems to be a common intrinsic mechanism for many types of cancer, and this should change our understanting of the means by which ‘hallmark cancer capabilities’ are acquired. This conceptualization of tumour reprogramming by oncogenes will change the way we investigate and treat cancer in the years to come.
This discovery will also force us to explore and answer fundamental questions in cancer biology, such as how cells acquire and maintain their tumour differentiation states. We hypothesize that the epigenetic reprogramming properties of (some) oncogenes work as a new type of gene–target cell interaction in which oncogene exposure targets the epigenome to induce cancer development. To add new layers of complexity, the effect of such epigenetic reprogramming may remain dormant until engaged in response to later adult events (genetic and/or environmental). Importantly, the installation of epigenetic programs that direct tumour-cell-specific differentiation during cancer development is unidirectional. Therefore, the potential exists for a brief exposure to an environmental agent to disrupt epigenetic programming during development, and reprogram the epigenome for life (Walker and Ho, 2012). The ability to generate tumour stem cells from specific diseases and mutations in vivo has opened prospects for studying how different disease states develop from the start. If we can understand the regulation of the oncogene–target cell interaction, and as a result we learn how to manipulate cellular states experimentally, we could unlock the potential to provide great advances in human cancer medicine.
Acknowledgments
We acknowledge the anonymous reviewers for their valuable contributions to this review. We are indebted to all members of lab 13 at IBMCC for useful discussions and for his critical reading of the manuscript. Research in ISG group is partially supported by FEDER and by MICINN (SAF2012-32810), by MEC OncoBIO Consolider-Ingenio 2010 (Ref. CSD2007-0017), by NIH grant (R01 CA109335-04A1), the ARIMMORA project (FP7-ENV-2011, European Union Seventh Framework Program) and by proyecto en red de investigación en celulas madre tumourales supported by Obra Social Kutxa y Conserjería de Sanidad de la Junta de Castilla y Leon. Research at CC’s lab was partially supported by FEDER, and from an institutional grant from the Fundacion Ramon Areces. All Spanish funding is co-sponsored by the European Union FEDER program. ISG is an API lab of the EuroSyStem project.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF (2009) Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Majeti R (2011) Surprise! HSC are aberrant in chronic lymphocytic leukemia. Cancer Cell 20: 135–136 [DOI] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J (2009) Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N (2008) The canonical Wnt/beta-catenin signalling pathway. Methods Mol Biol 468: 5–15 [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611 [DOI] [PubMed] [Google Scholar]
- Barnes DJ, Melo JV (2006) Primitive, quiescent and difficult to kill: the role of non-proliferating stem cells in chronic myeloid leukemia. Cell Cycle 5: 2862–2866 [DOI] [PubMed] [Google Scholar]
- Bhatla T, Wang J, Morrison DJ, Raetz EA, Burke MJ, Brown P, Carroll WL (2012) Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood 119: 5201–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoli E, Pulvirenti T, Oberstadt MC, Perna F, Wee B, Schultz N, Huse JT, Fomchenko EI, Voza F, Tabar V, Brennan CW, DeAngelis LM, Nimer SD, Holland EC, Squatrito M (2012) MEF promotes stemness in the pathogenesis of gliomas. Cell Stem Cell 11: 836–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Mayshar Y, Benvenisty N (2011) Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell 9: 97–102 [DOI] [PubMed] [Google Scholar]
- Blelloch RH, Hochedlinger K, Yamada Y, Brennan C, Kim M, Mintz B, Chin L, Jaenisch R (2004) Nuclear cloning of embryonal carcinoma cells. Proc Natl Acad Sci USA 101: 13985–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737 [DOI] [PubMed] [Google Scholar]
- Boxer RB, Jang JW, Sintasath L, Chodosh LA (2004) Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell 6: 577–586 [DOI] [PubMed] [Google Scholar]
- Chen L, Shen R, Ye Y, Pu XA, Liu X, Duan W, Wen J, Zimmerer J, Wang Y, Liu Y, Lasky LC, Heerema NA, Perrotti D, Ozato K, Kuramochi-Miyagawa S, Nakano T, Yates AJ, Carson WE 3rd, Lin H, Barsky SH et al. (2007) Precancerous stem cells have the potential for both benign and malignant differentiation. PLoS One 2: e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, Horner JW 2nd, Cordon-Cardo C, Yancopoulos GD, DePinho RA (1999) Essential role for oncogenic Ras in tumour maintenance. Nature 400: 468–472 [DOI] [PubMed] [Google Scholar]
- Chomel JC, Bonnet ML, Sorel N, Bertrand A, Meunier MC, Fichelson S, Melkus M, Bennaceur-Griscelli A, Guilhot F, Turhan AG (2011) Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood 118: 3657–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, McDonald T, Lin A, Chakraborty S, Huang Q, Snyder DS, Bhatia R (2011) Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood 118: 5565–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Sanchez-Garcia I (2009) B-cell acute lymphoblastic leukaemia: towards understanding its cellular origin. Bioessays 31: 600–609 [DOI] [PubMed] [Google Scholar]
- Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ (2011) Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest 121: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL (2003) Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev 17: 3029–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der CJ, Krontiris TG, Cooper GM (1982) Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci USA 79: 3637–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks PB (2008) Cancer’s source in the peripheral nervous system. Nat Med 14: 373–375 [DOI] [PubMed] [Google Scholar]
- Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L (2003) The case for early detection. Nat Rev Cancer 3: 243–252 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33 [DOI] [PubMed] [Google Scholar]
- Fialkow PJ, Jacobson RJ, Papayannopoulou T (1977) Chronic myelocytic leukemia: clonal origin in a stem cell common to the granulocyte, erythrocyte, platelet and monocyte/macrophage. Am J Med 63: 125–130 [DOI] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM (2012) Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 338: 1080–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CB, Shaffer CM, Alfaro MP, Smith AL, Sun J, Zhao Z, Young PP, VanSaun MN, Eid JE (2012) Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2. Oncogene 31: 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb M, Shimizu K, Perucho M, Wigler M (1982) Isolation and preliminary characterization of a human transforming gene from T24 bladder carcinoma cells. Nature 296: 404–409 [DOI] [PubMed] [Google Scholar]
- Graham SM, Jorgensen HG, Allan E, Pearson C, Alcorn MJ, Richmond L, Holyoake TL (2002) Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood 99: 319–325 [DOI] [PubMed] [Google Scholar]
- Guibal FC, Alberich-Jorda M, Hirai H, Ebralidze A, Levantini E, Di Ruscio A, Zhang P, Santana-Lemos BA, Neuberg D, Wagers AJ, Rego EM, Tenen DG (2009) Identification of a myeloid committed progenitor as the cancer-initiating cell in acute promyelocytic leukemia. Blood 114: 5415–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger A, Salmon SE (1977a) Primary bioassay of human myeloma stem cells. J Clin Invest 60: 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger A, Salmon SE (1977b) Primary bioassay of human tumor stem cells. Science 197: 461–463 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, Nicolini FE, Muller-Tidow C, Bhatia R, Brunton VG, Koschmieder S, Holyoake TL (2012) Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood 119: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hanna J, Carey BW, Jaenisch R (2008) Reprogramming of somatic cell identity. Cold Spring Harb Symp Quant Biol 73: 147–155 [DOI] [PubMed] [Google Scholar]
- Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, Graff J, Galindo RL, Olson EN (2012) A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell 22: 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Blelloch R, Brennan C, Yamada Y, Kim M, Chin L, Jaenisch R (2004) Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev 18: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S (2009) Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner CS, Zhang P, Van Etten RA, Tenen DG (2000) Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat Genet 24: 57–60 [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, Akashi K, Gilliland DG (2004) MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell 6: 587–596 [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA (2009) Epigenetic changes in cancer. Annu Rev Pathol 4: 229–249 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ (2009) Cancer statistics, 2009. CA Cancer J Clin 59: 225–249 [DOI] [PubMed] [Google Scholar]
- Joseph NM, Mosher JT, Buchstaller J, Snider P, McKeever PE, Lim M, Conway SJ, Parada LF, Zhu Y, Morrison SJ (2008) The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell 13: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G, Mori Y, Iino T, Yamauchi T, Eto T, Niiro H, Iwasaki H, Takenaka K, Akashi K (2011) Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell 20: 246–259 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, Golub TR, Armstrong SA (2006) Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442: 818–822 [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V, Lowe SW (2009) Stem cells: The promises and perils of p53. Nature 460: 1085–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Brendel C, Hochhaus A, Neubauer A, Burchert A (2012) Low BCR-ABL expression levels in hematopoietic precursor cells enable persistence of chronic myeloid leukemia under imatinib. Blood 119: 530–539 [DOI] [PubMed] [Google Scholar]
- Li L, Connelly MC, Wetmore C, Curran T, Morgan JI (2003) Mouse embryos cloned from brain tumors. Cancer Res 63: 2733–2736 [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F (eds.) (2013) Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman) [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD et al. (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Plath K (2013) Epigenetics of reprogramming to induced pluripotency. Cell 152: 1324–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LF, Tabin CJ, Shih C, Weinberg RA (1982) Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature 297: 474–478 [DOI] [PubMed] [Google Scholar]
- Perez-Caro M, Cobaleda C, Gonzalez-Herrero I, Vicente-Duenas C, Bermejo-Rodriguez C, Sanchez-Beato M, Orfao A, Pintado B, Flores T, Sanchez-Martin M, Jimenez R, Piris MA, Sanchez-Garcia I (2009) Cancer induction by restriction of oncogene expression to the stem cell compartment. EMBO J 28: 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulciani S, Santos E, Lauver AV, Long LK, Robbins KC, Barbacid M (1982) Oncogenes in human tumor cell lines: molecular cloning of a transforming gene from human bladder carcinoma cells. Proc Natl Acad Sci USA 79: 2845–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ (2008) Efficient tumour formation by single human melanoma cells. Nature 456: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells. Nature 414: 105–111 [DOI] [PubMed] [Google Scholar]
- Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suva D, Tercier S, Joseph JM, Guillou L, Stamenkovic I (2010) EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev 24: 916–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Camarero I, Jiang X, Natkunam Y, Lu X, Vicente-Duenas C, Gonzalez-Herrero I, Flores T, Luis Garcia J, McNamara G, Kunder C, Zhao S, Segura V, Fontan L, Martinez-Climent JA, Javier Garcia-Criado F, Theis JD, Dogan A, Campos-Sanchez E, Green MR, Alizadeh AA, Cobaleda C, Sanchez-Garcia I, Lossos IS (2013) Germinal centre protein HGAL promotes lymphoid hyperplasia and amyloidosis via BCR-mediated Syk activation. Nat Commun 4: 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia I (2009) The crossroads of oncogenesis and metastasis. N Engl J Med 360: 297–299 [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia I, Vicente-Duenas C, Cobaleda C (2007) The theoretical basis of cancer-stem-cell-based therapeutics of cancer: can it be put into practice? Bioessays 29: 1269–1280 [DOI] [PubMed] [Google Scholar]
- Santos E, Tronick SR, Aaronson SA, Pulciani S, Barbacid M (1982) T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature 298: 343–347 [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T (2011) In vitro generation of human cells with cancer stem cell properties. Nat Cell Biol 13: 1051–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, Rupec RA, Gerhard M, Schmid R, Barker N, Clevers H, Lang R, Neumann J, Kirchner T, Taketo MM, van den Brink GR et al. (2013) Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152: 25–38 [DOI] [PubMed] [Google Scholar]
- Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M (2011) Global DNA demethylation during mouse erythropoiesis in vivo. Science 334: 799–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C, Weinberg RA (1982) Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell 29: 161–169 [DOI] [PubMed] [Google Scholar]
- So CW, Karsunky H, Passegue E, Cozzio A, Weissman IL, Cleary ML (2003) MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell 3: 161–171 [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML (2006) Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 10: 257–268 [DOI] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engström PG, Carén H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, Smith A, Beck S, Pollard SM (2013) Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev 27: 654–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K (2009) Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Hernandez T, Vicente-Duenas C, Sanchez-Garcia I, Martin-Zanca D (2012) p53 restoration kills primitive leukemia cells in vivo and increases survival of leukemic mice. Cell Cycle 12: 1122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Duenas C, Fontan L, Gonzalez-Herrero I, Romero-Camarero I, Segura V, Aznar MA, Alonso-Escudero E, Campos-Sanchez E, Ruiz-Roca L, Barajas-Diego M, Sagardoy A, Martinez-Ferrandis JI, Abollo-Jimenez F, Bertolo C, Penuelas I, Garcia-Criado FJ, Garcia-Cenador MB, Tousseyn T, Agirre X, Prosper F et al. (2012a) Expression of MALT1 oncogene in hematopoietic stem/progenitor cells recapitulates the pathogenesis of human lymphoma in mice. Proc Natl Acad Sci USA 109: 10534–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Duenas C, Gonzalez-Herrero I, Cenador MB, Criado FJ, Sanchez-Garcia I (2012b) Loss of p53 exacerbates multiple myeloma phenotype by facilitating the reprogramming of hematopoietic stem/progenitor cells to malignant plasma cells by MafB. Cell Cycle 11: 203896–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Duenas C, Gutierrez de Diego J, Rodriguez FD, Jimenez R, Cobaleda C (2009a) The role of cellular plasticity in cancer development. Curr Med Chem 16: 3676–3685 [DOI] [PubMed] [Google Scholar]
- Vicente-Duenas C, Perez-Caro M, Abollo-Jimenez F, Cobaleda C, Sanchez-Garcia I (2009b) Stem-cell driven cancer: ‘hands-off’ regulation of cancer development. Cell Cycle 8: 1314–1318 [DOI] [PubMed] [Google Scholar]
- Vicente-Duenas C, Romero-Camarero I, Gonzalez-Herrero I, Alonso-Escudero E, Abollo-Jimenez F, Jiang X, Gutierrez NC, Orfao A, Marin N, Villar LM, Criado MC, Pintado B, Flores T, Alonso-Lopez D, De Las Rivas J, Jimenez R, Criado FJ, Cenador MB, Lossos IS, Cobaleda C et al. (2012c) A novel molecular mechanism involved in multiple myeloma development revealed by targeting MafB to haematopoietic progenitors. EMBO J 31: 3704–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CL, Ho SM (2012) Developmental reprogramming of cancer susceptibility. Nat Rev Cancer 12: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB (2002) Cancer. Addiction to oncogenes--the Achilles heel of cancer. Science 297: 63–64 [DOI] [PubMed] [Google Scholar]
- Wojiski S, Guibal FC, Kindler T, Lee BH, Jesneck JL, Fabian A, Tenen DG, Gilliland DG (2009) PML-RARalpha initiates leukemia by conferring properties of self-renewal to committed promyelocytic progenitors. Leukemia 23: 1462–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY (2008) Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2: 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef KK, Lapouge G, Bouvree K, Rorive S, Brohee S, Appelstein O, Larsimont JC, Sukumaran V, Van de Sande B, Pucci D, Dekoninck S, Berthe JV, Aerts S, Salmon I, del Marmol V, Blanpain C (2012) Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat Cell Biol 14: 1282–1294 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W et al. (2008) Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 3: 475–479 [DOI] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, dePinho RA (2008) Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol 73: 427–437 [DOI] [PubMed] [Google Scholar]
- Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ (2009) Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]