Abstract
Plasmids capable of undergoing genetic exchange in mitotically dividing Saccharomyces cerevisiae cells were used to measure the length of gene conversion events, to determine patterns of coconversion when multiple markers were present, and to correlate the incidence of reciprocal recombination with the length of conversion tracts. To construct such plasmids, restriction site linkers were inserted both within the HIS3 gene and in the flanking sequences, and two different his3- alleles were placed in a vector. Characterization of the genetic exchanges in these plasmids showed that most occur with the conversion of one his3- allele. Many of these events included coconversions in which more than one marker along the allelic sequence was replaced. The frequency of coconversion decreased with the distance between two markers such that markers further than 1 kilobase apart were infrequently coconverted. From these results the average length of conversion was determined to be approximately 0.5 kilobase. Examination of coconversions involving three or more markers revealed an almost obligatory, simultaneous coconversion pattern of all markers. Thus, when two markers which flank an intervening marker are converted, the intervening marker is 20 times more likely to be converted than to remain unchanged. The results of these studies also showed that the incidence of reciprocal recombination, which accompanies more than 20% of the conversion events, is more frequent when the conversion tract is longer than average.
Full text
PDF



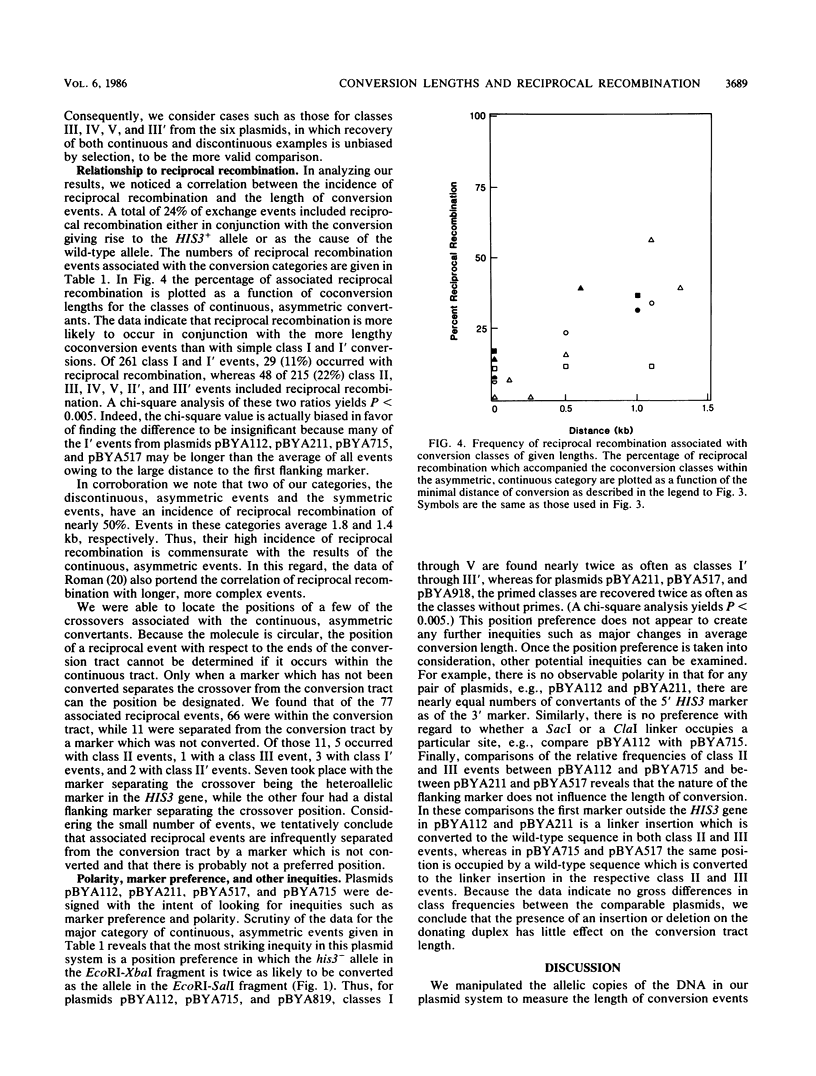
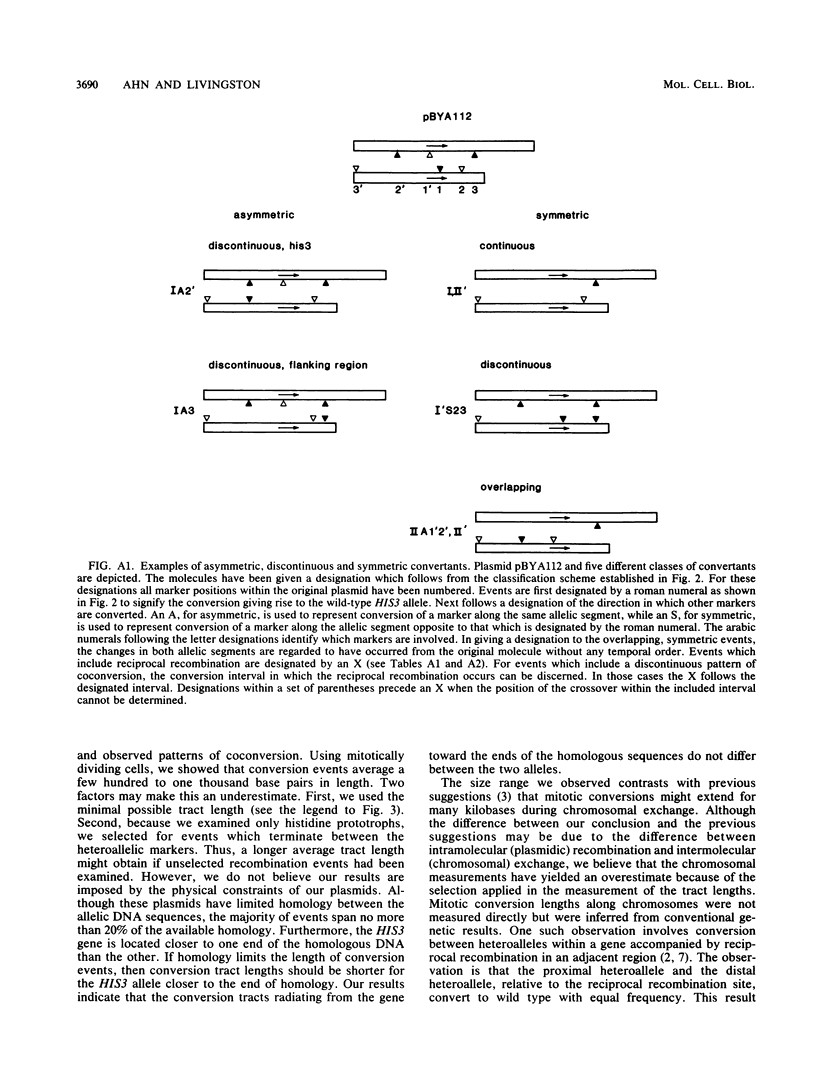



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Embretson J. E., Livingston D. M. A plasmid model to study genetic recombination in yeast. Gene. 1984 Sep;29(3):293–302. doi: 10.1016/0378-1119(84)90058-1. [DOI] [PubMed] [Google Scholar]
- Esposito M. S. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4436–4440. doi: 10.1073/pnas.75.9.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A. Gene conversion of deletions in the his4 region of yeast. Genetics. 1974 Jun;77(2):231–244. doi: 10.1093/genetics/77.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Golin J. E., Esposito M. S. Coincident gene conversion during mitosis in saccharomyces. Genetics. 1984 Jul;107(3):355–365. doi: 10.1093/genetics/107.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golin J. E., Esposito M. S. Mitotic recombination: mismatch correction and replicational resolution of Holliday structures formed at the two strand stage in Saccharomyces. Mol Gen Genet. 1981;183(2):252–263. doi: 10.1007/BF00270626. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Keil R. L., Roeder G. S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984 Dec;39(2 Pt 1):377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Sherman F., Jackson M., Gilmore R. A. Mapping and gene conversion studies with the structural gene for iso-1-cytochrome C in yeast. Genetics. 1975 Dec;81(4):615–629. doi: 10.1093/genetics/81.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. A physical, genetic and transcriptional map of the cloned his3 gene region of Saccharomyces cerevisiae. J Mol Biol. 1980 Jan 25;136(3):309–332. doi: 10.1016/0022-2836(80)90376-9. [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W. Production of a functional eukaryotic enzyme in Escherichia coli: cloning and expression of the yeast structural gene for imidazole-glycerolphosphate dehydratase (his3). Proc Natl Acad Sci U S A. 1977 Dec;74(12):5255–5259. doi: 10.1073/pnas.74.12.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- WHITEHOUSE H. L. A THEORY OF CROSSING-OVER BY MEANS OF HYBRID DEOXYRIBONUCLEIC ACID. Nature. 1963 Sep 14;199:1034–1040. doi: 10.1038/1991034a0. [DOI] [PubMed] [Google Scholar]



