Abstract
Diffusion magnetic resonance imaging has been used extensively to probe the white matter in vivo. Typically, the raw diffusion images are used to reconstruct a diffusion tensor image (DTI). The in-capacity of DTI to represent crossing fibers leaded to the development of more sophisticated diffusion models. Among them, multi-fiber models represent each fiber bundle independently, allowing the direct extraction of diffusion features for population analysis. However, no method exists to properly register multi-fiber models, seriously limiting their use in group comparisons. This paper presents a registration and atlas construction method for multi-fiber models. The validity of the registration is demonstrated on a dataset of 45 subjects, including both healthy and unhealthy subjects. Morphometry analysis and tract-based statistics are then carried out, proving that multi-fiber models registration is better at detecting white matter local differences than single tensor registration.
Keywords: Diffusion Imaging, Multi-Fiber Models, Registration, White Matter
1 Introduction
Diffusion magnetic resonance imaging offers the ability to investigate in vivo the white matter microstructure. The representation of the signal by diffusion tensor images (DTI) has proven useful for population analysis in two ways [14]. First, scalar features extracted from DTI, such as the fractional anisotropy (FA), may indicate the presence of brain diseases. Second, the use of DTI in registration improves the detection of morphometric differences, compared to scalar images.
The single tensor diffusion model has, however, proven inaccurate for two main reasons. First, it cannot represent the signal arising from multiple fibers with heterogeneous orientations in one voxel. Second, it does not account for the non-monoexponential decay observed when imaging at high b-values. Novel models addressing one or both of these issues have been introduced [8] : Q-ball imaging, spherical deconvolution, 4th order tensors, DOT, and others. Most of them focus on describing the general shape of the diffusion at each voxel. In contrast, mixture models represent each fiber bundle independently, keeping the interpretability of single fiber models while accounting for crossing fibers. Therefore, scalar quantities such as the fractional anisotropy (FA) can still be computed for each fiber independently. This property makes them very attractive for population analysis.
While the literature on the registration of complex diffusion models is growing (e.g., [4,12]), no method has been developed to register mixture models. This lack of a registration method limits the use of mixture models in population analysis despite their attractiveness. This issue has been previously reported, and has incited researches to register the raw diffusion weighted images instead [5].
The remaining of this paper is organized as follows. Section 2 introduces the diffusion mixture models. Section 3 presents a method to compute weighted average of mixture models. Section 4 develops a similarity metric for diffusion mixture images. Section 5 presents the integration of the developed methods in a registration algorithm and analyzes its complexity. Section 6 presents experimental results on the dataset of 45 subjects. Finally Section 7 concludes.
2 Diffusion Mixtures
The basic idea behind multi-fiber models is to fit a single fiber model to each of the fiber bundles present in the voxel. If Si(x) is a suitable model to represent the diffusion process in a single fiber, then,
| (1) |
is a multi-fiber model for N crossing fibers with relative volumetric occupancy given by fi. The assumption behind these models is that the exchange of water molecules between populations of fibers is negligible during the diffusion time [8].
The simplest multi-fiber model is the multi-tensor model in which Si = S0e−bgT Dig. More complex multi-fiber models have later been introduced [2]. Potentially, any single fiber model can be extended to a multi-fiber model by means of mixtures. One such model, the biexponential decay model [8], represents each fiber bundle by a Gaussian mixture to capture the non-monoexponential decay of the signal. The corresponding multi-fiber model would be a mixture of Gaussian mixtures which is itself a Gaussian mixture. A natural parameterization of diffusion Gaussian mixtures is the set of pairs (fraction, covariance matrix), that we write: {(f1, Σ1), …, (fN, ΣN)}. Alternatively, to connect with the tensor formalism, the inverse of the covariance matrix, , can be used.
3 Weighted Combination of Mixtures
Computing weighted combinations of voxel values is at the basics of interpolation (the value in one location is the weighted combination of the values in the neighborhood), smoothing (the value at a grid voxel is replaced by a weighted combination of the values in a neighborhood) and atlas construction (the value at one voxel is the average of the values in the aligned subjects').
Gaussian mixture simplification (GMS) was introduced to efficiently interpolate diffusion mixture models [11]. In this section, we underline the important aspects of this method. The idea behind GMS is that computing weighted combinations of mixtures would be trivial if the number of components of the result could be arbitrarily large. Indeed, the linear combination of K mixture models with N components is a mixture models with M = KN components:
| (2) |
We refer to this mixture as the complete mixture. GMS optimizes the parameters of a simplified mixture with N components that best approximates . The energy function to be minimized is the cumulative differential entropy:
| (3) |
where latent variables πi cluster the components of the complete mixture Si in N clusters each represented by a single component of the simplified mixture, Rj; πi = j means that Si is best represented by Rj. Following the recent developments in probabilistic clustering, an EM scheme is used to minimize (3). Banerjee et al showed that both the E-step and the M-step can be solved in closed form for mixtures of exponential distributions [3]. For Gaussian mixtures, the E-step consists in optimizing the latent variables πi by computing the Burg matrix divergence between the covariance matrices of each component of and each component of :
| (4) |
As for the M-step, it sums up to calculating:
| (5) |
Alternating (4) and (5) until convergence provides the parameters (hj and Rj) of the weighted combination of mixtures.
4 Generalized Correlation Coefficient for Mixtures
The correlation coefficient, invariant under linear transformations of the voxel intensities, is widely used in mono-modal image registration. The inter-subject variability of diffusivity values motivates the introduction of a generalized correlation coefficient, invariant under these differences. In DTI, this variability has been reported and partially accounted for in some registration methods [14]. The correlation coefficient between blocks F and G is defined as the scalar product of the normalized blocks:
where μF is the mean of the image values in the block. It is invariant if F (and/or G) is replaced by aF + b. It has been generalized to vector images by redefining the means μF and μG as the projection of the block onto a constant block T [9]:
The corresponding generalized correlation coefficient is invariant if F is replaced by aF + bT where T is now any constant vector block. The definition of a scalar product between two blocks of mixture models seems impractical if not impossible. We therefore further generalize the correlation coefficient by substituting the inner product by a more general scalar mapping, :
where is a generalization of the norm. This definition does not guarantee the invariance property of the metric for any scalar mapping. One can show that the invariance is preserved as long as the scalar mapping is linear with respect to the constant block :
| (6) |
To preserve the interpretability of ρ as a similarity metric, it needs to be symmetric, equal to one in case of perfect match and lower than one in any other case. These constraints on ρ translate into the following constraints on m:
| (7) |
| (8) |
| (9) |
The latter is a generalized form of the Cauchy-Schwartz inequality for inner products. Conditions (6–9), the choice of and the definition of the multiplication by a scalar and addition of the block , stand together as a model to generate correlation coefficients in potentially any space. For DTI, if is an isotropic tensor block , m is the log-Euclidean scalar product, and the log-Euclidean algebra is used, then ρ is invariant under linear transformations of the eigenvalues in the log-domain. For multi-tensor images, we fix , and we define the addition,, and the multiplication by a scalar, , component-wise in the log-domain. The scalar mapping is defined by pairing the tensors in each voxel to maximize the linear combination of pairwise scalar products. Let and defined on a domain Ω, we have:
where π is a pairing function associating one tensor of to each tensor of . This scalar mapping satisfies conditions (6–9). Interestingly, the resulting generalized correlation coefficient is invariant under any global (within the block) linear transformation of all eigenvalues in the log-domain. This similarity metric is therefore robust to the inter-subject variability of diffusivities.
5 Implementation and Complexity
The developed methods were integrated in the efficient block matching registration algorithm described in [6]. The parameters used were the following: 4 pyramid level, 10 iterations per level, block size: 5 × 5 × 5, outlier removal rate: 20%. The implementation was multi-threaded. On a 8 core workstation, with 220×220×176 two-fiber images, the entire registration takes 1.5 hour. All weighted combinations were computed until complete convergence of the soft clustering. The average number of iterations required for that convergence is 4.
6 Results
The registration was applied to a clinical dataset of 45 subjects, 13 controls and 32 patients with tuberous sclerosis complex (TSC), a rare genetic disease associated with impaired white matter integrity. A DTI and a multi-tensor model with three components (one isotropic and two anisotropic) were reconstructed for each subject [10].
6.1 Validation
An alternative to the method presented in this paper would be to select one of the two tensors in each voxel (e.g. the one with the highest FA) and to perform single tensor registration on this image. Here, we validate that our method works better than this simple alternative. The quality of the alignment is assessed by the sum of square differences of each eigenvalue after alignment of control subjects. Indeed, while the diffusivities can significantly differ in diseased brain, they are approximately equal for healthy subjects. We performed 26 randomly chosen registrations with four levels of regularization, totalizing 104 registrations. In each voxel, the eigenvalues were averaged between the two anisotropic components (weighted by the fractions). Results show that multi-fiber registration performs significantly better than single tensor registration (Fig. 1).
Fig. 1.
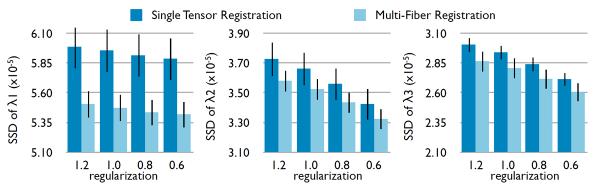
Comparison of the single tensor and multi-fiber registration in terms of the SSD between eigenvalues after alignment, for different regularization parameter [6]. Multi-fiber registration significantly improves the quality of the registration.
6.2 Atlas Construction
An atlas was constructed using our registration and alternating three steps: aligning all subjects to the current atlas (initially a randomly chosen subject), averaging the aligned subjects (using the weighted combination of mixtures), applying the mean inverse field to the resulting average [7]. This atlas remarkably shows areas where multiple fiber are consistently present in all subjects (Fig. 2).
Fig. 2.
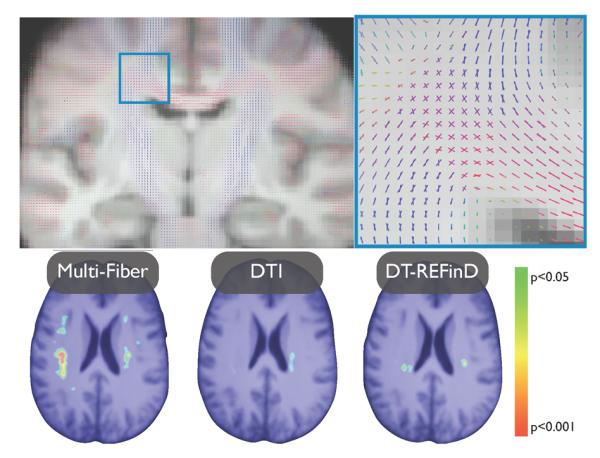
(top) The two-tensor atlas built by means of the developed registration method reveals crossing pathways common to all anatomies. (bottom) p-value maps of the white matter shrinkage after correction for multiple comparisons. Multi-fiber registration reveals more differences than single tensor registration and DT-REFinD
6.3 Morphometry
The clinical hypothesis according to which there is substantial white matter shrinkage in TSC subjects was tested by performing a one-tailed two sample t-test on the log-Jacobian of the deformation fields [1]. The subject classes were then randomly permuted 4000 times to assess the null distribution of extreme t-scores. The entire process was repeated with single tensor images. As a result, multi-fiber registration reveals more white matter differences (> 3800 significant voxels) than single tensor (< 1000 voxels) (Fig 2). The entire process was then repeated with DT-REFinD, a state-of-the art DTI registration algorithm [13], to test whether the improved detection of differences is truly due to the knowledge brought by multi-fiber models. Again, DT-REFinD did not capture all the differences detected by multi-fiber registration (< 1300 voxels) (Fig 2).
6.4 Tract-Based Statistics
Some structural subnetworks are believed to be impaired in TSC patients. To test this hypothesis, we analyzed the FA profile along the median tract of the arcuate fasciculus, generated on the atlas by a probabilistic tractography algorithm [5](Fig. 3(a)). A one-tailed two-sample t-test was performed at every location. A threshold t0 was then set to the t-statistics and the length of the contiguous supra-threshold segments were recorded. The null distribution of these lengths was assessed by randomly permuting the subjects classes 4000 times. The operation was repeated for a wide range of thresholds (1.5 ≤ t0 ≤ 4.5) to estimate the robustness of our findings. For t0 = 2.7 (p0 = 0.01), four significant clusters, together representing 15% of the tract were observed indicating a strong impairment of this subnetwork in TSC patients (Fig. 3(b)). These findings were robust to any threshold t0 ≤ 3:4. In contrast, single tensor registration only revealed one significant cluster representing 5% of the fiber, and was not robust outside the range 1:9 ≤ t0 ≤ 2:8.
Fig. 3.
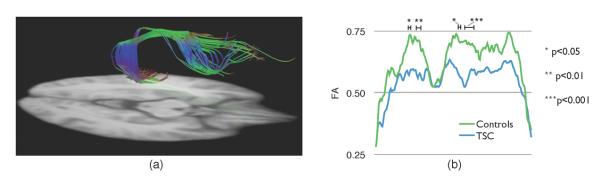
(a) Arcuate fasciculus, a set of fibers involved in language, on which tract based statistics was performed, (b) The FA profile in TSC patients shows significantly disrupted white matter fascicules in different clusters, indicated by the stars.
7 Conclusions
This paper introduced a registration and atlas construction method to align multi-fiber models. A proper interpolation method and a robust similarity metric were presented. Results in both morphometry and tract-based statistics demonstrate the advantage of multi-fiber models over single tensor DTI for population analysis. Therefore, we believe that this registration method opens new doors to understanding brain disorders based on multi-fiber models.
Acknowledgments
MT thanks the F.R.S.-FNRS, the BAEF and WBI.WORLD for their financial support. This investigation was supported in part by NIH grants R01 EB008015,R01 LM010033, R01 EB013248, and P30 HD018655 and by a research grant from the Boston Children's Hospital Translational Research Program.
References
- 1.Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K. Identifying global anatomical differences: deformation-based morphometry. Human Brain Mapping. 1998;6(5–6):348–357. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<348::AID-HBM4>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assaf Y, Basser P. Composite hindered and restricted model of diffusion (charmed) mr imaging of the human brain. Neuroimage. 2005;27(1):48–58. doi: 10.1016/j.neuroimage.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee A, Merugu S, Dhillon I, Ghosh J. Clustering with bregman divergences. The Journal of Machine Learning Research. 2005;6:1705–1749. [Google Scholar]
- 4.Barmpoutis A, Vemuri B. Groupwise registration and atlas construction of 4th-order tensor fields using the r+ riemannian metric. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2009. 2009:640–647. [PubMed] [Google Scholar]
- 5.Bergmann O, Kindlmann G, Peled S, Westin C. Two-tensor fiber tractography. Biomedical Imaging: From Nano to Macro, 2007. ISBI 2007. 4th IEEE International Symposium on; IEEE; 2007. pp. 796–799. [Google Scholar]
- 6.Commowick O, Arsigny V, Isambert A, Costa J, Dhermain F, Bidault F, Bondiau P, Ayache N, Malandain G. An efficient locally affine framework for the smooth registration of anatomical structures. MedIA. 2008;12(4):427–441. doi: 10.1016/j.media.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Guimond A, Meunier J, Thirion J. Average brain models: A convergence study. Computer vision and image understanding. 2000;77(2):192–210. [Google Scholar]
- 8.Minati L, Weglarz W. Physical foundations, models, and methods of diffusion magnetic resonance imaging of the brain: A review. Concepts in Magnetic Resonance Part A. 2007;30(5):278–307. [Google Scholar]
- 9.Ruiz-Alzola J, Westin C, Warfield S, Alberola C, Maier S, Kikinis R. Non-rigid registration of 3d tensor medical data. MedIA. 2002;6(2):143–161. doi: 10.1016/s1361-8415(02)00055-5. [DOI] [PubMed] [Google Scholar]
- 10.Scherrer B, Warfield S. Toward an accurate multi-fiber assessment strategy for clinical practice. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2011 IEEE; IEEE; 2011. pp. 2140–2143. [Google Scholar]
- 11.Taquet M, Scherrer B, Benjamin C, Prabhu S, Macq B, Warfield S. Interpolating multi-fiber models by gaussian mixture simplification. Biomedical Imaging: From Nano to Macro, 2012 IEEE International Symposium on.2012. [Google Scholar]
- 12.Yap P, Chen Y, An H, Yang Y, Gilmore J, Lin W, Shen D. Sphere: Spherical harmonic elastic registration of hardi data. NeuroImage. 2011;55(2):545–556. doi: 10.1016/j.neuroimage.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeo B, Vercauteren T, Fillard P, Peyrat J, Pennec X, Golland P, Ayache N, Clatz O. Dt-refind: Diffusion tensor registration with exact finite-strain differential. IEEE Trans. on Medical Imaging. 2009;28(12):1914–1928. doi: 10.1109/TMI.2009.2025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Avants B, Yushkevich P, Woo J, Wang S, McCluskey L, Elman L, Melhem E, Gee J. High-dimensional spatial normalization of diffusion tensor images improves the detection of white matter differences: an example study using amyotrophic lateral sclerosis. IEEE TMI. 2007;26(11):1585–1597. doi: 10.1109/TMI.2007.906784. [DOI] [PubMed] [Google Scholar]


