Abstract
Background
Results from genome-wide association studies (GWAS) represent a potential resource for etiological and treatment research. GWAS of obesity-related phenotypes have been especially successful. To translate this success into a research tool, we developed and tested a “genetic risk score” (GRS) that summarizes an individual’s genetic predisposition to obesity.
Methods
Different GWAS of obesity-related phenotypes report different sets of single nucleotide polymorphisms (SNPs) as the best genomic markers of obesity risk. Therefore, we applied a 3-stage approach that pooled results from multiple GWAS to select SNPs to include in our GRS: The 3 stages are (1) Extraction. SNPs with evidence of association are compiled from published GWAS; (2) Clustering. SNPs are grouped according to patterns of linkage disequilibrium; (3) Selection. Tag SNPs are selected from clusters that meet specific criteria. We applied this 3-stage approach to results from 16 GWAS of obesity-related phenotypes in European-descent samples to create a GRS. We then tested the GRS in the Atherosclerosis Risk in the Communities (ARIC) Study cohort (N=10,745, 55% female, 77% white, 23% African American).
Results
Our 32-locus GRS was a statistically significant predictor of body mass index (BMI) and obesity among ARIC whites (for BMI, r=0.13, p<1×10−30; for obesity, area under the receiver operating characteristic curve (AUC)=0.57 [95% CI 0.55–0.58]). The GRS improved prediction of obesity (as measured by delta-AUC and integrated discrimination index) when added to models that included demographic and geographic information. FTO- and MC4R-linked SNPs, and a non-genetic risk assessment consisting of a socioeconomic index (p<0.01 for all comparisons). The GRS also predicted increased mortality risk over 17 years of follow-up. The GRS performed less well among African Americans.
Conclusions
The obesity GRS derived using our 3-stage approach is not useful for clinical risk prediction, but may have value as a tool for etiological and treatment research.
INTRODUCTION
Genome-wide associations study (GWAS) results represent a potentially rich source of information for etiological and treatment research that builds bridges between genome science and clinical and public health practice 1,2. Given the large number of such studies, sufficient GWAS data exist to support such translational research for a number of common chronic health conditions, including obesity 3,4. Infrastructure is in place at the start of the translational pipeline with GWAS data banked and curated in continuously updated searchable databases 3,5. Likewise, at the other end of the pipeline, evidence from translational research is evaluated to establish the clinical utility of genomic information and to issue guidelines for clinical practice 6. However, significant gaps remain in the middle of the translational pipeline and approaches are needed to support research at this juncture, where population-based samples with rich environmental and phenotypic measurements can be used to follow-up disease markers identified in GWAS. Specifically, systematic approaches are needed to sift the results of numerous association studies and distill the most promising set of markers for further investigation. These approaches must be able to harness the power of existing resources and to flexibly accommodate new data produced by the fast pace of discovery in genome science.
A key hurdle for research using GWAS results is that risk SNPs identified in GWAS may not cause adverse health outcomes, but may instead be proxies for (correlated with) unmeasured disease-causing variation in the genome 7,8. GWAS methods exploit LD across the genome to leverage measurement of 100,000 – 1 million SNPs to capture variation in the 10 million plus SNPs the genome is estimated to contain. The very large sample sizes in GWAS permit detection of risk associations even when proxy SNPs are in imperfect LD with disease-causing variation (correlation<1). GWAS findings are generally applied to smaller samples designed to elucidate etiological and clinical correlates of discovered genes. When GWAS SNPs are translated to research using smaller samples, the measurement error resulting from imperfect LD with disease causing variants can attenuate associations below levels these samples are powered to detect. Genetic risk scores (GRSs) summarize risk-associated variation across the genome 9 by aggregating information from multiple risk SNPs (the simplest GRSs count disease-associated alleles). Because GRSs pool information from multiple SNPs, each individual SNP is less important to the summary measurement and the “signal” from the GRS is robust to imperfect linkage for any one SNP. For the same reason, GRSs are less sensitive to minor allele frequencies for individual SNPs. As the number of SNPs included in a GRS grows, the distribution of values approaches normality, even when individual risk alleles are relatively uncommon 10. Therefore, the GRS can be an efficient and effective means of constructing genome-wide risk measurements from GWAS findings.
Obesity is a public health problem that is well suited to risk assessment using a GRS. It is highly prevalent 11; it is a significant source of health-care costs, morbidity, and mortality 12–14; it is under strong genetic influence 15; and GWAS are beginning to elucidate its molecular genetic roots 16. Therefore, translational research in obesity genomics may ultimately help to address a public-health priority. A key challenge is that obesity’s genetic roots are diffuse, multifactorial, and non-deterministic; many variants scattered across the genome each contribute small risks for obesity 17. In other words, information from multiple genetic variants is needed to characterize genetic susceptibility to obesity. Thus, a GRS may be useful. A further challenge is uncertainty about the specific genetic variants to be included in an obesity GRS. Different GWAS identify different genomic loci and, when loci are replicated across GWAS, the specific SNPs identified may be different 18. To address this challenge, we developed a 3-stage approach to review GWAS results and select specific SNPs to include in a GRS. We devised our approach to be systematic and replicable and to leverage the discovery potential of GWAS while minimizing risk for including false-positive markers. In this article, we describe this 3-stage approach, apply it to develop a GRS for obesity, and test the GRS as a measure of obesity risk using data from the population-based Atherosclerosis Risk in the Communities (ARIC) Study.
METHODS
Sample
The ARIC sample is described elsewhere 19,20. Briefly, ARIC is a prospective epidemiologic cohort study sponsored by the National Heart, Lung, and Blood Institute to investigate the etiology of atherosclerotic disease. The study draws from 4 US communities: Minneapolis MN, Washington County MD, Forsyth County, NC, and Jackson MS. Participants were examined first during 1987–1989, and at 3 subsequent occasions (1990–1992, 1993–1995, and 1996–1998), with ongoing follow-up conducted annually by telephone. ARIC cohort genotype data from the Affymetrix Affy 6.0 Chip and selected phenotypes were obtained for this study from the NIH dbGaP.
The original ARIC sample includes 15,792 participants (27% African American, 55% female). The publicly available dataset obtained from dbGaP for this study includes genotype and phenotype data for 12,771 individuals. Of this sample, 1,212 participants had a missing call rate >2% for SNPs called successfully in ≥95% of the sample and were excluded from subsequent analyses per quality control recommendations of the GENEVA ARIC Project 21. In addition, although the ARIC study design did not aim to include relatives, genomic analysis by the ARIC investigators revealed familial relationships at the level of half-siblings or closer among 1,674 participants. One member was selected at random from each of the 105 “families” to form a sample of unrelated persons. After these exclusions, the sample consisted of 10,745 participants (23% African American, 55% female, hereafter the “analysis sample”).
Body Mass Index and Obesity
Body mass index (BMI: kg/m2) was calculated from measurements of weight to the nearest pound and height to the nearest centimeter. Obesity was defined according to U.S. Centers for Disease Control and Prevention Criteria as BMI≥30. Anthropometric measurements were collected from participants wearing a scrub suit and no shoes at the 4 in-person data collections.
Genotypes
Details on the genotyping of the ARIC sample are available through dbGaP and are described elsewhere 22. Briefly, genotyping was conducted by the Broad Institute using the Affymetrix Affy 6.0 SNP array and the Birdseed calling algorithm 23. Following guidelines for the use of genotypic data provided by the ARIC GWAS team, data were extracted for all SNPs with a sample-wide call rate ≥95%, fewer than 5 discordant calls across duplicated DNA samples in the quality control subsample (n=334), and in Hardy-Weinberg Equilibrium (p>0.001).
Genetic Risk Scores
Current mid-pipeline translational studies use either a “best guess” approach or a “top hits” approach to select genetic markers to include in GRSs. The “best guess” approach selects markers identified in association studies that are located in or near genes with plausible biological relationships to the pathophysiology of a phenotype or that demonstrate strong and replicable association signals 24–26. The “top hits” approach selects markers with the strongest association signals in a single GWAS, independent of their biological plausibility 27,28. Early studies have illustrated the promise of translational research with GWAS markers, but as the field moves forward, more systematic approaches are needed that can better integrate new information from the latest studies. Neither the top-hits nor the best-guess approach provides a systematic and replicable means of integrating results from multiple GWAS. Meta-analysis can accomplish this, but comprehensive meta-analyses are not always available. Moreover, the top-hits and best-guess approaches do not provide a means to select specific SNPs for follow-up, and this problem is not solved by meta-analysis. The approach of selecting the “lead” SNP at a locus, usually the SNP with the lowest p-value in the largest GWAS, is problematic because different GWAS can report different lead SNPs for the same locus because of differences in GWAS chips, genotyping quality, and data handling and analysis decisions. Thus, an approach is needed that facilitates systematic and replicable SNP selection from results of multiple GWAS.
Our 3-stage approach integrates public-access resources including continuously updated databases of GWAS results, web-based whole-genome analysis tools, and genome-wide data to identify the most promising set of single nucleotide polymorphisms (SNPs) for follow-up. Most importantly, the 3-stage approach addresses key limitations of the top-hits and best-guess approaches: It provides a systematic and replicable means of integrating findings across multiple GWAS and of selecting SNPs for follow-up in new samples. The 3 stages are:
Stage 1) Extraction: All SNPs associated with one of the selected phenotypes at a given significance threshold are “extracted” from each GWAS and retained for further analysis.
Stage 2) Clustering: Extracted SNPs are “clustered” according to patterns of linkage disquelibrium (LD) determined from a reference population that matches the population in the GWAS included in Stage 1. Clustering yields a set of “LD blocks.”
Stage 3) Selection: Statistical significance and replication are evaluated at the level of the LD block. The original GWAS results are used to assign a minimum p-value and a replication count for each LD block. The minimum p-value is the lowest p-value reported for any SNP in the LD block in any GWAS contributing data in Stage 1. The replication count is the number of GWAS that reported an association for any SNP in the LD block at the threshold defined in Stage 1.
We applied our 3-stage approach to construct two GRSs for obesity. First, we considered only GWAS published in print or online through December 31, 2008. We chose these GWAS because they were used in previous research that created “top-hits” and “best-guess” obesity GRSs. Thus, we used these GWAS to construct a GRS using our 3-stage approach and compared it to two published GRSs 29,30. Second, we considered all GWAS published through December 31, 2010. We applied our 3-stage approach to results from the full set of GWAS and compared the resulting GRS to a top-hits GRS generated from the largest meta-analysis of BMI GWAS published to date 31 and to a best-guess GRS generated from the full set of obesity-associated SNPs reported in the National Human Genome Research Institute (NHGRI) GWAS Catalog 18. The derivation of the GRS using the 3-stage approach is described in detail in the supplemental material. Analyses described in the supplemental material revealed that the 3-stage approach created GRSs that were at least as predictive of BMI and obesity as GRSs created with the top-hits and best-guess approaches. Further analyses to refine the 3-stage approach GRS yielded a final set of 32 SNPs (see supplemental material). We applied 2 weighting schemes to the 32 SNPs before summing them to create our obesity GRS: 1) equal weighting, under which the score was a simple count of BMI-increasing alleles; and 2) effect-size weighting, under which BMI-increasing alleles were weighted by the effect size reported for that locus in the GIANT Consortium 31 or DeCode 32 BMI GWAS. Effect-size weights were adjusted for LD between the SNP tested in the GWAS and the SNP genotyped in the ARIC sample. Each of the 32 SNPs in the GRS was missing for fewer than 1% of participants in any gender/ethnicity cell. GRSs were prorated by dividing the GRS by the number of SNPs contributing data and multiplying by 32. The SNPs included in the final obesity GRS, their BMI-increasing (“effect”) alleles, nearby genes, and weights are reported in Table 1.
Table 1. Single nucleotide polymorphisms included in the obesity genetic risk score (GRS).
Alleles are reported from the forward strand. The GRS was computed by counting the number of effect alleles at each SNP, multiplying that number by the SNP’s weight, and then summing the results across the set of 32 SNPs. Weights reflect per-allele changes in BMI estimated in the the GIANT Consortium GWAS meta-analysis 31 except for rs867559, for which the weight was estimated in the DeCODE GWAS meta-analysis 32.
| Chr | Nearby Gene | SNP | Effect Allele | Other Allele | Weight | Effect Alle Frequency (ARIC Sample) | |
|---|---|---|---|---|---|---|---|
| Whites | African Americans | ||||||
| 1 | NEGR1 | rs2815752 | G | A | 0.13 | 62% | 55% |
| TNNI3K | rs1514175 | A | G | 0.07 | 43% | 68% | |
| PTBP2 | rs1555543 | A | C | 0.06 | 58% | 43% | |
| SEC16B | rs543874 | G | A | 0.22 | 20% | 25% | |
|
| |||||||
| 2 | FANCL | rs759250 | A | G | 0.10 | 29% | 8% |
| LRP1B | rs2121279 | T | C | 0.08 | 14% | 3% | |
| TMEM18 | rs2867123 | G | C | 0.30 | 83% | 88% | |
| RBJ | rs10182181 | G | A | 0.14 | 54% | 16% | |
|
| |||||||
| 3 | CADM2 | rs12714640 | A | C | 0.10 | 19% | 6% |
| ETV5/DGKG | rs1516728 | T | A | 0.11 | 77% | 48% | |
|
| |||||||
| 4 | GNPDA2 | rs12641981 | T | C | 0.18 | 43% | 23% |
| SLC39A8 | rs13114738 | T | C | 0.13 | 8% | 1% | |
|
| |||||||
| 5 | POC5 FLJ35779 | rs10057967 | C | T | 0.10 | 63% | 51% |
| ZNF608 | rs6864049 | A | G | 0.07 | 54% | 81% | |
|
| |||||||
| 6 | TFAP2B | rs734597 | A | G | 0.13 | 17% | 9% |
|
| |||||||
| 9 | LING02 LRRN6C | rs1412235 | C | G | 0.11 | 31% | 16% |
| LMX1B | rs867559 | G | A | 0.24 | 20% | 32% | |
|
| |||||||
| 11 | RPL27A | rs2028882 | C | A | 0.06 | 50% | 34% |
| BDNF | rs10501087 | C | T | 0.18 | 79% | 93% | |
| MTCH2 | rs12419692 | A | C | 0.05 | 36% | 9% | |
|
| |||||||
| 12 | BDCDIN3D, FAIM2 | rs7138803 | A | G | 0.12 | 38% | 17% |
|
| |||||||
| 13 | MTIF3, GRF3A | rs1475219 | C | T | 0.09 | 21% | 22% |
|
| |||||||
| 14 | PRKD1 | rs1440983 | A | G | 0.15 | 5% | 23% |
| NRXN3 | rs7144011 | T | G | 0.13 | 22% | 24% | |
|
| |||||||
| 15 | MAP2K5 | rs28670272 | G | A | 0.13 | 77% | 59% |
|
| |||||||
| 16 | GPR5B | rs11639988 | G | A | 0.17 | 85% | 76% |
| ATXN2L, TUFM, SH2B1 | rs12443881 | T | C | 0.15 | 39% | 9% | |
| FTO | rs9939609 | A | T | 0.38 | 41% | 48% | |
|
| |||||||
| 18 | MC4R | rs12970134 | A | G | 0.21 | 26% | 13% |
|
| |||||||
| 19 | KCTD15 | rs11084753 | A | G | 0.04 | 67% | 64% |
| QPCTL | rs11083779 | C | T | 0.07 | 96% | 89% | |
| ZC3H4 TMEM160 | rs7250850 | G | C | 0.09 | 71% | 20% | |
Evaluation of the Obesity GRS
Associations between the GRS and obesity-related traits (BMI, weight, waist circumference, obesity) were tested with linear and logistic regression models. These and subsequent models were adjusted for demographic and geographic control variables: age was specified as a linear and a quadratic term; a product term was included for the interaction between age and sex to account for sex differences in BMI and obesity distributions at different ages; the 4 ARIC Study Centers where participants were enrolled in the study were entered as a series of dummy variables (this collection of variables is referred to hereafter and elsewhere in the manuscript as demographics and geography). Predictiveness of the GRS was evaluated using 3 metrics that are established tools for evaluating risk markers in general 33 as well as for the specific case of genetic risk scores 34: 1) R2, the proportion of variation explained in BMI. R2 was estimated using demographics and geography-adjusted linear regression models. 2) AUC, the area under the receiver operating characteristic curve for obesity, also known as the discrimination index. The AUC corresponds to the probability that a randomly selected obese case will have a higher GRS as compared to a randomly selected non-obese control. A marker that discriminates no better than chance has an AUC of 0.50. A marker that discriminates perfectly has an AUC of 1. A related metric is the partial AUC (PAUC). The PAUC sets a specificity threshold and calculates an AUC-like statistic specific to that specificity. Analyses of PAUC for the GRS set specificity at 80% (the bottom 5th of the ROC curve). AUC and PAUC analyses were stratified by ARIC Study Center using Pepe’s method 35. To determine whether the GRS improved discrimination over and above demographic and geographic information, we calculated a second set of statistics, delta AUC and delta PAUC. Probit regression models were used to generate predicted probabilities of obesity for each ARIC participant using a baseline model that included demographic and geographic information and a test model that also included the GRS. AUCs and were calculated using these predicted probabilities as “risk scores” 36, and estimates of the differences between the baseline and test models were bootstrapped to obtain confidence intervals. AUC analyses were conducted using the Stata package “comproc” 37. 3) IDI, the integrated discrimination index for obesity. The IDI evaluates the added predictiveness of a marker by comparing predictions made using a baseline set of risk markers to predictions that also include information about the new risk marker:
where “Prob” is the average predicted probability for a particular group from a particular model. The IDI measures change in model sensitivity net of change in model specificity and is a more sensitive measure than delta AUC 38. An IDI of zero indicates that the test model performs comparably to the baseline model. Positive IDI values index net improvement in model sensitivity. Baseline and test models for IDI analyses were identical to those used in delta AUC analyses.
We tested differences between the predictiveness metrics for different risk scores by bootstrapping confidence intervals around the R2 and AUC metrics (comparing the difference in estimated metric values across 1,000 random samples drawn with replacement from the ARIC database 37) and by applying Pencina’s method 38 to test change in the IDI metric. Comparisons were as follows: Un-Weighted GRS vs. Weighted GRS; Weighted GRS vs. Simple Genetic Risk Assessment (the sum of risk alleles at the two best-replicated obesity loci, in the gene FTO and downstream of the gene MC4R, rs9939609 and rs12970134, respectively); Weighted GRS vs. Socioeconomic Index (Educational attainment measured in 5 categories: grade-school or less, some high school, high school graduate, vocational school, college, graduate/professional school, Supplementary Table 8).
RESULTS
Obesity risk-allele distributions were similar for males and females, but were different for whites and African Americans. The variance of the un-weighted GRS was greater for whites as compared to African Americans (SD= 3.50 as compared to 3.25, p<0.001 using Brown and Forsythe’s method 39), as was the mean (M=28.80 as compared to 24.87, p<0.001 using t-test for unequal variances; see also Supplementary Figure 1) This difference reflected lower frequencies of BMI-increasing alleles for several GRS SNPs among African American ARIC participants (Table 1). Subsequent analyses were stratified by race.
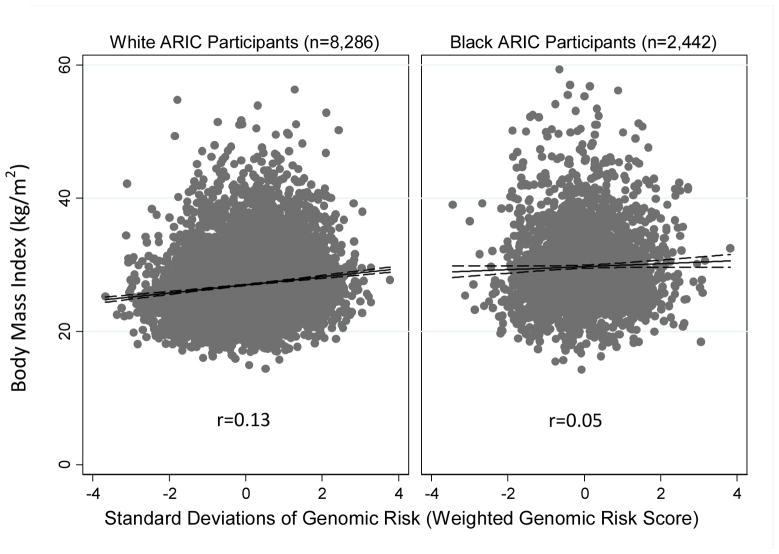
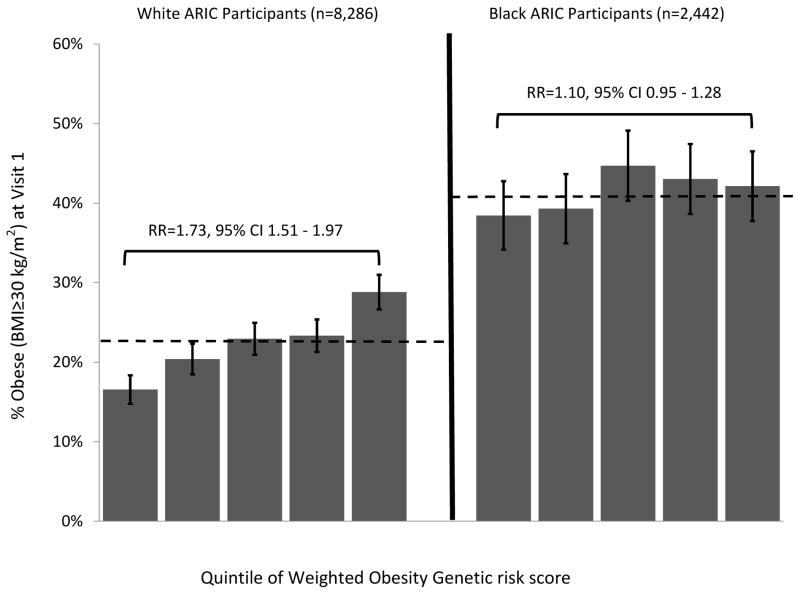
The obesity GRSs were weakly but consistently associated with BMI and the probability of being obese among whites and African Americans, but associations were weaker among African Americans (Figure 1). Among whites, after adjusting for age, sex, and geography, the un-weighted GRS was associated with BMI at r=0.12 and the weighted GRS was associated with BMI at r=0.13 (p<1×10−26 for both). This effect size corresponded to a 0.60 unit increase in BMI per standard-deviation increase in the GRS. For each standard-deviation increase in their un-weighted and weighted GRSs, a white ARIC participant’s risk for obesity increased by 19.35% and 20.51%, respectively (p<1×10−18 for both). Among African Americans, the weighted and un-weighted GRSs were associated with BMI at r=0.05 (p<0.05 for both). For each standard deviation increase in their un-weighted and weighted GRSs, an African American ARIC participant’s risk for obesity increased by 3.54% (p=0.059) and 4.92% (p=0.017), respectively. Results were substantively unchanged when control variables were removed from the models. To determine whether population substructure influenced our estimates of GRS-BMI or GRS-obesity associations, we repeated our analyses of the white and black subsamples, including as covariates the first 5 principal components derived for each ethnic group using the method described by Patterson et al.40 (principal components derived for the white and black subsamples were included as part of the ARIC database obtained from dbGaP41) Adjustment for these principal components is a valid method of controlling for population stratification in genetic association analyses.42 Inclusion of principal components as covariates in regression analyses did not change results.
Figure 1.
Panel A. BMI for African American and White ARIC Participants Plotted Against the Weighted Obesity Genetic Risk Score Dashed outlines represent 95% confidence intervals. Pearson correlations (r) were adjusted for gender, age and ARIC Study Center where data were collected. Removal of outliers (not shown) did not alter correlation estimates at the third decimal point. Correlations were statistically significant for white (p<1×10−30) and African American (p=0.014) ARIC participants.
Panel B. Percentage White and African American ARIC Participants Who Were Obese (BMI≥30 kg/m2) at the First Study Visit, by Quintile of Genetic Risk Score. Quintiles were determined separately for whites and African Americans. Error bars represent 95% confidence intervals. Risk ratios are for comparisons of highest to lowest quintiles of genomic risk and were estimated with adjustment for gender, age, and ARIC study center where data were collected. Dashed lines represent sample means. Among white ARIC participants, all quintile to quintile differences are statistically significant (p<0.01), with the exception of the 3rd and 4th quintiles. Among African American ARIC participants, the percent obese in the lowest quintile was lower than in the third and fourth quintiles (p<0.05).
We conducted a series of additional sensitivity analyses to evaluate heterogeneity in GRS associations (described in detail in the Supplement). These analyses supported a linear association between the GRS and BMI; showed that GRS-BMI associations were similar to GRS-weight and GRS-waist circumference associations; and revealed no sex or age differences in GRS-BMI associations.
The obesity GRSs performed similarly on the 3 predictiveness metrics (Table 2). The top panel of Table 3 addresses clinical validity. It presents the 3 metrics for the un-weighted and weighted GRSs. Among whites, weighted and un-weighted obesity GRSs explained small, but statistically significant proportions of the variance in BMI (R2), discriminated obese from non-obese participants modestly better than chance (AUC), and contributed small net improvements to the sensitivity of an obesity prediction model over and above demographic and geographic information (IDI). Among African Americans, the GRS did not contribute to the explanation of variance in BMI over and above demographic and geographic information, to the discrimination of obese from non-obese participants, or to the net sensitivity of the obesity prediction model. Use of weights derived from BMI GWAS improved the performance of the GRS among whites and African Americans, but this improvement was not statistically significant (p>0.10 for all comparisons).
Table 2. Predictiveness Metrics for the 3-Stage Approach Obesity Genetic Risk Score and Comparison Measures of Risk for Obesity.
The simple genetic risk score is a component of the weighted obesity genetic risk scores. Values of R2 were estimated using linear regression models adjusted for demographics and geography. Percentile-based confidence intervals were generated using the bootstrap method. AUCs and percentile-based confidence intervals were estimated from ROC curves constructed for raw values (i.e. actual values of the measures tested rather than predicted values generated from a regression model) and were adjusted for the ARIC Study Center where data were collected. IDIs and test statistics were estimated for comparisons of a baseline model including demographic and geographic information to a test model that included this information and the GRS.
| White ARIC Participants (n=8,286) | Black ARIC Participants (n=2,442) | |||||
|---|---|---|---|---|---|---|
| R2 (95% CI) | AUC (95% CI) | IDI (p-value) | R2 (95% CI) | AUC (95% CI) | IDI (p-value) | |
|
Panel A. Predictivness of the un-weighted and weighted obesity GRSs
| ||||||
| Un-Weighted GRS | 1.39% (0.94% – 1.89%) | 0.565 (0.550 – 0.581) | 0.009 (4.65E-18) | %0.11 (−%0.04 – %0.57) | 0.515 (0.491 – 0.540) | 0.001 (0.067) |
|
| ||||||
| Weighted GRS | 1.57% (1.11% – 2.10%) | 0.570 (0.554 – 0.584) | 0.010 (8.25E-20) | %0.14 (−%0.03 – %0.65) | 0.521 (0.497 – 0.544) | 0.002 (0.152) |
|
| ||||||
|
Panel B. Predictiveness of comparison risk measures
| ||||||
| Simple Genetic Risk Assessment: FTO & MC4R-linked SNPs only | 0.59% (0.31% – 0.97%) | 0.543 (0.528 – 0.557) | 0.004 (3.54E-09) | −%0.02 (−%0.04 – %0.25) | 0.516 (0.493 – 0.539) | 0.001 (0.149) |
|
| ||||||
| Socioeconomic Index: 5-category measure of educational attainment | 0.57% (0.29% – 0.87%) | 0.532 (0.517 – 0.546) | 0.003 (7.83E-07) | %1.06 (%0.42 – %1.99) | 0.561 (0.538 – 0.584) | 0.016 (2.71E-11) |
|
| ||||||
|
Panel C. Predictiveness of model-based risk assessments (including demographic and geographic information)
| ||||||
| Simple Genetic Risk Assessment | 3.88% | 0.550 | 5.35% | 0.607 | ||
|
| ||||||
| Weighted GRS | 4.88% | 0.574 | 5.52% | 0.609 | ||
|
| ||||||
| Change in predictiveness with addition of weighted GRS to model | 1.00% (0.58%–1.42%) | 0.024 (0.012–0.036) | 0.006 (7.81E-13) | 0.17% (−%0.15–%0.51) | 0.002 (−0.005–0.009) | 0.001 (0.055) |
|
| ||||||
| Socioeconomic Status | 4.70% | 0.550 | 7.70% | 0.643 | ||
|
| ||||||
| Socioeconomic Status + weighted GRS | 6.20% | 0.586 | 7.92% | 0.645 | ||
|
| ||||||
| Change in predictiveness with addition of weighted GRS to model | 1.50% (1.00%–1.99%) | 0.036 (0.023–0.050) | 0.010 (5.46E-19) | 0.22% (−%0.14–%0.55) | 0.002 (−0.003–0.008) | 0.002 (0.012) |
The bottom panel of Table 3 addresses research utility. It presents predictiveness metrics for two comparison measures of obesity risk: the simple genetic risk assessment (weighted combinations of rs9939609 in FTO and rs12970134 downstream of MC4R) and the socioeconomic index (a 5-category measure of educational attainment). The FTO and MC4R loci and socioeconomic status are robust correlates of BMI and obesity in adult samples 43,44. Comparison of the 32-locus GRS to a two-locus risk assessment can illustrate whether the GRS offers value added over a simpler genetic risk assessment. Comparison of the GRS to socioeconomic status can illustrate how the predictiveness of the GRS compares to the predictiveness of a social determinant of obesity that is not easily changed but that is understood to be important in etiological research 45. Among whites, the genetic risk scores performed better than the comparison measures of obesity risk on all 3 metrics (p<0.01 for all comparisons). Among African Americans, the GRSs performed no differently from the simple genetic risk assessment (p>0.10) and performed less well as compared to the socioeconomic index (p=0.021). When combined with the comparison risk measures and with demographic and geographic information, the GRS improved predictiveness for whites but not for African Americans (Supplementary Table 9).
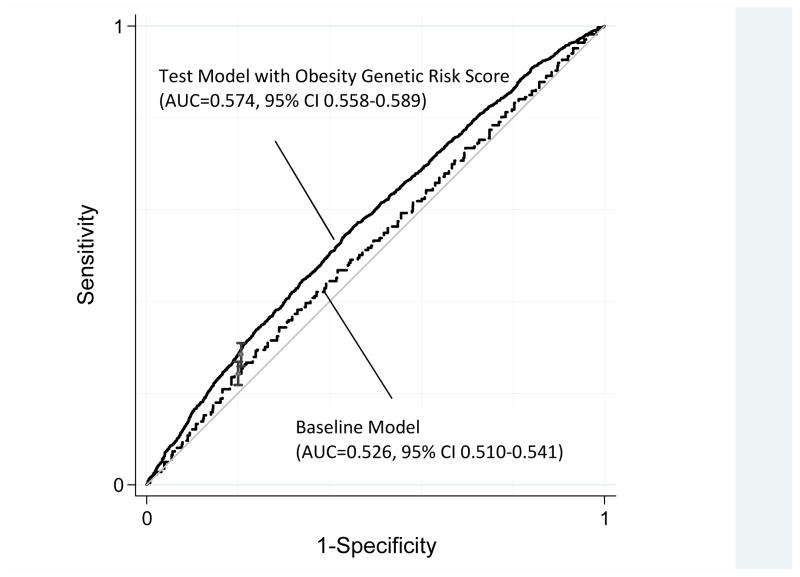
Figure 2 shows the model-based receiver operating characteristic curves for a baseline model that included demographic and geographic information and a test model that also included the weighted GRS. The change in AUC from the baseline model to the test model was greater than zero (Delta AUC=0.048, 95% CI 0.313–0.658, p<10−7), indicating that the GRS improved discrimination of obese cases. This improvement in discrimination was concentrated at low specificities, but extended to the portion of the ROC curve of greatest interest to clinicians. At a specificity of 0.8, the test model including the GRS was marginally more sensitive as compared to the baseline model (Delta Partial AUC=0.007, 95% CI <0.0003–0.010, p=<0.001). Results for African Americans are presented in Supplementary Figure 2.
Figure 2. Receiver Operating Characteristic Curves for Obesity Among White ARIC Participants (n=8,286).
Baseline Model = gender, age (quadratic), gender x age interaction, ARIC study center; Test Model = baseline model + weighted obesity genetic risk score. ROC Curves were constructed using predicted values from probit regressions of obesity (BMI≥30) on the model terms. Delta AUC (AUCTest − AUCBaseline) = 0.048, 95% CI 0.031–0.066, p<1×10−7. Delta Partial AUC at 80% specificity=0.007, 95% CI 0.003–0.010, p<0.001. AUCs, partial AUCs, and delta AUCs were estimated using Pepe’s method 35,37.
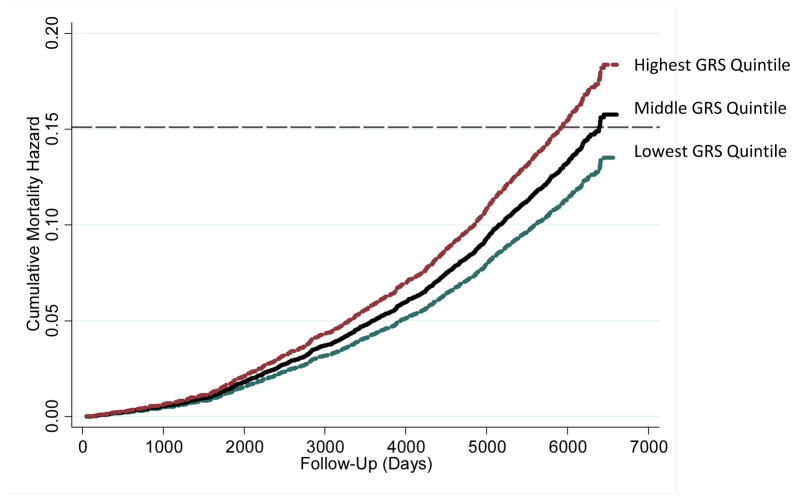
As a final analysis, we asked whether the obesity GRS was associated with mortality risk. The ARIC study conducted follow-up with participants through December 31, 2004 to determine whether study members had died. Mortality follow-up data were available for 8,284 of the 8,286 white participants in our analysis sample. 15% of this sample (n=1,253 individuals) died during the 17 years of follow-up from the first study visit. We analyzed mortality risk using Cox proportional hazard models to adjust for demographic and geographic factors. Independent of demographics and geography, individuals with higher genetic risk scores were more likely to die during the follow up period (Hazard Ratio=1.12, 95% CI [1.04–1.15]). Consistent with analyses of BMI and obesity, the GRS was not associated with mortality among African Americans. Figure 3 presents cumulative mortality hazards for white ARIC participants in the top, middle, and bottom quintiles of the genetic risk distribution. The mortality hazard associated with the GRS did not depend on individuals’ BMIs. Adjustment of the mortality hazard model for BMI only slightly reduced the mortality hazard associated with genetic risk (Hazard Ratio=1.10 [1.04–1.17]).
Figure 3. Cumulative Mortality Hazards for White ARIC Participants in the Highest, Middle, and Lowest Genetic Risk Score (GRS) Qunitiles.
Hazards were estimated from a Cox proportional hazard model adjusted for age, sex, the age-sex interaction, and the ARIC Study Center where data were collected. The dashed line represents sample-wide mortality at the end of follow-up (15%). By the end of follow-up, unadjusted mortality was 12.17% in the lowest GRS quintile, 15.48% in the middle GRS quintile, and 17.32% in the highest GRS quintile.
DISCUSSION
We used a 3-stage approach to construct an obesity GRS from GWAS results. Our tests of this obesity GRS in the population-based ARIC cohort revealed it to be a highly statistically significant predictor of BMI measured at 4 time points across 10 years, of weight and waist-circumference, and of obesity. In terms of value added, the GRS improved prediction of BMI and obesity over and above demographic and geographic information, FTO and MC4R genotypes, and information about socioeconomic status. Thus, the GRS provides a measure of genetic predisposition to obesity that could inform etiological and treatment research. Finally, the GRS was associated with mortality risk. Interestingly, higher mortality risk for individuals with higher GRSs did not depend on their BMI.
The research utility of the GRS is likely limited to samples of European descent. GRS-BMI and GRS-obesity associations in African American ARIC participants were much smaller than comparable associations in white ARIC participants. Although the sample included fewer African Americans than whites, power to detect effects of equal size to those observed in whites was well over 80% in the African American sample. Moreover, effect-size measures (r, R2, relative risk, AUC, IDI) showed little evidence that the GRS predicted BMI or obesity among African Americans. These results suggest caution in using GWAS of European-descent populations to derive GRSs for African Americans. Our analyses indicated the GRS performed similarly among men and women. However, emerging evidence for gene-sex interactions in obesity 46,47 suggests that future obesity GRSs may require sex-specific construction.
Our results have implications for theory, research, and clinical practice. With respect to theory, our results are consistent with the hypothesis that genetic risk for obesity is quantitatively distributed and can be operationalized in a GRS 48. With respect to research methods, our findings illustrate one approach to operationalize quantitative genetic risk. A systematic and replicable approach to selecting SNPs from association studies to follow-up in etiological and treatment research will be especially important with the advent of next-generation sequencing approaches. Next generation sequencing is likely to uncover many new disease-associated loci for obesity and for other phenotypes of interest to clinicians and researchers. These variants, though rarer in the population, may have higher penetrance and thus greater clinical relevance. Future research can also make use of the GRS derived in this study as a measure of inherited obesity risk. With respect to clinical practice, results indicate that, for persons in middle age, GWAS SNP-based approaches to obesity risk assessment offer little in the absence of more detailed information about lifestyle and environment. Although genetic information reliably predicted risk for obesity over and above demographics and geography, the magnitude of this additional risk was insufficient to recommend our score for use in clinical risk assessments. This result is especially important in the context of questions over consumer genomics services 49. Our 3-stage approach derived a more comprehensive genetic risk assessment for obesity than those currently used by companies marketing genomics services directly to consumers. The very modest risk information furnished by our GRS recommends caution on the part of health professionals in interpreting risk information provided by consumer genomics companies. The standard of evidence used here—multi-method assessment of predictiveness in large, population-based samples--should be considered a minimum standard for the validity of such risk information.
Results should be considered in light of the following limitations: First, some ARIC participants were included in the samples of some of the GWAS used to construct the GRS. However, these ARIC participants represented a minority of the GWAS samples and results in the ARIC sample are similar to results from samples not included in any of the GWAS 29,30. Second, some risk loci identified by our 3-stage approach could only be genotyped in the ARIC sample using relatively weak proxies. Given the small improvement to predictiveness associated with each additional SNP included in the GRS, it is unlikely that this limitation influenced the substance of our results, but it is possible that our GRS is moderately more predictive than analyses in the ARIC cohort suggest. Third, our analyses were limited to African American and white Americans. The ARIC cohort does not contain Asian-descent or Hispanic individuals. It remains unclear whether the relatively greater similarity between these and European populations 50 would support the generalization of our GRS. However, GWAS of Asian and Hispanic samples 28,51 suggest that a European-descent population-derived GRS may omit important risk loci for these populations. As more GWAS of non-European populations become available, our 3-stage approach can be used to derive additional population-specific GRSs. Fourth, there is mounting evidence that many genetic factors predisposing individuals to obesity are sex specific 52 and that GWAS that fail to model such sex specificity may not detect important risk variants 53. Results from GWAS modeling gene-by-sex interaction support this hypothesis 47,54,55. As more such GWAS become available, our 3-stage approach can be used to derive sex-specific GRSs for obesity. Finally, the ARIC sample is limited to individuals in middle age. There is evidence that genetic risk for obesity has dynamic consequences across development 56,57. It will be important in subsequent investigations to evaluate our obesity GRS in longitudinal cohorts that capture a broader section of the life course, and particularly in young people, as they are a key prevention target 58.
We constructed a GRS for obesity and showed that it predicted BMI and obesity in a population-based sample of middle-aged adults. We further showed that this GRS was longitudinally associated with mortality risk. These associations suggest that future research into obesity etiology and treatment can make use of genetic information. However, our analyses do not support the use of genetic testing for individual-level obesity-risk prediction. Future research with this GRS should characterize the expression of genetic risk across the life course and particularly during childhood, when intervention to prevent the development of obesity may be most effective.
Supplementary Material
Acknowledgments
This research received support from UK Medical Research Council grants G0100527 and G0601483, US-National Institute on Aging grant AG032282, and US-NIMH grant MH077874. Additional support was provided by the Jacobs Foundation. Mr. Belsky is supported in part by a fellowship from the Agency for Healthcare Research and Quality (1R36HS020524-01). The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. ARIC data (phs000090.v1.p1) were obtained from dbGaP.
References
- 1.Janssens A. Is the time right for translation research in genomics? Eur J Epidemiol. 2008 Nov;23(11):707–710. doi: 10.1007/s10654-008-9293-8. [DOI] [PubMed] [Google Scholar]
- 2.Khoury MJ, McBride CM, Schully SD, et al. The Scientific Foundation for Personal Genomics: Recommendations from a National Institutes of Health-Centers for Disease Control and Prevention Multidisciplinary Workshop. Genetics in Medicine. 2009 Aug;11(8):559–567. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009 Jun 9;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007 Oct;17(10):1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008 Feb;40(2):124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 6.Khoury MJ, Feero WG, Reyes M, et al. The genomic applications in practice and prevention network. Genet Med. 2009 Jul;11(7):488–494. doi: 10.1097/GIM.0b013e3181a551cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson G, Goldstein DB. Human genetics: The hidden text of genome-wide associations. Curr Biol. 2007 Nov;17(21):R929–R932. doi: 10.1016/j.cub.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 8.Orozco G, Barrett JC, Zeggini E. Synthetic associations in the context of genome-wide association scan signals. Hum Mol Genet. 2010 Oct 15;19(R2):R137–144. doi: 10.1093/hmg/ddq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horne BD, Anderson JL, Carlquist JF, et al. Generating genetic risk scores from intermediate phenotypes for use in association studies of clinically significant endpoints. Annals of Human Genetics. 2005 Mar;69:176–186. doi: 10.1046/j.1529-8817.2005.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher R. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- 11.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006 Apr 5;295(13):1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 12.Adams KF, Schatzkin A, Harris TB, et al. Overweight, obesity, and mortality in a large prospective cohort of persons 50 to 71 years old. N Engl J Med. 2006 Aug 24;355(8):763–778. doi: 10.1056/NEJMoa055643. [DOI] [PubMed] [Google Scholar]
- 13.Allender S, Rayner M. The burden of overweight and obesity-related ill health in the UK. Obesity Reviews. 2007 Sep;8(5):467–473. doi: 10.1111/j.1467-789X.2007.00394.x. [DOI] [PubMed] [Google Scholar]
- 14.Trogdon JG, Finkelstein EA, Hylands T, Dellea PS, Kamal-Bahl SJ. Indirect costs of obesity: a review of the current literature. Obesity Reviews. 2008 Sep;9(5):489–500. doi: 10.1111/j.1467-789X.2008.00472.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang WJ, Kelly T, He J. Genetic epidemiology of obesity. Epidemiologic Reviews. 2007;29:49–61. doi: 10.1093/epirev/mxm004. [DOI] [PubMed] [Google Scholar]
- 16.O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009 Nov;462(7271):307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010 Dec 9;363(24):2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 18.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. [Accessed April 30, 2010];A Catalog of Published Genome-Wide Association Studies. http://www.genome.gov/gwastudies/
- 19.Folsom AR, Chambless LE, Ballantyne CM, et al. An assessment of incremental coronary risk prediction using C-reactive protein and other novel risk markers: the atherosclerosis risk in communities study. Arch Intern Med. 2006 Jul 10;166(13):1368–1373. doi: 10.1001/archinte.166.13.1368. [DOI] [PubMed] [Google Scholar]
- 20.The ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989 Apr;129(4):687–702. [PubMed] [Google Scholar]
- 21.GENEVA ARIC Project. ARIC Quality Control Report. 2009. [Google Scholar]
- 22.Psaty BM, O’Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009 Feb;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008 Oct;40(10):1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyssenko V, Jonsson A, Almgren P, et al. Clinical Risk Factors, DNA Variants, and the Development of Type 2 Diabetes. N Engl J Med. 2008 Nov;359(21):2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 25.Morrison AC, Bare LA, Chambless LE, et al. Prediction of coronary heart disease risk using a genetic risk score: The atherosclerosis risk in communities study. American Journal of Epidemiology. 2007 Jul;166(1):28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 26.Talmud PJ, Hingorani AD, Cooper JA, et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. Br Med J. 2010 Jan;340:b4838. doi: 10.1136/bmj.b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demirkan A, Penninx BW, Hek K, et al. Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Mol Psychiatry. 2010 Jun 22;16(7):773–783. doi: 10.1038/mp.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He MA, Cornelis MC, Franks PW, Zhang CL, Hu FB, Qi L. Obesity Genotype Score and Cardiovascular Risk in Women With Type 2 Diabetes Mellitus. Arteriosclerosis Thrombosis and Vascular Biology. 2010 Feb;30(2):327–U370. doi: 10.1161/ATVBAHA.109.196196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Zhao JH, Luan J, et al. Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am J Clin Nutr. 2010 Jan;91(1):184–190. doi: 10.3945/ajcn.2009.28403. [DOI] [PubMed] [Google Scholar]
- 30.Peterson RE, Maes HH, Holmans P, et al. Genetic risk sum score comprised of common polygenic variation is associated with body mass index. Hum Genet. 2011 Feb;129(2):221–230. doi: 10.1007/s00439-010-0917-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010 Nov;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009 Jan;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 33.McGeechan K, Macaskill P, Irwig L, Liew G, Wong TY. Assessing new biomarkers and predictive models for use in clinical practice: a clinician’s guide. Archives of Internal Medicine. 2008 Nov 24;168(21):2304–2310. doi: 10.1001/archinte.168.21.2304. [DOI] [PubMed] [Google Scholar]
- 34.Mihaescu R, van Zitteren M, van Hoek M, et al. Improvement of risk prediction by genomic profiling: reclassification measures versus the area under the receiver operating characteristic curve. American Journal of Epidemiology. 2010 Aug 1;172(3):353–361. doi: 10.1093/aje/kwq122. [DOI] [PubMed] [Google Scholar]
- 35.Janes H, Pepe MS. Adjusting for covariate effects on classification accuracy using the covariate-adjusted receiver operating characteristic curve. Biometrika. 2009 Jun;96(2):371–382. doi: 10.1093/biomet/asp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pepe MS, Cai TX, Longton G. Combining predictors for classification using the area under the receiver operating characteristic curve. Biometrics. 2006 Mar;62(1):221–229. doi: 10.1111/j.1541-0420.2005.00420.x. [DOI] [PubMed] [Google Scholar]
- 37.Pepe M, Longton G, Janes H. Estimation and Comparison of Receiver Operating Characteristic Curves. Stata J. 2009 Mar 1;9(1):1. [PMC free article] [PubMed] [Google Scholar]
- 38.Pencina MJ, D’Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat Med. 2008 Jan;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 39.Brown MB, Forsythe AB. Robust tests for equality of variances. Journal of the American Statistical Association. 1974;69:364–367. [Google Scholar]
- 40.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. Plos Genetics. 2006 Dec;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.GENEVA ARIC Project. ARIC Quality Control Report. 2009. [Google Scholar]
- 42.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006 Aug;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.Hardy R, Wills AK, Wong A, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010 Feb;19(3):545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ford ES, Mokdad AH. Epidemiology of obesity in the Western Hemisphere. J Clin Endocrinol Metab. 2008 Nov;93(11 Suppl 1):S1–8. doi: 10.1210/jc.2008-1356. [DOI] [PubMed] [Google Scholar]
- 45.Drewnowski A. Obesity, diets, and social inequalities. Nutr Rev. 2009 May;67 (Suppl 1):S36–39. doi: 10.1111/j.1753-4887.2009.00157.x. [DOI] [PubMed] [Google Scholar]
- 46.Benjamin AM, Suchindran S, Pearce K, et al. Gene by sex interaction for measures of obesity in the framingham heart study. J Obes. 2011;2011:329038. doi: 10.1155/2011/329038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010 Nov;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nature Reviews Genetics. 2009 Dec;10(12):872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 49.Evans JP, Meslin EM, Marteau TM, Caulfield T. Genomics. Deflating the genomic bubble. Science. 2011 Feb 18;331(6019):861–862. doi: 10.1126/science.1198039. [DOI] [PubMed] [Google Scholar]
- 50.Jorde LB, Wooding SP. Genetic variation, classification and ‘race’. Nature Genetics. 2004 Nov;36(11 Suppl):S28–33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 51.Norris JM, Langefeld CD, Talbert ME, et al. Genome-wide association study and follow-up analysis of adiposity traits in Hispanic Americans: the IRAS Family Study. Obesity. 2009 Oct;17(10):1932–1941. doi: 10.1038/oby.2009.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCarthy JJ, Meyer J, Moliterno DJ, Newby LK, Rogers WJ, Topol EJ. Evidence for substantial effect modification by gender in a large-scale genetic association study of the metabolic syndrome among coronary heart disease patients. Hum Genet. 2003 Dec;114(1):87–98. doi: 10.1007/s00439-003-1026-1. [DOI] [PubMed] [Google Scholar]
- 53.McCarthy JJ. Gene by sex interaction in the etiology of coronary heart disease and the preceding metabolic syndrome. Nutr Metab Cardiovasc Dis. 2007 Feb;17(2):153–161. doi: 10.1016/j.numecd.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Benjamin AM, Suchindran S, Pearce K, et al. Gene by sex interaction for measures of obesity in the framingham heart study. J Obes. 2011;2011:329038. doi: 10.1155/2011/329038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiu YF, Chuang LM, Kao HY, et al. Sex-specific genetic architecture of human fatness in Chinese: the SAPPHIRe Study. Hum Genet. 2010 Nov;128(5):501–513. doi: 10.1007/s00439-010-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sovio U, Mook-Kanamori DO, Warrington NM, et al. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: The complex nature of genetic association through growth and development. Plos Genetics. 2011;7(2):e1001307. doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elks CE, Loos RJ, Sharp SJ, et al. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med. 2010;7(5):e1000284. doi: 10.1371/journal.pmed.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dietz WH. Overweight in childhood and adolescence. N Engl J Med. 2004 Feb 26;350(9):855–857. doi: 10.1056/NEJMp048008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.