Abstract
A total of 134 Egyptian liver flukes were collected from different definitive hosts (cattle, sheep, and buffaloes) to identify them via the use of PCR-RFLP and sequence analysis of the first nuclear ribosomal internal transcribed spacer (ITS1). Specimens of F. hepatica from France, as well as F. gigantica from Cameroon were included in the study for comparison. PCR products of ITS1 were subjected for digestion by RsaI restriction enzyme and visualized on agarose gel. According to RFLP pattern, Egyptian flukes were allocated into two categories. The first was identical to that of French hepatica flukes to have a pattern of 360, 100, and 60 (bp) band size, whereas the second resembled to that of Cameroonian gigantica worms to have a profile of 360, 170, and 60 bp in size. Results of RFLP analysis were confirmed by sequence analysis of representative ITS1 amplicons. No hybrid forms were detected in the present study. Taken together, this study concluded that both species of Fasciola are present in Egypt, whereas the hybrid form may be not very common.
Keywords: Fasciola gigantica, Fasciola hepatica, Egypt, PCR-RFLP
Abstract
Des douves égyptiennes (134 spécimens) provenant de différents hôtes définitifs (bovins, buffles et moutons) ont été analysées par PCR-RFLP et l’étude de la séquence ITS-1 pour identifier les espèces locales de Fasciola (F. hepatica, F. gigantica, ou les formes hybrides entre ces deux espèces). Des douves provenant de France (F. hepatica) et du Cameroun (F. gigantica) ont été utilisées comme références. Deux types de bandes ont été trouvés dans les fragments de la séquence ITS-1 : le premier type est identique à celui des F. hepatica de France (trois bandes de 60, 100 et 360 bp) tandis que le second ressemble à celui des F. gigantica du Cameroun (trois bandes de 60, 170 et 360 bp). De plus, le séquençage des amplicons ITS-1 confirme cette différence en montrant la présence de six sites nucléotidiques variables, ce qui permet de discriminer F. hepatica de F. gigantica. Aucune forme intermédiaire entre les deux Fasciola n’a été trouvée dans les spécimens analysés. Pris ensemble, cette étude permet de conclure que les deux espèces de Fasciola sont présentes en Égypte, alors que la forme hybride pourrait être pas très commune.
Keywords: Fasciola gigantica, Fasciola hepatica, Égypte, PCR-RFLP
The species of genus Fasciola (Platyhelmintha: Digenea) are the common liver flukes of a wide range of animals (Mas-Coma et al., 2005). Severe negative economic impact due to fascioliasis has been estimated in ruminants (Urquhart et al., 1996; Spithill & Dalton, 1998). In addition, human fascioliasis represents a significant health problem, as the estimated number of infected people is between 2.4 and 17 million and the number of those at the risk is more than 180 million throughout the world (World Health Organization, 1995; Mas-Coma et al., 1999; Ishii et al., 2002; Mas-Coma et al., 2009). Although, liver flukes have a global geographical distribution, the disease probably exerts most of its impact on developing countries. In Africa, Fasciola infection has been recognized as a major constraint for public health and animal farm industry (Mekroud et al., 2006; Pfukenyi et al., 2006; Phiri et al., 2007).
In Egypt, fascioliasis has serious veterinary and medical impacts, as it provokes the mortality of livestock and affects people at all ages (Haseeb et al., 2002; Curtale et al., 2003; Esteban et al., 2003). Coexistence of Egyptian F. hepatica and F. gigantica, the causative agents of fascioliasis, was determined by Lofty et al. (2002) using morphometric and isoelectric focusing techniques. In addition, Marcilla et al. (2002) used a PCR-RFLP assay, based on 28S ribosomal DNA, to distinguish between both fasciolids collected from different countries including Egypt. Notably, the occurrence of F. hepatica/F. gigantica intermediate forms was confirmed in Egypt morphologically (Periago et al., 2008) and molecularly based on ribosomal and mitochondrial gene markers (Amer et al., 2011). Latter findings suggested a possible hybridization between these two species of Fasciola, as demonstrated in Japanese, Korean, Chinese and Vietnamese flukes (Itagaki & Tsutsumi, 1998; Agatsuma et al., 2000; Lin et al., 2007; Itagaki et al., 2009). Such hybridization of both fluke species may result in messing up chromosomal numbers with the appearance of diploid and triploid forms with spermic and aspermic (with no or a few spermatozoa in the seminal vesicle) features.
The present study aimed to spot light on speciation of Fasciola population in Egypt. Fasciola spp. collected from different hosts and localities in Egypt were identified by PCR-RFLP based on ITS1 fragment using RsaI restriction endonuclease. Results of RFLP analysis were confirmed by sequence analysis of representative ITS1 amplicons.
Materials and Methods
A total of 134 adult Fasciola spp. (Table I) were collected from livers of naturally infected buffaloes, cattle and sheep at Al Basateen (Cairo, Egypt), and Tanta (about 90 Km north of Cairo, Egypt) abattoirs during 2008. F. hepatica specimens, collected from cattle at the slaughterhouse of Limoges (France), and F. gigantica, obtained from cattle at Yaoundé (Cameroon), were included for comparison. Fresh worms were washed extensively in physiological saline and fixed in 70° ethanol.
Table I.
Total number of analyzed liver flukes from different definitive hosts.
| Distribution of collected flukes |
|||||
|---|---|---|---|---|---|
| Species | Origin | Cattle | Sheep | Buffaloes | Total number |
| Fasciola hepatica | Cairo | 31 | 15 | – | 46 |
| Tanta | 32 | 15 | – | 47 | |
| Limoges | 3 | – | – | 3 | |
| Fasciola gigantica | Cairo | 14 | 15 | 12 | 41 |
| Yaoundé | 10 | – | – | 10 | |
| Total number | 90 | 45 | 12 | 147 | |
DNA Extraction and PCR-RFLP Analysis
Genomic DNA was extracted from the adult flukes using QIAamp DNA Mini Kits (Qiagen, USA) following manufacturer’s recommendations. The ITS1 fragment was amplified by PCR (Itagaki et al., 2005), using a set of 5’-TTGCGCTGATTACGTCCCTG-3’ and 5’-TTGGCTGCGCTCTTCATCGAC- 3’ as forward and reverse primers, respectively. The reaction was done in a total volume of 25 μl containing of 12.5 μl Qiagen Multiplex, 1 μl of each primer (0.3 μM), 1 μl genomic DNA, and 9.5 μl H2O. Reaction cycles consisted of an initial denaturation step at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 90 sec, annealing at 53 °C for 30 sec and extension at 72 °C for 60 sec, followed by a final extension step at 72 °C for 10 min. RFLP reaction was performed using RsaI restriction enzyme as described by Ichikawa and Itagaki (2010). The digested fragments were electrophoresed on 2% agarose gel and visualized by ethidium bromide using GELSMART 7.0 software (Clara Vision). The size of each band was determined by a 100-bp plus ladder molecular weight marker.
ITS1 Sequencing
For confirmation of RFLP reaction, ITS1 amplicons of representative samples of Egyptian as well as French and Cameroonian flukes were subjected to sequence analysis. PCR products of ITS1 were purified using rapid PCR purification systems (Marligen Bioscience, Inc., USA) according to manufacturer’s instructions. The sequencing reaction was performed utilizing the same PCR primers using ABI Big Dye kit on 3130xl genetic analyzer (Applied Biosystems, France). Accuracy of data was confirmed by two-directional sequencing. Representative sequences were deposited in the GenBank under the accession numbers of JF294998, JF294999, JF295000, and JF295001.
Results
PCR amplification of ITS1 resulted in a fragment size of 680 bp (Fig. 1). The products were subsequently subjected to digestion by RsaI restriction enzyme. Electrophoresis of the digested products revealed to two different bands patterns (Fig. 2), regardless of their geographical origins. The first pattern composed of three bands of 360, 100, and 60 bp in size, whereas the second was 360, 170, and 60 bp in size.
Fig. 1.

Agarose gel electrophoresis of amplified ITS1 ribosomal region. Lanes 1-10 denote to different fluke samples amplified as a single band of 680 bp; lane 11, negative control; lane M, 100 bp ladder molecular weight marker.
Fig. 2.
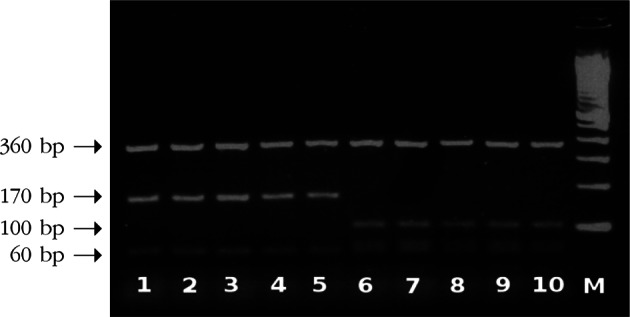
RsaI restriction enzyme digestion products of ITS1 fragment. Lanes 1-5 denote to those of Fasciola gigantica isolated from cattle at Yaoundé, Cameron (1) and Cairo, Egypt (2, 3); as well as from sheep at Cairo, Egypt (4, 5), respectively. Lanes 6-10 denote to those of Fasciola hepatica isolated from sheep and cattle (6, 7) at Cairo, Egypt, and sheep and cattle (8, 9) at Tanta, Egypt, respectively, as well as from cattle at Limoges, France (10).
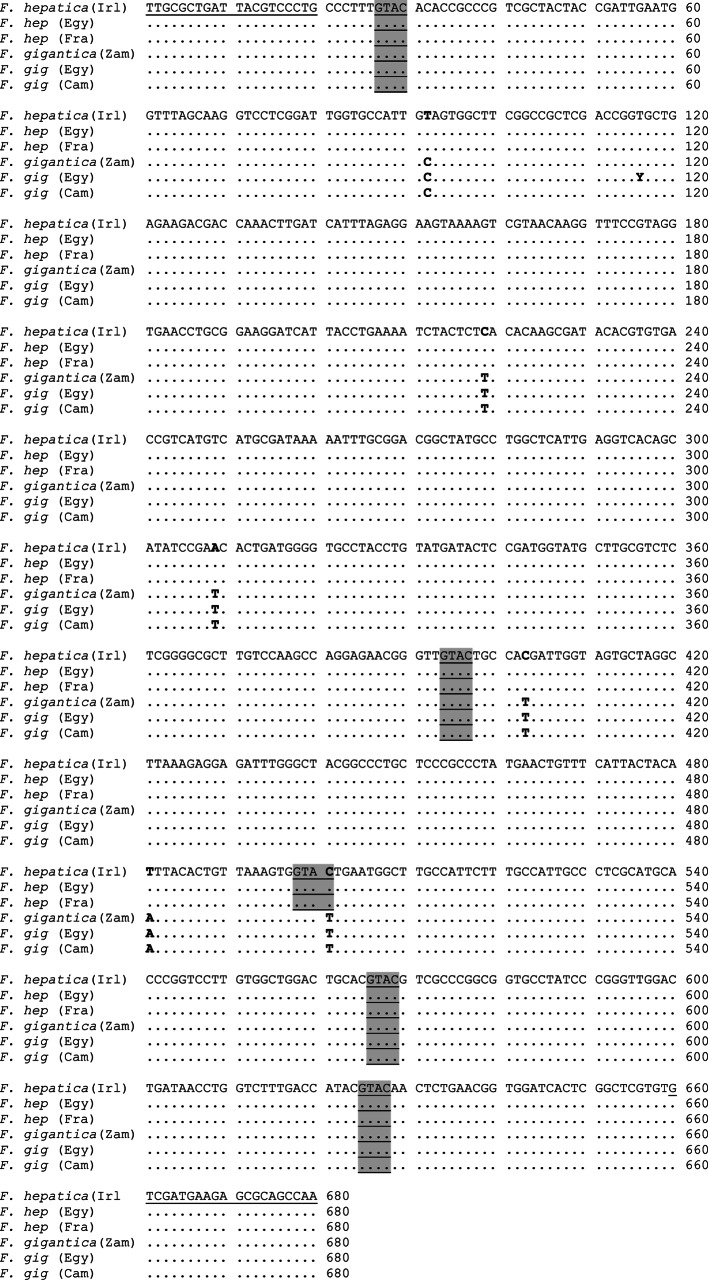
Alignment of obtained sequences revealed that the members of the first pattern including French flukes belonged to F. hepatica type, whereas those of the second pattern including Cameroonian flukes was belonging to F. gigantica type (Fig. 3). Six variable nucleotide positions of 92, 219, 309, 403, 481, and 501 were discriminating between nucleotide sequence of both patterns of Fasciola. In addition, five restriction sites RsaI enzyme (GTAC) were recognized at positions 27, 394, 498, 566 and 625 in case of F. hepatica sequences, whereas four restriction sites were detected at positions 27, 394, 566 and 625 in case of F. gigantica sequences. Interestingly, a single nucleotide difference between ITS1 sequences of Cameroonian and Egyptian F. gigantica was detected at position 116. At this site, the nucleotide was (T) in the former isolate, whereas it showed a combination of both C and T in the latter one. Collectively, Table I illustrates the identified liver flukes in relation to their origins and their definitive hosts. Out of 134 analyzed specimens from Egypt, 93 (69.4%) flukes were identified as F. hepatica and 41 (30.6%) as F. gigantica. The latter species was only detected in animals slaughtered at Cairo abattoir, while F. hepatica was found in cattle and sheep sacrificed in both abattoirs of Cairo and Tanta.
Fig. 3.
Nucleotide sequence alignment of ITS1 fragments of Fasciola species from Egypt (Egy) and four other countries: Cameroon (Cam), France (Fra), Ireland (Irl), and Zambia (Zam). The restriction sites of RsaI endonuclease are underlined and shaded in gray. The nucleotide differences between the two Fasciola species are denoted in bold. Dots indicate that the nucleotides are identical to those in the upper line. ‘Y’ represents a nucleotide indicating the presence of a double peak, C and T. (Irl), GenBank accession number: AB514850 for Fasciola hepatica from Ireland. (Zam), GenBank accession number: AB514855 for Fasciola gigantica from Zambia. Underline denote to primer position.
Discussion
PCR-RFLP based on ITS1 is a reliable tool to differentiate F. hepatica from F. gigantica (Ichikawa & Itagaki, 2010; Rokni et al., 2010). In the present study, results showed the presence of two different ITS1-RFLP patterns corresponding to F. hepatica (360, 100, and 60 bp band size) and F. gigantica (360, 170, and 60 bp band size) as confirmed by PCR-sequence analysis. These results come in full agreement with those described by Ichikawa & Itagaki (2010) for Fasciola from several geographical regions. Although nucleotide composition reveals five (hepatica) and four (gigantica) restriction sites in the affiliated sequences, only three bands in each case were recognized. The band of 60 bp was thought to include the three fragments of 68 bp (only F. hepatica), 59 and 54 bp, and the small fragment of 28 bp was not detected on agarose gels (Ichikawa & Itagaki, 2010). Although the intermediate form of Fasciola sp. has been reported in Egypt (Amer et al., 2011), the present study did not detect the discriminating pattern (360, 170, 100, and 60 bp band size as described by Ichikawa and Itagaki, 2010) by RFLP reaction nor by sequence analysis. This may indicate that the hybrid form of Fasciola is not very common in Egypt and could be detected on sporadic bases. Of note, several PCR-RFLP techniques were developed to differentiate between F. hepatica and F. gigantica as described by El-Gozamy and Shoukry (2009) using 18S rRNA, Marcilla et al. (2002) using 28S rRNA and Rokni et al. (2010) using ITS1 utilizing different array of endonucleases. Although nucleotide sequence analysis was consistent with the results of RFLP reaction, Egyptian F. gigantica proved heterogeneity at position 116 compared to Cameroonian gigantica flukes. Such heterogeneity was reported in Egyptian flukes (Amer et al., 2011). Moreover, nucleotide variability was reported in Fasciola flukes (Vara-Del Río et al., 2007, Walker et al., 2011) based on sequence of different genetic markers.
The percentage of F. hepatica (69.4%) noted among the slaughtered animals in the present study gave an idea about the adaptation of this fluke to the local environment in Egypt. Occurrence of this digenean, which may have be introduced into this country from Europe through imported infected animals (Lofty et al., 2002; Mas-Coma et al., 2005), was supported by a recent report suggesting the role of Radix natalensis (the principal snail host of F. gigantica) as a potential intermediate host for F. hepatica (Dar et al., 2010). The other fasciolid, F. gigantica present in 30.7% in the analyzed samples, was considered as an indigenous species of Fasciola found in Egypt (Farag, 1998), as it was recorded in nearly all governorates, especially those of the Nile Delta in Lower Egypt (Lotfy & Hillyer, 2003; Dar et al., 2005).
Acknowledgments
The authors are grateful to Dr D. Rondelaud, University of Limoges, Faculty of Medicine, France, for precise comments on the article and to F. Djuikwo Teukeng, University of Limoges, Faculty of Pharmacy, for bringing samples of Fasciola gigantica from Cameroon.
References
- Agatsuma T., Arakawa Y., Iwagami M., Honzako Y., Cahyaningish U. & Kang S.Y.Molecular evidence of natural hybridization between Fasciola hepatica and Fasciola gigantica. Parasitology International, 2000, 49, 231–238 [DOI] [PubMed] [Google Scholar]
- Amer S., Dar Y., Ichikawa M., Fukuda Y., Tada C., Itagaki T. & Nakai Y.Identification of Fasciola species isolated from Egypt based on sequence analysis of genomic (ITS1 and ITS2) and mitochondrial (NDI and COI) gene markers. Parasitology International, 2011, 60, 5–12 [DOI] [PubMed] [Google Scholar]
- Curtale F., Abd-El Wahab Hassanein Y., El Wakeel A., Mascoma S. & Montresore A.Distribution of human fascioliasis by age and gender among rural population in the Nile Delta, Egypt Journal of Tropical Pediatry 2003, 49, 264–268 [DOI] [PubMed] [Google Scholar]
- Dar Y., Djuikwo Teukeng F.F., Vignoles P., Dreyfuss G.& Rondelaud D.Radix natalensis, a potential intermediate host of Fasciola hepatica in Egypt. Parasite, 2010, 17, 251–256 [DOI] [PubMed] [Google Scholar]
- Dar Y., Rondelaud D. & Dreyfuss G.Update of fasciolosis- transmitting snails in Egypt (review and comment). Journal of Egyptian Society of Parasitology, 2005, 35, 477–490 [PubMed] [Google Scholar]
- El-Gozamy B.R. & Shoukry N.M.Identification of Egyptian Fasciola species by PCR and restriction endonuclease digestion of the nuclear small subunit ribosomal RNA gene. Journal of Egyptian Society of Parasitology, 2009, 39, 429–439 [PubMed] [Google Scholar]
- Esteban J.G., Gonzalez C., Curtale F., Munoz-Antoli C., Valero M.A., Bargues M.D., El-Sayed M., El-Wakeel A.A., Abdel-Wahab Y., Montresor A., Engles D., Savioli L. & Mas-Coma S.Hyper-endemic fascioliasis associated with schistosomiasis in villages in the Nile Delta of Egypt. American Journal of Tropical Medicine and Hygiene, 2003, 69, 429–437 [PubMed] [Google Scholar]
- Farag H.F.Human fascioliasis in some countries of eastern Mediterranean region. Eastern Mediterranean Health Journal, 1998, 4, 156–160 [Google Scholar]
- Haseeb A.N., El-Shazly A.M., Arafa M.A. & Morsy A.T.A review on fascioliasis in Egypt. Journal of Egyptian Society of Parasitology, 2002, 32, 317–354 [PubMed] [Google Scholar]
- Ichikawa M. & Itagaki T.Discrimination of the ITS1 types of Fasciola spp. based on a PCR-RFLP method. Parasitology Research, 2010, 106, 757–761 [DOI] [PubMed] [Google Scholar]
- Ishii Y., Nakamura-Uchiyama F. & Nawa Y.A praziquantel ineffective fascioliasis case successfully treated with triclabendazole. Parasitology International, 2002, 51, 205–209 [DOI] [PubMed] [Google Scholar]
- Itagaki T. & Tsutsumi K.Triploid form of Fasciola in Japan: genetic relationships between Fasciola hepatica and Fasciola gigantica determined by ITS-2 sequence of nuclear rDNA. International Journal of Parasitology, 1998, 28, 777–781 [DOI] [PubMed] [Google Scholar]
- Itagaki T., Kikawa M., Terasaki K., Shibahara T. & Fukuda K.Molecular characterization of parthenogenic Fasciola sp. in Korea on the basis of DNA sequences of ribosomal ITS1 and mitochondrial NDI gene. Journal of Veterinary Medical Science, 2005, 67, 1115–1118 [DOI] [PubMed] [Google Scholar]
- Itagaki T., Sakaguchi K., Terasaki K., Sasaki O., Yoshihara S. & Dung T.V.Occurrence of spermic diploid and aspermic triploid forms of Fasciola in Vietnam and their molecular characterization based on nuclear and mitochondrial DNA. Parasitology International, 2009, 58, 81–85 [DOI] [PubMed] [Google Scholar]
- Lin R.Q., Dong S.J., Nie K., Wang C.R., Song H.Q., Li A.X., Huang W.Y. & Zhu X.Q.Sequence analysis of the first internal transcribed spacer of rDNA supports the existence of the intermediate Fasciola between F. hepatica and F. gigantica in mainland China. Parasitology Research, 2007, 101, 813–817 [DOI] [PubMed] [Google Scholar]
- Lotfy W.M. & Hillyer G.V.Fasciola species in Egypt. Experimental Pathology and Parasitology, 2003, 6, 9–22 [Google Scholar]
- Lotfy W.M., El-Morshedy H.N., Abou El-Hoda M., El-Tawila M.M., Omar E.A. & Farag H.F.Identification of the Egyptian species of Fasciola. Veterinary Parasitology, 2002, 103, 323–332 [DOI] [PubMed] [Google Scholar]
- Marcilla A., Bargues M.D. & Mas-Coma S.A PCR-RFLP assay for the distinction between Fasciola hepatica and Fasciola gigantica. Molecular Cellular Probes, 2002, 16, 327–333 [DOI] [PubMed] [Google Scholar]
- Mas-Coma S., Bargues M.D. & Valero M.A.Fascioliasis and other plant-borne trematode zoonoses. International Journal of Parasitology, 2005, 35, 1255–1278 [DOI] [PubMed] [Google Scholar]
- Mas-Coma S., Esteban J.G. & Bargues M.D.Epidemiology of human fascioliasis: a review and proposed new classification. Bulletin of the World Health Organization, 1999, 77, 340–346 [PMC free article] [PubMed] [Google Scholar]
- Mas-Coma S., Valero M.A. & Bargues M.D.Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Chapter 2. Advances in Parasitology, 2009, 69, 41–146 [DOI] [PubMed] [Google Scholar]
- Mekroud A., Titi A., Benakhla A. & Rondelaud D.The proportion of liver excised in Algerian abattoirs is not a good indicator of Fasciola hepatica infections in local cattle breeds. Journal of Helminthology, 2006, 80, 319–321 [PubMed] [Google Scholar]
- Periago M.V., Valero M.A., Sayed M., Ashrafi K., Wakeel A., Mohamed M.Y., Desquesnes M., Curtale F. & Mas-Coma S.First phenotypic description of Fasciola hepatica/Fasciola gigantica intermediate forms from human endemic area of the Nile Delta Egypt. Infection, Genetics and Evolution, 2008, 8, 51–58 [DOI] [PubMed] [Google Scholar]
- Pfukenyi D.M., Mukaratirwa S., Willingham A.L. & Monrad J.Epidemiological studies of Fasciola gigantica infections in cattle in the highveld and lowveld communal grazing areas of Zimbabwe. Onderstepoort Journal of Veterinary Research, 2006, 73, 37–51 [PubMed] [Google Scholar]
- Phiri A.M., Phiri I.K., Chota A. & Monrad J.Trematode infections in freshwater snails and cattle from the Kafue wetlands of Zambia during a period of highest cattle-water contact. Journal of Helminthology, 2007, 81, 85–92 [DOI] [PubMed] [Google Scholar]
- Rokni M.B., Mirhendi H., Mizani A., Mohebali M., Sharbatkhori M., Kia E.B., Abdoli H. & Izadi S.Identification and differentiation of Fasciola hepatica and Fasciola gigantica using a simple PCR-restriction enzyme method. Experimental Parasitology, 2010, 124, 209–213 [DOI] [PubMed] [Google Scholar]
- Spithill T.W. & Dalton J.P.Progress in development of liver fluke vaccines. Parasitology Today, 1998, 14, 224–228 [DOI] [PubMed] [Google Scholar]
- Urquhart G.M., Armour J., Duncan J.L., Dunn A.M. & Jennings F.M.Veterinary parasitology, 2nd edit.Blackwell Science: Oxford: 1996 [Google Scholar]
- Vara-Del Río M.P., Villa H., Martinez-Valladares M. & Rojovázquez F.A.Genetic heterogeneity of Fasciola hepatica isolates in the northern of Spain. Parasitology Research, 2007, 101, 1003–1006 [DOI] [PubMed] [Google Scholar]
- Walker S., Johnston C., Hoey E., Fairweather I., Borgsteede F., Gaasenbeek C., Prodohl P. & Trudgett A.Population dynamics of the liver fluke, Fasciola hepatica: the effect of time and spatial separation on the genetic diversity of fluke populations in the Netherlands. Parasitolology, 2011, 138, 215–223 [DOI] [PubMed] [Google Scholar]
- World Health Organization Control of foodborne trematode infections. World Health Organization, Technical Reports Series, 1995, 849, 1–157 [PubMed] [Google Scholar]