Abstract
In order to better understand the epidemiology of Human and Animal trypanosomiasis that occur together in sleeping sickness foci, a study of prevalences of animal parasites (Trypanosoma vivax, T. congolense “forest type”, and T. simiae) infections was conducted on domestic animals to complete the previous work carried on T. brucei gambiense prevalence using the same animal sample. 875 domestic animals, including 307 pigs, 264 goats, 267 sheep and 37 dogs were sampled in the sleeping sickness foci of Bipindi, Campo, Doumé and Fontem in Cameroon. The polymerase chain reaction (PCR) based method was used to identify these trypanosome species. A total of 237 (27.08%) domestic animals were infected by at least one trypanosome species. The prevalence of T. vivax, T. congolense “forest type” and T. simiae were 20.91%, 11.42% and 0.34% respectively. The prevalences of T. vivax and T. congolense “forest type” differed significantly between the animal species and between the foci (p < 0.0001); however, these two trypanosomes were found in all animal species as well as in all the foci subjected to the study. The high prevalences of T. vivax and T. congolense “forest type” in Bipindi and Fontem-Center indicate their intense transmission in these foci.
Keywords: T. vivax, T. congolense “forest type”, T. simiae, PCR, pig, goat, sheep, dog
Abstract
Afin de mieux comprendre l’épidémiologie des trypanosomoses humaines et animales sévissant ensemble dans les foyers de maladie de sommeil, une étude des prévalences des infections par Trypanosoma vivax, T. congolense “type forêt” et T. simiae a été menée chez les animaux domestiques pour compléter celle ayant porté sur la prévalence de T. b. gambiense. L’étude a concerné 875 animaux domestiques dont 307 porcs, 264 chèvres, 267 moutons et 37 chiens échantillonnés dans les foyers de maladie du sommeil de Bipindi, Campo, Fontem et Doumé au Cameroun. L’identification spécifique de ces trypanosomes a utilisée la technique de PCR. Au total, 237 (27,08 %) animaux domestiques étaient infectés par au moins une espèce de trypanosome. Les prévalences de T. vivax, T. congolense “type forêt” et T. simiae étaient de 20,91 %, 11,42 % et 0,34 % respectivement. Les prévalences de T. vivax et T. congolense “type forêt” diffèrent significativement entre les espèces animales et entre les foyers (p < 0,0001); toutefois, ces deux trypanosomes ont été identifiés chez les quatre espèces animales et dans tous les foyers soumis à l’étude. Les prévalences les plus élevées de T. vivax et T. congolense “type forêt” à Bipindi et Fontem-Centre indiquent une transmission intense de ces trypanosomes dans ces foyers.
Keywords: T. vivax, T. congolense “type forêt”, T. simiae, PCR, porc, chèvre, mouton, chien
Introduction
Animal trypanosomosis constitutes a serious handicap to animal husbandry in all regions of sub- Saharan Africa infested by tsetse flies (Vaucel et al., 1963). The infection agents are protozoans of the genus Trypanosoma. In tsetse infected areas, species and subspecies T. congolense, T. vivax, T. simiae and to a less extend T. b. brucei are pathogenic parasites to animals, especially Suidae and domestic ruminants (Omeke, 1994; Reifenberg et al., 1997). Most of these trypanosome species pathogenic to animals are transmitted by tsetse flies. However, T. vivax can be mechanically transmitted by Tabanids or Stomoxys in Africa, and it is presumed to be transmitted only mechanically in areas not infested by tsetse flies (Gardiner & Wilson, 1987; Gardiner, 1989; D’Amico et al., 1996). Moreover, experimental studies have demonstrated the possibility of mechanical transmission of T. congolense and T. brucei species by tabanids and Stomoxys (Mihok et al., 1995; Sumba et al., 1998). However, the importance of such transmission under natural conditions is still under debate (Desquesnes & Dia, 2003).
In animals, infection with trypanosome may result in a chronic, debilitating, emaciating and often fatal disease but the outcome of the infection differs substantially between trypanosome species or subspecies, between livestock species and within a livestock species among breeds depending on the challenge and virulence of the strains (Connor & van Den Bossche, 2004). Due to their frequencies, pathogenicity and consequence on productivity, T. congolense and T. vivax are the principal trypanosomes in domestic ruminants (Wellde et al., 1983; Trail et al., 1991).
Desoxyribonucleic acids (DNA) probes have allowed the identification of four different sub-species of T. congolense in different ecological zones: T. congolense “forest type”, T. congolense “savannah type”, T. congolense “Kilifi type” and T. congolense “Tsavo” (Majiwa et al., 1985; Masiga et al., 1992; Clausen et al., 1998; Majiwa et al., 1993). These variants of T. congolense are pathogenic and develop high parasitaemia accompanied by anemia and leucopenia (Sidibe et al., 2002; Bengaly et al., 2002a, b). However, there are clear differences in the pathogenicity among the various types of T. congolense and even within one single type. For example, experimental studies comparing the virulence of one strain of each subgroup in mice and cattle have shown differences between the subgroups with the T. congolense strain of the Savannah subgroup being the most virulent (Bengaly et al., 2002a, b). Moreover, substantial differences in the virulence of T. congolense strains of the “savannah” subgroup, isolated in one geographical area from a single host species have also been recently reported (Masumu et al., 2006).
For T. vivax infections, it is well known that animals infected by this trypanosome support better the infection because their genetic diversity is more limited than that of T. congolense and T. brucei (Authié et al., 1999). However, fulminant and hemorrhagic forms of T. vivax that cause death or abortion have been described (Hudson, 1944; Olubayo et al, 1985). T. congolense and T. vivax species have been shown to cause serious infections in horses and asses (Faye et al., 2001; Dhollander et al., 2006). T. simiae has been reported as a parasite of suidae (Stephen, 1966).
During herd control by veterinary services, T. vivax, T. congolense or T. b. brucei were found in pigs, ruminants and equines in West Africa (Mattioli et al., 1994; Omeke et al., 1994; Solano et al., 1999; Faye et al., 2001; Dhollander et al., 2006). In the Central African region, limited work has been conducted on animal trypanosomosis, especially in sleeping sickness foci where Trypanosoma brucei gambiense is the causative agent of the disease. Indeed, in the forest area of southern Cameroon where studies were undertaken in the historic sleeping sickness foci of Fontem and Mbam, T. congolense, T. vivax and T. simiae were identified in pigs and small domestic ruminants (Asonganyi et al., 1986; Asonganyi et al., 1990; Simo et al., 2006). Moreover, the presence of T. brucei s.l., T. congolense and T. vivax were reported in cattle of the pastoral zone of Adamaoua (North-Cameroon) (Mamadou et al., 2006). However, most of these works were performed either on one species of domestic animals or used parasitological and immunological methods. Given the low sensitivity and specificity of these methods, it is obvious that the prevalence of different trypanosomes species or subspecies were considerably underestimated. With the development of molecular biology during the last decades, specific DNA sequences of different trypanosome species and subspecies have been identified and several PCR based methods were developed to improve the detection of various parasites. Applied in human and animal trypanosomosis, PCR appeared as a reliable, sensitive and specific techniques enabling to detect different trypanosome species and subspecies in vertebrate hosts as well as in tsetse flies (Masiga et al., 1992; Desquesnes & Tresse, 1996; Penchenier et al., 2000; Mugutu et al., 2001; Picozzi et al., 2002; Geysen et al., 2003; Delespaux et al., 2003; Gonzales et al., 2005; Cox et al., 2005).
The present study reports the level of infections with T. vivax, T. congolense “forest type” and T. simiae in four species of domestic animals commonly found in sleeping sickness foci of forest areas of southern Cameroon.
Materials and Methods
Study area
The study was carried from 2002 to 2004 in four sleeping sickness foci of the forest region of Southern-Cameroon (Fig. 1):
Fig. 1.
Study area in Cameroon.
M: Molecular weight markers (100 bp); C + : T. vivax DNA (150 bp); C-: negative control; 1-9 samples; 2; 4; 7 and 8: negative samples; 1; 3; 5; 6 and 9: positive samples.
Bipindi (3°2’N, 10°22’E), is a historic sleeping sickness focus discovered in 1920 which has been in recrudescence recently (Grébaut et al., 2001). It lies between Lolodorf and Kribi, at 75 Km from the Atlantic Ocean. Though, Glossina palpalis palpalis is the dominant tsetse fly species, other tsetse fly species like G. pallicera, G. caliginea and G. nigrofusca can be also found in this focus (Morlais et al., 1998; Simo et al., 2008). The domestic animals were bled in the villages Bijouka, Lambi, Memel, Bipindi-Centre and Ebimingbang.
Campo (2°20’N, 9°52’E), is a hypo-endemic focus where a few sleeping sickness patients are detected each year. It lies along the River Ntem which separates Cameroon and Equatorial Guinea and flows into the Atlantic Ocean. Several tsetse fly species including G. p. palpalis, and to a lesser extent, G. pallicera, G. caliginea and G. nigrofusca are encountered in this sleeping sickness focus (Morlais et al., 1998; Simo et al., 2008). Samples were taken from domestic animals in the villages Akak, Mabiogo, Ipono, and Campo center.
Fontem (5°40’N, 9°55’E) is a sleeping sickness focus known since 1949 where the prevalence of sleeping sickness has reduced considerably. It has a much varied topography with numerous hills and valleys through which high speed rivers flow in the South Western Region of Cameroon. G. p. palpalis is the only tsetse fly species found in this area (Morlais et al., 1998; Simo et al., 2008). The focus is divided into three sub-foci (North, Center and South). In the Center sub-focus where the samples were taken (villages of Menji, Fotabong, Soko, Azi), few sleeping sickness patients were detected during the last decade. In the Northern villages (Bechati, Folepi, Besali), no patients have been detected during the last 20 years (Ebo’o Eyenga, personal communication), although in pigs, the prevalence of T. b. gambiense group 1 infections was 15% in 1999 (Nkinin et al., 2002).
Doumé (4°16’N, 13°25’E) is an old sleeping sickness focus in the Eastern Region of Cameroon where very few sleeping sickness patients were detected during the last decade. Glossina fuscipes is the main tsetse fly species found in this focus (Mbida Mbida et al., 2009). Doumé is a degraded forest zone with many rivers and vast areas of wetlands. Samples were collected from domestic animals in the villages Medim, Paki, Baillon and Loumbou.
In these four sleeping sickness foci where cattle is rare, pigs, sheep, goats and chickens are kept to meet dietary, ceremonial and commercial needs. Dogs serve as pets and hunting companions.
Blood collection from animals
Domestic animals were sampled during five field surveys: one was done at Doumé (October 2002), one at Bipindi (July 2003), one at Fontem (October, 2003) and two at Campo (April 2003 and June 2004). During each survey, about one in three animals that has spent at least three months in the focus was sampled and bled with the cooperation of the owners, but some dogs did not cooperate. Goats and sheep were bled from the jugular vein; pigs from the sub-clavicular vein and dogs from the cephalic vein. The blood was put in EDTA coated tubes, labeled and preserved at 4 °C for molecular analyses.
All pigs and dogs sampled in this study were of a local breed, originating from a mixture of different breeds. The sheep and goats are Dwarf breeds (Djallonke West-African Dwarf for sheep and Guinea goat), which are known to be trypanotolerant.
Extraction and amplification of DNA
DNA was extracted from the samples using the kit “Ready Amp Genomic DNA purification system” (PROMEGA) essentially as described by Penchenier et al., (2000). The supernatant containing DNA was stored at -20 °C or used directly for PCR.
Amplification of trypanosome DNA was conducted using the primer pairs TCF1 (GGACACGCCAGAAGGTACTT) / TCF2 (GTTCTCGCACCAAATCCAAC), TVW1 (CTGAGTGCTCCATGTCCCAC) / TVW2 (CCACCAGAACACCAACCTGA), and TSM1 (CCGGTCAAAAACGCATT) / TSM2 (AGTCGCCCGGAGTCGAT) specific for T. congolense “forest type”, T. vivax and T. simiae respectively (Masiga et al., 1992). The amplifications were conducted in a total volume of 25 μl containing 2.5 μl of PCR buffer 10X [10 mM Tris – HCl (pH 9.0), 50 mM KCl, 3 mM MgCl2], 15 picomoles of each primer, 200 μl of deoxynucleotide-triphosphate (dNTP), one unit of Taq DNA polymerase (Appligene-Oncor, USA), sterile water and 3 μl DNA extract. Amplification involved pre-denaturation at 94 °C for 3 min 30 s followed by 40 cycles of denaturation at 94 °C (30 s), hybridization of primers at 60 °C and elongation at 72 °C for 1 minute, then final elongation at 72 °C for 5 min. The amplification products were resolved on 2% agarose gel containing ethidium bromide (0.3 μg/ml). DNA bands were visualized under ultraviolet (UV) light.
Statistical analyses
The proportions of animals infected by different trypanosome species were compared between animals and localities using Chi-square (χ2) test of the Statistix Computer program.
Results
Fig. 2 shows an example of profiles obtained after resolution of PCR products from the amplification of T. vivax DNA. Tables I and II report the number of animals analyzed and the PCR-positive samples by animal species and localities. Although only the results of three trypanosomes species (T. vivax, T. congolense “forest type” and T. simiae) are reported in this study, T. brucei s.l. and T. b. gambiense group 1 (human infective trypanosome) were also investigated and the results are reported in Njiokou et al. (2010). These authors showed that 19.88% and 3.08% of the animals were infected by T. brucei s.l. and T. b. gambiense respectively. T. b. gambiense were harboured by pigs, already known to be reservoir hosts, but also by goats and sheep, pointing out their contribution to the epidemiology of HAT. The prevalences significantly varied according to the animal species and the focus, in connection with the level of endemicity of HAT. In this study, 27.08% (237/875) of animals analyzed were infected by at least one of the three trypanosome species (Tables I and II). The levels of infection differed significantly between animal species (χ2 = 77.92; p < 0.0001) and between localities (χ2 = 52.89; p < 0.0001). T. vivax was the most predominant trypanosome in animals with a global infection rate reaching 20.91% and more precisely 36.15% in pigs, 18.18% in goats, 8.61% in sheep and 2.7% in dogs. T. congolense “forest type” was identified in 11.42% of the animals, and 19.86%, 10.22%, 8.1% and 3.37% for pigs, goats, dogs and sheep respectively. T. simiae was rare (0.34%) and was diagnosed only in two (0.75%) goats and one (0.37%) sheep. The frequency of T. vivax and T. congolense “forest type” differed significantly between animal species and between localities (p < 0.0001). The comparison of T. vivax infection rates in different pairs of animal species showed significant differences, except for the sheep/dog pair. Significant differences were also observed for the T. congolense “forest type” infection rates for the pig/goat, pig/sheep and goat/sheep pairs.
Fig. 2.
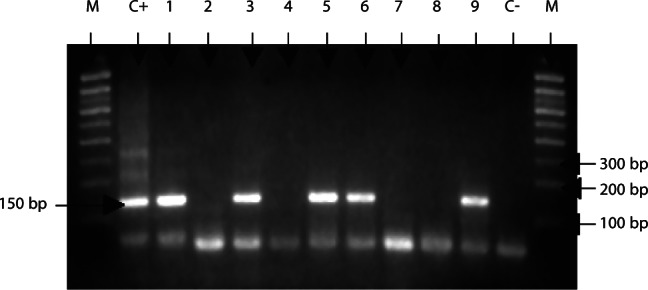
Electrophoretic profile showing the resolution of T. vivax specific DNA.
Table I.
Number and percentage of animals infected with T. congolense, T. vivax and T. simiae.
| Number of positive PCR (percentage) |
|||||
|---|---|---|---|---|---|
| Animal species | NE | TCF | TVW | TSM | NP |
| Pigs | 307 | 61 (19.86) | 111 (36.15) | 0 (0) | 133 (43.32) |
| Goats | 264 | 27 (10.22) | 48 (18.18) | 2 (0.75) | 69 (26.13) |
| Sheep | 267 | 9 (3.37) | 23 (8.61) | 1 (0.37) | 31 (11.61) |
| Dogs | 37 | 3 (8.1) | 1 (2.7) | 0 (0) | 4 (10.81) |
| Total | 875 | 100 (11.42) | 183 (20.91) | 3 (0.34) | 237 (27.08) |
| χ2 | 39,5 | 76,15 | 77,92 | ||
| p | 0.0001 | 0.0001 | 0.0001 | ||
The number of positive PCR are given with their percentage in the brackets; TCF: Trypanosoma congolense “forest type”; TVW: Trypanosoma vivax, TSM: Trypanosoma simiae; NE: number of animals examined; NP: number of animals parasite-positive (having DNA of at least one species), χ2: Chi-square test; p: p value.
Table II.
Number and percentage of animals infected with T. congolense, T. vivax and T. simiae in various localities.
| Number of positive PCR (percentage) |
|||||
|---|---|---|---|---|---|
| Locality | NE | TCF | TVW | TSM | NP |
| Bipindi | 204 | 30 (14.7) | 65 (31.86) | 0 (0) | 80 (39.21) |
| Campo | 310 | 32 (10.32) | 65 (20.96) | 2 (0.64) | 82 (26.45) |
| Fontem-center | 154 | 28 (18.18) | 44 (28.57) | 0 (0) | 55 (35.71) |
| Doumé | 207 | 10 (4.83) | 9 (4.34) | 1 (0.48) | 20 (9.66) |
| Total | 875 | 100 (11.42) | 183 (20.91) | 3 (0.34) | 237 (27.08) |
| χ2 | 18.3 | 54.6 | 52.89 | ||
| p | 0.0001 | 0.0001 | 0.0001 | ||
The number of positive PCR are given with their percentage in the brackets; TCF: Trypanosoma congolense “forest type”; TVW: Trypanosoma vivax, TSM: Trypanosoma simiae; NE: number of animals examined; NP: number of animals parasite-positive (having DNA of at least one species), χ2: Chi-square test; p: p value.
The comparison by locality showed that animals from Bipindi and Fontem-Center are more frequently infected with T. vivax and T. congolense “forest type”. Howerver, the level of infection of these trypanosome species does not differ significantly between these two localities (χ2 = 0.44; p = 0.5) and (χ2 = 0.78; p = 0.37) respectively. Animals from Doumé were significantly less infected by T. vivax and T. congolense “forest type” than those of the other localities. Significant differences were observed between the T. vivax infection rates in Bipindi and Campo (χ2 = 7.72; p = 0.005) as well as between the T. congolense “forest type” infection rates in Fontem-Center and Campo (χ2 = 5.64; p = 0.017). However, animals from Bipindi and Fontem-Center are more infected by T. vivax and T. congolense “forest type” respectively.
Mixed infections were found in 32 animals including 26 pigs and 6 goats carrying both T. congolense “forest type” and T. vivax; one sheep carried both T. vivax and T. simiae; and one goat carried triple infections of T. congolense “forest type”, T. vivax and T. simiae (Table III). When taking into account the occurrence of T. b. gambiense revealed in Njiokou et al., (2010), three other double infections with T. b. gambiense group 1 and T. vivax are identified in one sheep and one goat while T. b. gambiense group 1 and T. congolense “forest type” was identified in one sheep. Furthermore, one goat was infected with four trypanosomes including T. b. gambiense group 1, T. vivax, T. congolense “forest type” and T. simiae.
Table III.
Type, nature and number of infections revealed by PCR in different domestic animal species.
| Number of single and mixed infections |
||||||
|---|---|---|---|---|---|---|
| Type of infections | Trypanosome identified | Pigs | Goats | Sheep | Dogs | Total |
| Single | TCF | 70 | 31 | 11 | 1 | 113 |
| TVW | 19 | 17 | 6 | 2 | 44 | |
| TSM | 0 | 1 | 0 | 0 | 1 | |
| Double | TCF et TVW | 26 | 6 | 0 | 0 | 32 |
| TVW et TSM | 0 | 0 | 1 | 0 | 1 | |
| Triple | TCF, TVW et TSM | 0 | 1 | 0 | 0 | 1 |
TCF: Trypanosoma congolense “forest type”; TVW: Trypanosoma vivax, TSM: Trypanosoma simiae.
Discussion
In this study, PCR revealed infections with T. congolense “forest type”, T. vivax and T. simiae in the domestic animals, thus confirming the circulation of these parasites in the sleeping sickness foci of southern Cameroon as previously reported in tsetse flies (Morlais et al., 1998), wild animals (Herder et al., 2002; Njiokou et al., 2004; Njiokou et al., 2006) and pigs (Penchenier et al., 1996; Simo et al., 2006). The infection rate of 27.08% is in line with results obtained in pigs, small domestic ruminants, and dogs in the Central and West African regions (Asonganyi et al., 1986, 1990; Makumyaviri et al., 1989; Omeke, 1994; Penchenier et al., 1996; Dadah et al., 1997; Kalu et al., 2001), also out of any sleeping sickness areas.
The high prevalence of T. vivax corroborates other findings in domestic animals (Bourzat & Gouteux, 1990; Dadah et al., 1997; Kalu et al., 2001) as well as wild animals (Komoin-Oka et al., 1994; Truc et al., 1997; Herder et al., 2002; Njiokou et al., 2004; Njiokou et al., 2006) of the Central and West Africa regions. This high prevalence of T. vivax may result from the level of pathogenicity of this trypanosome, which is generally low and better controlled by animals (Stephen, 1970; Authié et al., 1999). It may result also from the mechanical transmission, which has not been reported in the other species studied here, except in certain extend T. congolense.
The identification of T. vivax in pigs is in accord with results obtained by Ng’ayo et al. (2005) in East Africa. Using specific primers, these authors showed that T. vivax infections are frequent in pigs and goats. Penchenier et al. (1996) and Simo et al. (2006) also found numerous T. vivax infections in pigs from Cameroon, by PCR. These results need to be confirmed by other techniques because it is generally accepted that pigs are refractory to infections with T. vivax (Taylor & Authié, 2004). The fact that T. vivax is detected only by PCR based methods and not by parasitological techniques could be explained by ‘‘transient’’ infections. Moreover, with the development of new genotyping tools that do not require isolation of trypanosomes, it is urgent to characterize T. vivax circulating in pigs as well as in other vertebrate hosts in order to investigate if some strains might be infective to pigs and others non-infective. Indeed, previous studies have suggested genetic variation in T. vivax populations that renders some isolates more pathogenic than others (Mwongela et al., 1981; Wellde et al., 1983; Roeder et al., 1984).
The lower prevalence of T. congolense “forest type” with respect to T. vivax in domestic animals may result from higher parasitemia in T. congolense infections, accompanied by serious anemia, which leads to the rapid death of the host animal (Sidibe et al., 2002; Bengaly et al., 2002a, b).
The very low prevalence of T. simiae as already reported in pigs (Penchenier et al., 1996; Simo et al., 2006) and wild animals (Herder et al., 2002; Njiokou et al., 2004) of several sleeping sickness foci indicates a low transmission of this parasite in various localities of Cameroon. The absence of T. simiae in pigs of various areas of Cameroon is likely due to its high pathogenicity because pigs infected with this trypanosome species would probably not survive. Our results corroborate those obtained by Simo et al. (2006) in pigs of the Fontem sleeping sickness focus of Cameroon.
Our results also showed a significant difference in the prevalence of T. vivax and T. congolense “forest type” between the different animal species. Pigs are more infected than goats, sheep and dogs; this could either be indicative of a higher susceptibility of pigs to T. vivax and T. congolense, or of a higher frequency of contact with the tsetse fly vector. Indeed, it has been shown that G. p. palpalis takes more blood meals from pigs than from goats and sheep and rarely on carnivores (Dagnogo et al., 1985; Sané et al., 2000; Späth, 2000; Simo et al., 2008). The keeping of pigs in sties near habitations exposes them to more contacts with peri-domestic tsetse flies (Frézil et al., 1980) than animals that roam freely. Finally, the highest prevalences of T. vivax and T. congolense “forest type” in domestic animals from Bipindi and Fontem-Centre respectively, confirm the results reported in wild animals from Bipindi (Herder et al., 2002; Njiokou et al., 2004) and in domestic animals from Fontem (Asonganyi et al., 1990) and indicate their intense transmission in these sleeping sickness foci.
The presence of T. vivax and T. congolense in all the localities studied is not only indicative of the ubiquity of the trypanosomes, but also of the presence of an appropriate vector in the foci. Their prevalence in Bipindi and Fontem where G. p. palpalis represents about 100% is higher than in Campo where G. p. palpalis represents only 56% and Doumé where G. fuscipes is the only tsetse fly species found (Mbida Mbida et al., 2009).
Our results on mixed infections in pigs corroborate those of Jamonneau et al., (2004) who reported a high proportion of mixed infection in pigs of the Bonon sleeping sickness focus in Côte d’Ivoire and those of Simo et al., (2006) in pigs of the Fontem sleeping sickness focus in Cameroon. These results are in line with those obtained in tsetse flies (Morlais et al., 1998; Masiga et al., 1996; Reifenberg et al., 1997; Malele et al., 2003). Indeed, entomological studies showed that infected tsetse flies from the sleeping sickness foci of Cameroon (Morlais et al., 1998) and other zones of Africa (Masiga et al., 1996; Reifenberg et al., 1997; Malele et al., 2003) frequently harbour more than one trypanosome species. This demonstrates the high probability of tsetse flies to transmit several trypanosome species to vertebrate hosts. Up till now, the interaction and the evolution of different trypanosome species or subspecies in the infected hosts remain not yet well understood. Experimental studies have shown that primary infection with T. congolense prevents the establishment of a second strain of T. congolense (Morrison et al., 1982) while an animal already infected with T. congolense becomes refractory to T. brucei s.l. infections (second infection), but not T. vivax (Dwinger et al., 1989). Results of our study, especially the low proportion of mixed infection including T. congolense “forest type” and T. b. gambiense (see also Njiokou et al., 2010 data) and the considerable numbers of mixed infections involving T. vivax and T. congolense “forest type” play in favor of these experimental studies. However, in the field conditions where tsetse flies are infected by several trypanosome species, it is obvious that several trypanosome species or subspecies are transmitted to the same hosts. In such conditions, the prevalence of each species of trypanosome in the mixed infections may reflect probably the prevalence of this trypanosome species in the locality. Our results on the mixed infections of several trypanosomes species in vertebrate hosts suggest further investigations on the establishment and the evolution of different trypanosomes species or subspecies in the vertebrate hosts. Such investigations may enable to understand some aspects of the Human African Trypanosomiasis as well as Animal African Trypanosomiasis. For example, these investigations may enable to know if some trypanosome species can prevent the establishment and the evolution of T. b. gambiense and if genetic exchanges between the same trypanosome species can occur in vertebrate hosts.
Acknowledgments
This study received IRD funds through JEA and UMR 177. The laboratory phase was hosted by OCEAC and University of Yaoundé I. The authors thank Justin Menounga and Ernest Mooh for technical assistance.
References
- Asonganyi T., Sede M.J. & Ngu J.L.Trypanosomiasis in Mbam division (Cameroon). Parasitological and immunodiagnostic examination of the domestic animal population. Annales Universitaires des Sciences de la Santé, 1986, 3 (3), 181–189 [Google Scholar]
- Asonganyi T., Suh S. & Tetuh M.D.Prevalence of domestic animal trypanosomiasis in the Fontem sleeping sickness focus, Cameroon. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 1990, 43 (1), 69–74 [PubMed] [Google Scholar]
- Authié E., Bringaud F., Bakalara N., Tetaud E. & Baltz T.Trypanosomoses humaines et animales : Maladie du sommeil et Nagana. Annales de l’Institut Pasteur, 1999, 10 (1), 27–50 [Google Scholar]
- Bengaly Z., Sidibé I., Boly H., Sawadogo L. & Desquesnes M.Comparative pathogenicity of three genetically distinct Trypanosoma congolense-types in inbred Balb/c mice. Veterinary Parasitology, 2002a, 105 (2), 111–118 [DOI] [PubMed] [Google Scholar]
- Bengaly Z., Sidibé I., Ganaba R., Desquesnes M., Boly H. & Sawadogo L.Comparative pathogenicity of three genetically distinct types of Trypanosoma congolense in cattle: clinical observation and haematological changes. Veterinary Parasitology, 2002b, 108, 1–19 [DOI] [PubMed] [Google Scholar]
- Bourzat D., Gouteux J.P.Données préliminaires sur le contact glossines-petits ruminants dans le massif forestier de Mayombé, Congo. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 1990, 43 (2), 199–206 [Google Scholar]
- Clausen P.H., Wiemann A., Patzlt R., Kakaire D., Poetzsch C., Peregrine A. & Mehlitz D.Use of PCR assay for the specific and sensitive detection of Trypanosoma spp in naturally infected dairy cattle in peri-urban Kampala, Uganda. Annales of New York Academic Science, 1998, 29, 21–31 [DOI] [PubMed] [Google Scholar]
- Connor R.J. & van Den Bossche P.African animal trypanosomosis, in: Infectious diseases of livestock. 2nd edition, Coetzer J.A.W. & Tustin R.C. (Eds), Oxford University Press, 2004, 1, 251–296 [Google Scholar]
- Cox A., Tilley A., Mc Odimba F., Fyfe J., Eisler M., Hide G. & Welbrum S.A PCR based assay for detection and differenciation of African trypanosome species in blood. Experimental Parasitology, 2005, 111, 24–29 [DOI] [PubMed] [Google Scholar]
- D’Amico F., Gouteux J.P., Le Gall F. & Cuisance D.Are stable flies (Diptera, Stomoxynae) vectors of Trypanosoma vivax in the Central African Republic? Veterinary Research, 1996, 27, 161–170 [PubMed] [Google Scholar]
- Dadah A.J., Duhlinska-Popova D.D., Daniel A.D. & Dede P.M.Trypanosomosis among sheep and goat at slaughter in Jos abattoir, Nigeria. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 1997, 50 (3), 214–216 [Google Scholar]
- Dagnogo M., Lohuiligno K. & Gouteux J.P.Comportement des populations de Glossina palpalis (Robineau-Desvoidy) et Glossina tachnoides Westwood du domaine guinéen de Côte d’Ivoire. Cahiers ORSTOM, Séries Entomologie Médicale et Parasitologie, 1985, 23, 3–8 [Google Scholar]
- Delespaux V., Ayral F., Geysen D. & Geerts S.PCR-RFLP using Ssu-rDNA amplification: applicability for the diagnosis of mixed infections with different trypanosome species in cattle. Veterinary Parasitology, 2003, 117 (3), 185–193 [DOI] [PubMed] [Google Scholar]
- Desquesnes M. & Dia M.L.Mechanical transmission of Trypanosoma congolense in cattle by the African tabanid Atylotus agrestis. Experimental Parasitology, 2003, 105, 226–231 [DOI] [PubMed] [Google Scholar]
- Desquesnes M. & Tresse L.Evaluation of sensitivity of PCR for detecting DNA of Trypanosoma vivax with several methods of blood sample preparations. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 1996, 49, 322–327 [PubMed] [Google Scholar]
- Dhollander S., Jallow A., Mbodge K., Kora S., Sanneh M., Gaye M., Bos J., Leak S., Berkvens D. & Geerts S.Equine trypanosomiasis in the Central River Division of Gambia: A study of veterinary gate-clinic consultation records. Preventive Veterinary Medecine, 2006, 75, 152–162 [DOI] [PubMed] [Google Scholar]
- Dwinger R.H., Luckins A.G., Murray M., Raie P. & Moloo S.K.Interference in the establishment of tsetse-transmitted Trypanosoma congolense, T. brucei or T. vivax superinfections in goats already infected with T. congolense or T. vivax. Veterinary Parasitology, 1989, 30 (3), 177–189 [DOI] [PubMed] [Google Scholar]
- Faye D., de Almeida P., Goossen B., Ossaer S., Ndao M., Berkevens D., Speybroeck N., Nieberding F. & Geerts S.Prevalence and incidence of trypanosomiasis in horses and donkeys in the Gambia. Veterinary Parasitology, 2001, 101 (2), 101–114 [DOI] [PubMed] [Google Scholar]
- Frézil J.L., Eouzan J.P., Alary J.C., Malonga J.R. & Ginoux P.Y.Epidémiologie de la trypanosomiase humaine en République Populaire du Congo. II. Le foyer du Niari. Cahiers ORSTOM, Séries Entomologie Médicale et Parasitologie, 1980, 18, 329–346 [Google Scholar]
- Gardiner P.R. & Wilson A.J.Trypanosoma (Duttonella) vivax. Parasitology Today, 1987, 3, 49–52 [DOI] [PubMed] [Google Scholar]
- Gardiner P.R.Recent studies on the biology of Trypanosoma vivax. Advance in Parasitology, 1989, 28, 229–317 [DOI] [PubMed] [Google Scholar]
- Geysen D., Delespaux V. & Geerts S.PCR-RFLP using SsurDNA amplification as an easy method for species-specific diagnosis of trypanosome species in cattle. Veterinary Parasitology, 2003, 110, 171–180 [DOI] [PubMed] [Google Scholar]
- Gonzales J.L., Loza A. & Chocon E.Sensitivity of different Trypanosoma vivax primers for the diagnosis of livestock trypanosomiasis using different DNA extraction methods. Veterinary Parasitology, 2005, 136, 119–126 [DOI] [PubMed] [Google Scholar]
- Grébaut P., Wang S., Bodo J.M., Ebo’o Eyenga V., Binzouli J.J., Ndong Ngoe C., Nomo E., Njiokou F., Ollivier G., Foumane V. & Bureau P.Aspects épidémiologiques d’un foyer de la maladie du sommeil mal connu: le foyer de Bipindi au Cameroun. Bulletin de Liaison et de Documentation de l’OCEAC, 2001, 33, 16–20 [Google Scholar]
- Herder S., Simo G., Nkinin S.W. & Njiokou F.Identification of trypanosomes in wild animals from Southern Cameroon using the polymerase chain reaction (PCR). Parasite, 2002, 9, 345–349 [DOI] [PubMed] [Google Scholar]
- Hudson J.R.Acute and subacute trypanosomiasis in cattle caused by Trypanosoma vivax. Journal of Pathology, 1944, 54, 108–119 [Google Scholar]
- Jamonneau V., Ravel S., Koffi M., Kaba D., Zeze D.G., Ndri L., Sane B., Coulibary B., Cuny G. & Solano P.Mixed infections of trypanosomes in tsetse and pigs and their epidemiological significance in sleeping sickness focus in Côte d’Ivoire. Parasitology, 2004, 129, 693–702 [DOI] [PubMed] [Google Scholar]
- Kalu A.U., Oboegbulem S.I. & Uzoukwu M.Trypanosomose chez les petits ruminants maintenue par une faible population de glossines ripicoles dans la région centrale du Nigeria. Small Ruminant, 2001, 40, 109–115 [DOI] [PubMed] [Google Scholar]
- Komoin-Oka C., Truc P., Bengaly Z., Formenty P., Duvallet G., Lauginie F., Raath P.P., N’Depo A.E. & Leforban Y.Étude de la prévalence des infections à trypanosomes chez différentes espèces d’animaux sauvages du parc national de la Comoé en Côte d’Ivoire : résultats préliminaires sur la comparaison de trois méthodes de diagnostic. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 1994, 47, 189–194 [PubMed] [Google Scholar]
- Majiwa P.A., Matthyssens G., Williams R.O. & Hamers R.Cloning and analysis of Trypanosoma (Nannomonas) congolense IL Nat 2.1 VSG gene. Molecular Biochemestry and Parasitology, 1985, 16 (1), 97–108 [DOI] [PubMed] [Google Scholar]
- Majiwa P.A.O., Maina M., Waitumbi J.N., Mihok S. & Zweygarth E.Trypanosoma (Nannomonas) congolense: molecular characterization of a new genotype from Tsavo Kenya.. Parasitology, 1993, 106, 151–162 [DOI] [PubMed] [Google Scholar]
- Makumyaviri A., Mehlitz D., Kageruka P., Kazyumba G.L. & Molisho D.Le réservoir animal de Trypanosoma brucei gambiense au Zaïre : infections trypanosomiennes dans deux foyers du Bas-Zaïre. Tropical Medecine and Parasitology, 1989, 40, 259–262 [PubMed] [Google Scholar]
- Malele I., Craske L., Knight C., Ferris V., Njiru Z., Hamilton P., Lehane S., Lehane M. & Gibson W.The use of specific and generic primers to identify trypanosome infections of wild tsetse flies in Tanzania by PCR. Infection, Genetics and Evolution, 2003, 3, 271–279 [DOI] [PubMed] [Google Scholar]
- Mamadou A., Zoli A., Mbahin N., Tanenbe C., Bourdanne Clausen P.-H., Marcotty T., Van Den Bossche P. & Geerts S.Prevalence and incidence of bovine trypanosomiasis on the Adamaoua plateau in Cameroon 10 years after the tsetse eradication campaign. Veterinary Parasitology, 2006, 142, 16–22 [DOI] [PubMed] [Google Scholar]
- Masiga D.K., McNamara J.J., Laveissière C., Truc P. & Gibson W.C.A high prevalence of mixed infections in tsetse flies in Sinfra, Côte d’Ivoire, detected by DNA amplification. Parasitology, 1996, 112, 75–80 [DOI] [PubMed] [Google Scholar]
- Masiga D.K., Smyth A.J., Hayes P., Bromidge T.J. & Gibson W.C.Sensitive detection of trypanosomes in tsetse flies by DNA amplification. International Journal of Parasitology, 1992, 22, 909–918 [DOI] [PubMed] [Google Scholar]
- Masumu J., Marcotty T., Geysen D., Geerts S., Vercruysse J., Dorny P. & Van Den Bossche P.Comparison of the virulence of Trypanosoma congolense strains isolated from cattle in trypanosomiasis endemic area of eastern Zambia. International Journal for Parasitology, 2006, 36, 497–501 [DOI] [PubMed] [Google Scholar]
- Mattioli R.C., Zinsstag J. & Pfister K.Frequency of trypanosomiasis and gastrointestinal parasites in draught donkeys in Gambia in relation to animal husbandry. Tropical Animal Health Production, 1994, 26, 102–108 [DOI] [PubMed] [Google Scholar]
- Mbida Mbida J.A., Mimpfoundi R., Njiokou F., Manga L. & Laveissière C.Distribution et écologie des vecteurs de la trypanosomiase humaine africaine de type savanicole en zone de forêt dégradée au sud du Cameroun. Bulletin de la Société de Pathologie Exotique, 2009, 102, 101–105 [PubMed] [Google Scholar]
- Mihok S., Maramba O., Munyoki E. & Kagoiya J.Mechanical transmission of Trypanosoma spp. by African Stomoxynae (Diptera: Muscidae). Tropical Medecine and Parasitology, 1995, 46 (2), 103–105 [PubMed] [Google Scholar]
- Morlais I., Grébaut P., Bodo J.M., Djoha S., Cuny G. & Herder S.Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Tropica, 1998, 70, 109–117 [DOI] [PubMed] [Google Scholar]
- Morrison W.I., Wells P.W., Moloo S.K., Paris J. & Murray M.Interference in the establishment of super-infections with Trypanosoma congolense in cattle. Journal of Parasitology, 1982, 68 (5), 755–764 [PubMed] [Google Scholar]
- Mugutu K.N., Silayo R.S., Majiwa P.A., Kimbita E.K., Mutayoba B.M. & Maselle R.Application of PCR and DNA probes in the characterization of trypanosomes in the blood of cattle in farms in Morogoro Tanzania. Veterinary Parasitology, 2001, 94, 177–189 [DOI] [PubMed] [Google Scholar]
- Mwongela G.N., Kovatch R.M. & Frézil R.M.Acute Trypanosoma vivax in dairy cattle in coast Province Kenya. Tropical animal Health Production, 1981, 13, 63–69 [DOI] [PubMed] [Google Scholar]
- Ng’Ayo G.M., Njiru Z.K., Kenya E.U., Muluvi G.M., Osia E.O. & Masiga D.K.Detection of trypanosomes in small ruminants and pigs in western Kenya: important reservoirs in the epidemiology of sleeping sickness? Kinetoplastid Biology and Disease, 2005, 14, 4–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njiokou F., Laveissière C., Simo G., Grébaut P., Cuny G. & Herder S.Wild fauna as probable animal reservoir for Trypanosoma brucei gambiense in Cameroon. Infection, Genetics and Evolution, 2006, 6, 147–153 [DOI] [PubMed] [Google Scholar]
- Njiokou F., Nimpaye H., Simo G., Njitchouang G.R., Asonganyi T., Cuny G. & Herder S.Domestic animals as potential reservoir hosts of Trypanosoma brucei gambiense in sleeping sickness foci in Cameroon. Parasite, 2010, 17, 61–66 [DOI] [PubMed] [Google Scholar]
- Njiokou F., Simo G., Nkinin S.W., Laveissière C. & Herder S.Infection rate of Trypanosoma brucei s.1, T. vivax, T. congolense “forest type” and T. simiae in small wild vertebrate in south Cameroon. Acta Tropica, 2004, 92, 139–146 [DOI] [PubMed] [Google Scholar]
- Nkinin S.W., Njiokou F., Penchenier L., Grébaut P., Simo G. & Herder S.Characterization of Trypanosoma brucei s.l subspecies by izoenzymes in domestic pigs from the Fontem sleeping sickness focus of Cameroon. Acta Tropica, 2002, 81, 225–232 [DOI] [PubMed] [Google Scholar]
- Olubayo R.O. & Mugera G.M.Pathogenesis of haemorrhagies in Trypanosoma vivax infection in cattle. Disseminated intravascular coagulation. Bulletin of Animal Health Production of Africa, 1985, 33, 211–217 [Google Scholar]
- Omeke B.C.O.Pig trypanosomiasis prevalence and significance in the endemic Middle Belt zone of South of Nigeria. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 1994, 47 (4), 381–386 [PubMed] [Google Scholar]
- Penchenier L., Bodo J.M., Bureau P.H., Morlais I., Grébaut P., Djoha S. & Herder S.Utilisation de la PCR sur sang pour le diagnostic des trypanosomoses porcines. Bulletin de Liaison et de Documentation de l’OCEAC, 1996, 29, 50–53 [Google Scholar]
- Penchenier L., Simo G., Grébaut P., Nkinin S.W., Laveissière C. & Herder S.Diagnosis of human trypanosomiasis due to Trypanosoma brucei gambiense in Central Africa, by the polymerase chain reaction. Transaction of the Royal Society of Tropical Medecine and Hygiene, 2000, 94, 392–394 [DOI] [PubMed] [Google Scholar]
- Picozzi K., Telley A., Fèvre E.M., Coleman P.G., Magona J.W., Odiit M., Eisler M.C. & Welbrum S.C.The diagnosis of trypanosome infections: applications of novel technology for reducing disease risk. African Journal of Biotechenology, 2002, 1, 39–45 [Google Scholar]
- Reifenberg J.M., Solano P., Bauer B., Kaboré I., Cuny G., Duvallet G. & Cuisance D.Apport de la technique PCR pour une meilleure compréhension de l’épizootiologie des trypanosomoses bovines : exemple de la zone d’aménagement pastorale de Yalé au Burkina Faso. Revue d’Élevage et de Médecine Vétérinaire des Pays Tropicaux, 1997, 50 (1), 14–22 [Google Scholar]
- Roeder P.L., Scott J.M. & McIntyure W.I.M.Acute Trypanosoma vivax infection of Ethiopian cattle in the apparent absence of tsetse. Tropical Animal Health Production, 1984, 16, 141–147 [DOI] [PubMed] [Google Scholar]
- Sané B., Laveissière C. & Meda H.A.Diversité du régime alimentaire de Glossina palpalis palpalis en zone forestière de Côte d’Ivoire : relation avec la prévalence de la trypanosomiase humaine africaine. Tropical Medecine International Health, 2000, 5 (1), 73–78 [DOI] [PubMed] [Google Scholar]
- Sidibe I., Bengaly Z., Boly H., Ganaba R., Desquesnes M. & Sawadogo L.Differential pathogenicity of Trypanosoma congolense subgroup: implication for the strategic control of trypanosomiasis. Newsletter on Integrated Control of Pathogenic Trypanosomes and their Vectors (I.C.P.T.V), 2002, (6), 33–35 [Google Scholar]
- Simo G., Asonganyi T., Nkinin S.W., Njiokou F. & Herder S.High prevalence of Trypanosoma brucei gambiense group 1 in pigs from the Fontem sleeping sikness focus in Cameroon. Veterinary Parasitology, 2006, 139, 57–66 [DOI] [PubMed] [Google Scholar]
- Simo G., Njiokou F., Mbida Mbida J.A., Njitchouang G.R., Herder S., Asonganyi T. & Cuny G.Tsetse fly host preference from sleeping sickness foci in Cameroon. Epidemiological implications. Infection, Genetics and Evolution, 2008, 8, 34–39 [DOI] [PubMed] [Google Scholar]
- Solano P., Michel J.F., Lefrançois T., De La Rocque S., Sidibe I., Zoungrana A. & Cuisance D.Polymerase chain reaction as a tool detecting trypanosomes in naturally infected cattle in Burkina Faso. Veterinary Parasitology, 1999, 86, 95–103 [DOI] [PubMed] [Google Scholar]
- Späth J.Feeding patterns of tree sympatric tsetse species (Glossina spp) (Diptera: Glossinidae) in the preforest zone of Côte d’Ivoire. Acta Tropica, 2000, 75, 109–118 [DOI] [PubMed] [Google Scholar]
- Stephen L.E.Pig trypanosomiasis in Africa. Commonwelth Bureau for Animal Health, Farnham House, United Kingdom, 1966, 29–36, (Reviews Series No 8). [Google Scholar]
- Stephen L.E.Clinical manifestations of the trypanosomiasis in livestock and other domestic animals, in: The African Trypanosomiases. Mulligan H.W. (ed.), London Allen and Unwin, 1970, 774–794 [Google Scholar]
- Sumba L.A., Mihok S. & Oyieke F.A.Mechanical transmission of Trypanosoma evansi and T. congolense by Stomoxys niger and S. taeniatus in a laboratory mouse model. Medecine Veterinary and Entomology, 1998, 12, 417–422 [DOI] [PubMed] [Google Scholar]
- Taylor K.A. & Authié E.Pathogenesis of animal trypanosomiasis, The trypanosomiasis. Maudlinet al. (eds), CABI Publishing, 2004, 331–353 [Google Scholar]
- Trail J.M.C., D’Ietern G.D.M., Feron A., Kankiese O., Mulungo M., & Pelo M.Effect of trypanosome infection, control of parasitaemia and control of anaemia development on productivity of N’dama cattle. Acta Tropica, 1991, 48, 37–45 [DOI] [PubMed] [Google Scholar]
- Truc P., Formenty P., Diallo P.B., Komoin-Oka C. & Lauginie F.Identification of trypanosomes isolated by KIVI from wild animals in Côte d’Ivoire: diagnostic, taxonomic and epidemiological consideration. Acta Tropica, 1997, 67, 1–10 [DOI] [PubMed] [Google Scholar]
- Vaucel M.A., Waddy B.B., de Andrade da Silva M.A. & Pons V.E.Répartition de la trypanosomiase africaine chez l’homme et les animaux. Bulletin de l’Organisation Mondiale de la Santé, 1963, 28, 545–594 [PMC free article] [PubMed] [Google Scholar]
- Wellde B.T., Chumo D.A., Adoyo M., Kovatch R.M., Mwongela G.N. & Opiyo E.A.Hemorrhagic syndrome in cattle associated with Trypanosoma vivax infection. Tropical Animal Health Production, 1983, 15 (2), 95–102 [DOI] [PubMed] [Google Scholar]


