Abstract
Polymeric synthetic mimics of antimicrobial peptides (SMAMPs) have recently demonstrated similar antimicrobial activity as natural antimicrobial peptides (AMPs) from innate immunity. This is surprising, since polymeric SMAMPs are heterogeneous in terms of chemical structure (random sequence) and conformation (random coil), in contrast to defined amino acid sequence and intrinsic secondary structure. To understand this better, we compare AMPs with a ‘minimal’ mimic, a well characterized family of polydisperse cationic methacrylate-based random copolymer SMAMPs. Specifically, we focus on a comparison between the quantifiable membrane curvature generating capacity, charge density, and hydrophobicity of the polymeric SMAMPs and AMPs. Synchrotron small angle x-ray scattering (SAXS) results indicate that typical AMPs and these methacrylate SMAMPs generate similar amounts of membrane negative Gaussian curvature (NGC), which is topologically necessary for a variety of membrane-destabilizing processes. Moreover, the curvature generating ability of SMAMPs is more tolerant of changes in the lipid composition than that of natural AMPs with similar chemical groups, consistent with the lower specificity of SMAMPs. We find that, although the amount of NGC generated by these SMAMPs and AMPs are similar, the SMAMPs require significantly higher levels of hydrophobicity and cationic charge to achieve the same level of membrane deformation. We propose an explanation for these differences, which has implications for new synthetic strategies aimed at improved mimesis of AMPs.
Introduction
Natural antimicrobial peptides (AMPs) from the innate immunity of eukaryotic organisms comprise a class of membrane-active antimicrobial agents, which are promising anti-infective therapeutics especially for antibiotic-resistant bacterial pathogens1. The therapeutic development of AMPs is, however, hindered largely by their liability to proteases in vivo and expensive manufacturing cost2. Synthetic mimetics of AMPs (SMAMPs) including non-natural peptides3–10, peptoids11, and abiotic oligomers12–15 and polymers16–22, are therefore developed. Being cationic and amphiphilic, these SMAMPs are designed to simultaneously capture the two main structural motifs shared by most AMPs.
AMPs can destabilize the bacterial cytoplasmic membranes via processes such as pore formation, membrane micellization, blebbing, and budding1, 23–28. AMPs have well defined sequences, and can participate in signaling, DNA-binding, and other functions besides membrane permeabilization. Moreover, AMPs often also adopt secondary structures near a membrane surface that give them a specific geometric presentation of their cationic and hydrophobic components. For example, the AMP magainin adopts an α-helical conformation upon binding to membranes, in which cationic and hydrophobic residues are segregated to opposite phases of the helix. In contrast, polymeric SMAMPs are based on synthetic polymers such as polymethacrylate, which have heterogeneous sequences and no specific secondary structures. Much recent work has focused on the similarities between AMPs and SMAMPs, and has demonstrated that polymeric SMAMPs have in vitro antimicrobial activity similar to natural AMPs16–22, 29, 30. Clearly, both AMPs and polymeric mimics meet the minimal structural requirements for cell specific membrane permeabilization activity despite their distinctive difference in structural and conformational homogeneity/heterogeneity. In this work, we focus on their structural and resultant functional differences with respect to natural AMPs, and investigate the role of homogeneous vs. heterogeneous structures in their antimicrobial mechanisms by comparing AMPs with cationic amphiphilic random copolymers, a demonstrated family of polymeric SMAMPs17–22, 29. Specifically, we used a ‘minimal‘ family of three polydisperse cationic methacrylate-based polymeric SMAMPs as heterogeneous models that are inactive, preferentially active against bacteria over human cells, and non-specifically active, respectively. We show that bacterial inner membrane permeabilization results are consistent with those from synchrotron small angle x-ray scattering (SAXS), which is used to quantify the negative Gaussian curvature (NGC) deformations induced in model membranes by these random copolymers, as NGC is topologically necessary for a variety of membrane destabilization processes. We find that, although the amount of NGC generated by these SMAMPs and AMPs are similar, the SMAMPs require significantly higher levels of hydrophobicity and cationic charge to achieve the same quantitative level of membrane deformation. We propose an explanation for these similarities and differences, which has implications for new synthetic strategies aimed at improved mimesis of AMPs.
Materials and Methods
1. Copolymer preparation and characterization
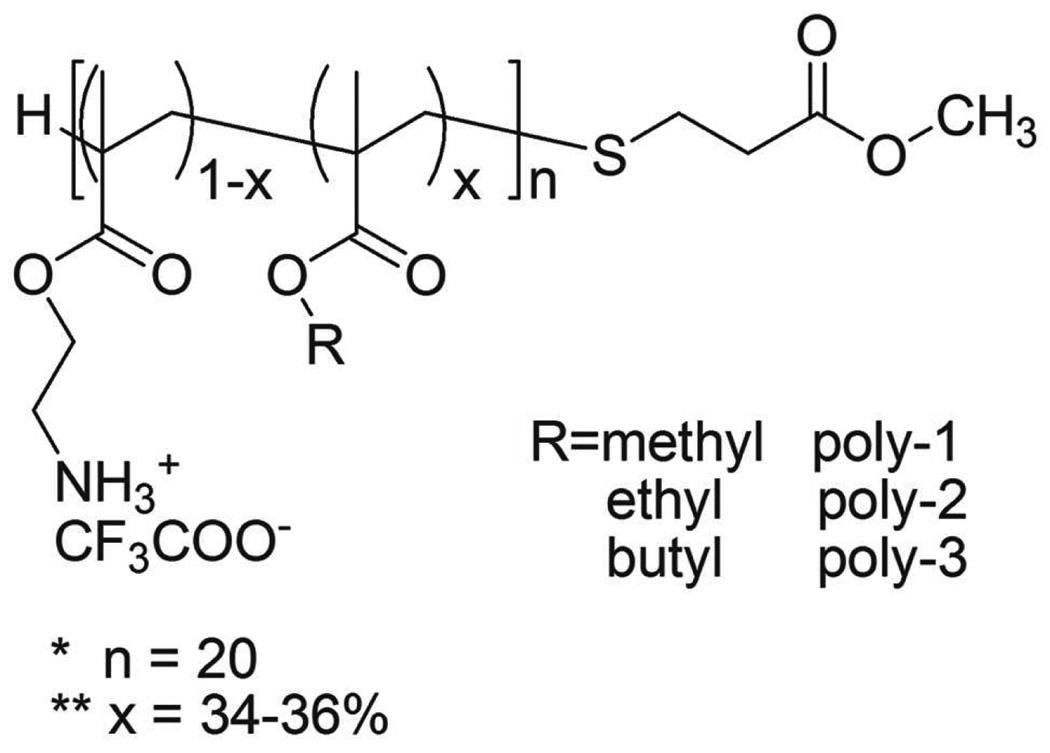
The SMAMPs are random copolymers composed of methacrylate and aminoethyl methacrylate (MA-co-AEMA) (Figure 1), which were prepared via free radical copolymerization and characterized using 1H NMR as described previously31. The following are 1H NMR spectra data (Methanol-d4, 400 MHz, Bruker-400 NMR spectrometer)) of the three as-prepared copolymers after de-protection. Poly-1: δ 4.4–4.1 (bs, 26.10H), 3.8-3.5 (bm, 22.85H), 2.8-2.5 (m, 7.00H), 2.3-1.8 (m, 32.51H), 1.7-0.8 (m, 56.51H); poly-2: δ 4.4–4.1 (bs, 26.30H), 4.1-3.9 (bs, 14.21H), 3.698 (s, 3.00H), 2.8-2.5 (m, 7.57H), 2.3-1.8 (m, 30.40H), 1.7-0.8 (m, 81.70H); poly-3: δ 4.4–4.1 (bs, 25.19H), 4.1-3.9 (bs, 14.33H), 3.699 (s, 3.00H), 2.8-2.5 (m, 7.99H), 2.3-1.8 (m, 31.04H), 1.7-0.8 (m, 111.41H). Gel permeation chromatography (GPC) characterization was performed on the three copolymers collected after de-protection. The GPC spectra (acetic acid/acetate buffer supplemented with 20% acetonitrile, calibrated with polystyrene standard) were obtained from a Waters 515 Pump and Waters 2410 Refractive Index Detector. Growth inhibition assays against E. coli (ATCC 25922) and hemolysis assays against human red blood cells were described in details previously31.
Figure 1.
Structures of the SMAMPs used in this study. The copolymers’ average degree of polymerization (DP) and average mole fraction of methacrylate (falkyl) are fixed but the side-group of MA is varied (as indicated by R = methyl, ethyl, or butyl) to modify the copolymer hydrophobicity.
2. Inner membrane permeability assays
The inner membrane permeability was characterized by measuring the β-galactosidase activity of E. coli ML-35 cells with o-nitrophenyl-β-D-galactoside (ONPG) as substrate32–35. The E. coli ML-35 strain was a kind gift from Professor Andrè J. Ouellette at the University of Southern California. ONPG was purchased from Sigma-Aldrich (Shanghai, China). ML-35 cells were grown in tryptic soy broth (TSB) at 37°C for 18 h to stationary growth phase. A 100-µL culture was subsequently diluted with fresh TSB by 100-fold and regrown at 37°C to mid-log growth phase (OD600 = 0.5–0.7, measured using Eppendorf BioPhotometer). ML-35 cells at mid-log phase were collected and washed for 3 times with 10 mM PBS (10mM sodium phosphate, 150mM NaCl, pH 7.4) and resuspended into 10mM PBS to approximately 1*108 CFU/mL. Into a 96-well microplate, we inoculated 15 µL of the adjusted bacterial inoculum to each well. Thus each well had 1*107 CFU/mL ML-35 cells suspended in 10 mM PBS (10mM sodium phosphate, 150mM NaCl, pH 7.4) supplemented with 1% TSB and 2.5mM ONPG, and copolymers at expected concentrations. The β-galactosidase activity was monitored by measuring OD400 at 37°C with an interval of 1 min for 130 min using Microplate Reader (VarioskanFlash, Thermo Scientific), and copolymer stock solutions were added at time t in-between the 9th and 10th minutes during an experiment.
A set of blank controls containing no copolymer were measured at same experimental conditions, which provide baselines for data analysis. Contribution of blank control was subtracted from raw data, which yielded delta(OD400). Each inner membrane permeabilization trial was carried out in triplicate, and the reported results are the averages of two independent trials.
3. Liposome Preparation
All lipids used in this work were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Small unilamellar vesicles (SUVs) were prepared using sonication method. Binary mixtures of DOPG (1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)](sodium salt)) with DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) were used as first-order model for bacterial membranes. Ternary membranes composed of DOPG, DOPE, and DOPC (1,2-dioleoyl-sn-glycero-3-phosphatidylcholine) were used as a more realistic model for the heterogeneous distribution of lipids in real bacterial membranes. These ternary membranes also allow us to isolate the membrane curvature from the membrane surface charge density. DOPG stock solution was mixed with DOPC and/or DOPE stock solutions; all stock solutions were at 20 mg/mL in chloroform. The mixture was dried under N2, desiccated under vacuum overnight, rehydrated with Millipore water to a final lipid concentration of 20 mg/mL at 37°C overnight, sonicated to clarity, and extruded through a 0.2 µm Nucleopore filter (Whatman).
4. Small angle X-ray Scattering experiments
Copolymers dissolved in Millipore water at ~0.8 mg/mL were mixed with SUVs at specific polymer to lipid molar ratios (P/L) and NaCl concentrations, and sealed in quartz capillaries for SAXS experiments. Synchrotron SAXS experiments were performed at the Stanford Synchrotron Radiation Laboratory (SSRL, BL4-2), the Advanced Light Source (ALS, beamline 7.3.3), and the Shanghai Synchrotron Radiation Facility (SSRF, BL16B1), using monochromatic X-rays with energies of 9 keV, 10 keV, and 10 keV, respectively. The scattered radiation was collected using a Rayonix MX225-HE detector (pixel size, 73.2µm) at SSRL, a Pilatus 1M detector (pixel size, 172 µm) at ALS, and a MAR Research CCD area detector (pixel size, 79 µm) at SSRF. No radiation damage was observed for the exposure times used. 2D SAXS powder patterns were integrated using the FIT2D.
5. Comparison on <hydrophobicity> and cationicity of SMAMPs with those of AMPs
To directly compare methacrylate polymer hydrophobicities with natural AMP hydrophobicities, we used published octanol-H2O partition coefficients, Log P's, for the methacrylate monomers36–39 and the Wimley-White whole-residue hydrophobicity scale for the free energies of transfer from water to octanol, ΔGwoct, for the AMPs.40, 41 Since, ΔG = −2.3 R T Log P, where R is the gas constant and T is the temperature,42 the MMA, EMA, and BMA Log P values can be converted into ΔG values.
ΔG for aminoethyl methacrylate has not been measured. To determine its value we assumed that methacrylate monomer hydrophobicities are the sum of the contributions from the side chain and backbone. Note that the ΔGwoct values from the Wimley-White whole residue hydrophobicity scale are also the sum of the contributions from the side chains and the backbone. For the methacrylates these contributions were decoupled by a linear least squares fit of the published Log P values as a function of side chain length, L, (i.e. LMMA = 1, LEMA = 2, LnPMA = 3, LnBMA = 4). The resulting linear trendline has the form Log P = slope*L + intercept, where the intercept represents the contribution of the backbone, and the slope provides the increase in Log P from increasing the side chain length. Excellent Log P versus L relationships were observed over the side chain lengths used in this study (Figure S2). The extracted backbone Log P was converted into a ΔG, and then added to the side chain value for lysine from the Wimley-White scale.
AMP sequences were obtained from the antimicrobial peptide database.43 Since arginine and lysine are known to interact differently with membranes,44, 45 AMP sequences which exclusively contain the cationic amino acid lysine were used. This yielded 335 lysine-rich AMPs. The average hydrophobicity for the jth AMP was calculated via:
where wi is the Wimley-White scale ΔGwoct value and n is the length of jth AMP. A histogram of the average hydrophobicity values of the 335 AMPs was constructed by placing them into 50 bins. The average hydrophobicity values for methacrylate polymers containing methyl, ethyl, and butyl side chains were determined using the same equation. For each amphiphilic methacrylate copolymer the average degree of polymerization was 20, and falkyl was fixed at 35%, implying Namine = 13, and Nalkyl = 7. Average hydrophobicity values for methyl, ethyl, and butyl side chain copolymers were superimposed over the histogram for lysine-rich AMPs.
Results and Discussions
The SMAMPs are random copolymers composed of methacrylate and aminoethyl methacrylate (MA-co-AEMA) (Figure 1), which have been previously characterized31. AEMA has a primary ammonium group and serves as a mimic of lysine. The copolymers’ average degree of polymerization (DP) and average mole fraction of AEMA (fAEMA) are fixed (DP = 20, and fAEMA = 1 − falkyl = 0.65 ± 0.01)31, but the alkyl length of side-group of MA is varied (as indicated by R = methyl, ethyl, or butyl) to modify the copolymer hydrophobicity (Figure 1). The resultant homologues have approximately equivalent average polymer length and closely resembling side chain composition but tunable balance between hydrophobicity and cationic charge. Growth inhibition and hemolysis assays have demonstrated that these homologous polymer SMAMPs have activity profiles spanning the whole activity range—from inactive (poly-1, when R = methyl), over preferentially active against bacteria over human erythrocytes (poly-2, when R = ethyl), to unselectively active against both bacteria and human erythrocytes (poly-3, when R = butyl)31—ideal models for investigating the membrane-interactions of SMAMPs systematically.
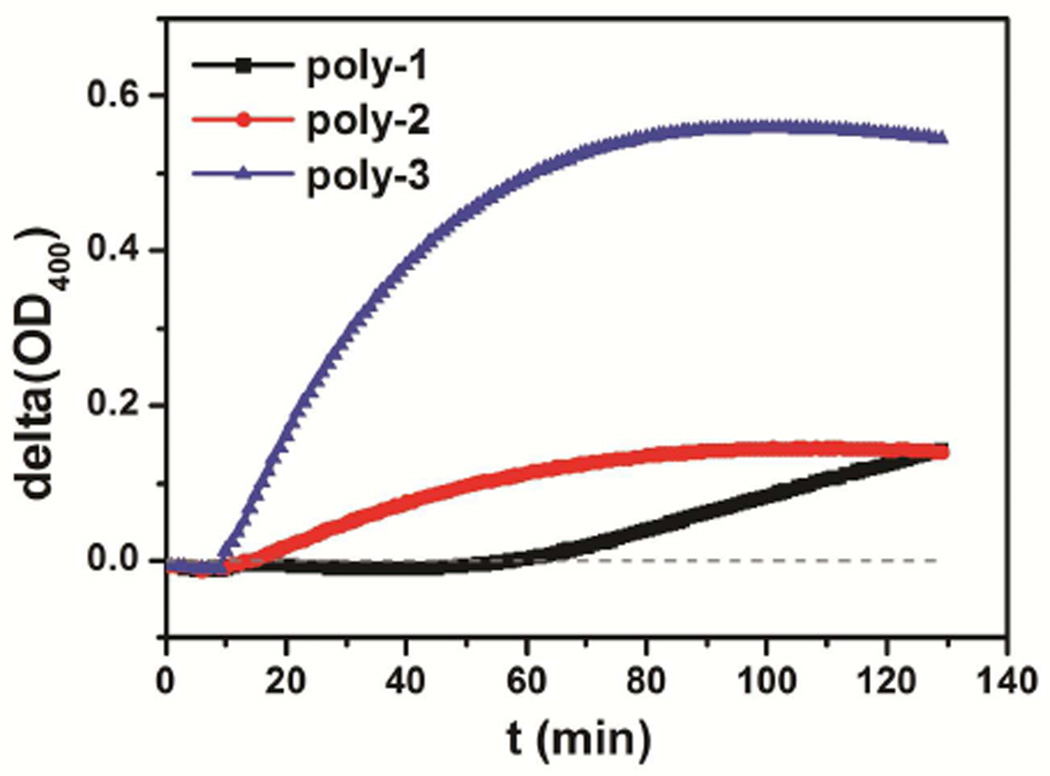
To test whether the differences in polymer activity against bacteria is correlated with permeabilization of their cell membranes we examined inner membrane permeability of E. coli ML-35 by measuring the β-galactosidase activity with o-nitrophenyl-β-D-galactoside (ONPG) as substrate32–35. The mutant E. coli ML-35 (i−, y−, z+) has constitutive cytoplasmic β-galactosidase activity but no lactose permease. Intact E. coli ML-35 cells therefore cannot hydrolyze ONPG until their cytoplasmic membranes are permeabilized and allow release of β-galactosidase into the solution or diffusion of ONPG into the cytoplasm. o-nitrophenol (ONP), the product of ONPG hydrolysis by β-galactosidase, shows strong absorption at 400nm, whereas ONPG does not. Because the hydrolysis of ONPG catalyzed by β-galactosidase is monitored by measuring optical density at 400nm (OD400), the rate of increase in OD400 reflects the rate of ONP production and, as a result, the capability of β-galactosidase to be released from bacteria or ONPG to cross the inner membrane. Such inner membrane permeabilization assays have been performed to assess the membrane destabilization capability of AMPs32–35. Interestingly, all three of our homologues induced inner membrane permeabilization at 100 µg/mL. However, the OD400 signal varied considerably in time and in magnitude depending on the copolymer activity profiles (Figure 2), indicating significant kinetic differences for these different copolymers. Poly-1, the nominally inactive one, required a 48 min incubation with ML-35 cells before inducing a low level OD400 increase. In contrast, within 5 min after addition of poly-2 or immediately after addition of poly-3, OD400 started increasing dramatically and, after 48 min incubation, increased to levels significantly higher than that of poly-1. Poly-3, the most active homologue, caused the largest increase in OD400. These in vitro bacterial assays are consistent with biomimetic vesicle leakage experiments31 which showed that all the three homologues caused fluorescent dye leakage from bacterial membrane mimicking vesicles with the homologue with the lowest MIC inducing the greatest leakage. Our results indicate that the ability of these homologues for E. coli membrane permeabilization track with their inhibitory activity against E. coli.
Figure 2.
Membrane permeation assays showed that, although all the three copolymers (at 100 µg/mL) permeabilized the inner membranes of E. coli ML-35 cells, the OD400 signal varied considerably in time and in magnitude for different copolymer activity profiles. The assays were performed in PBS-TSB (10mM Na3PO4-Na2HPO4, 150mM NaCl, pH 7.4, supplemented with 2.5 mM ONPG and 1% TSB). Copolymers were added at t in-between 9th and 10th min. Contribution of blank control was subtracted from raw data, which yielded delta(OD400). The reported data are average of two independent trials each performed in triplicate.
To examine the microscopic origins of bacterial membrane permeabilization by the random copolymers we used synchrotron small angle x-ray scattering (SAXS) to characterize the curvature deformations they induced in model cell membranes. Specifically, we assayed the ability of each homologue to induce negative Gaussian curvature (NGC) deformations. Generation of negative Gaussian curvature is a common physical feature for a broad range of membrane-destabilization processes45, 46. It has been shown that the amino acid content of AMPs can be related to the need for generating NGC. It will therefore be interesting to examine the similarities and differences in membrane deformation capacity between natural AMPs with defined sequence and conformation and these random polymer SMAMPs.
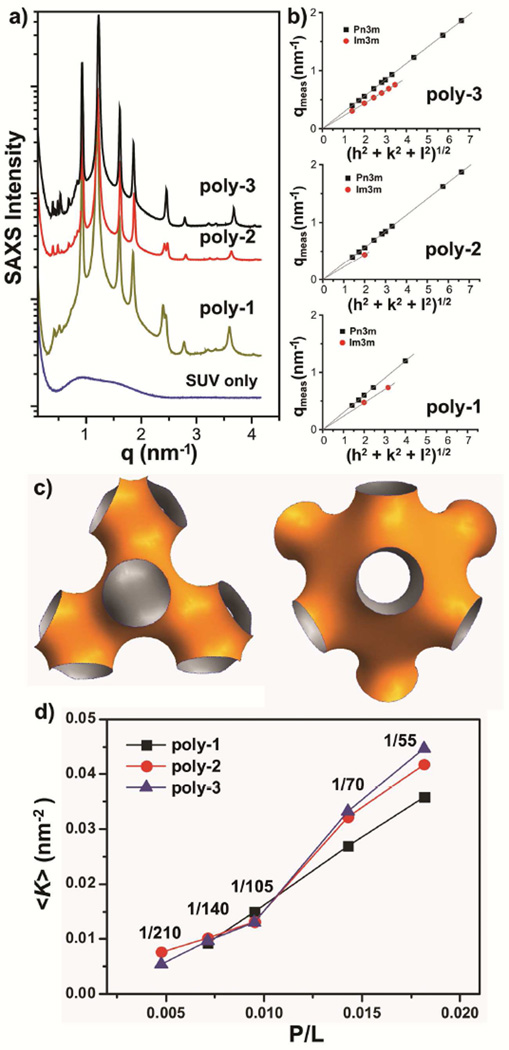
Small unilamellar vesicles composed of 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt) (DOPG) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) at ratio of DOPG/DOPE = 20/80 are used as first order model membranes for gram-negative bacteria. Before exposure to a copolymer, SAXS profile shows a broad form factor expected for unilamellar vesicles (Figure 3a, bottom curve). After exposure to the copolymers (P/L = 1/105), the SUVs underwent drastic topological transformation as indicated by the appearance of scattering peaks on SAXS profile (Figure 3a). For the most active homologue poly-3 (Figure 3a, top curve), the observed scattering peaks can be indexed to coexisting Im3m and Pn3m inverse cubic lattices according to relationships of the measured q positions. A 1st set of scattering peaks, starting at q = 0.306 nm−1, have the measured q positions satisfy ratios of √2: √4: √6: √8: √10 characteristic of an inverse cubic ‘Im3m’ lattice which has a calculated lattice parameter aIm3m = 28.85 nm (Figure 3b, top, red circle). A 2nd set of scattering peaks, starting at q = 0.396 nm−1, have the measured q positions satisfy ratios of √2: √3: √4: √6: √8: √9: √11 characteristic of an inverse cubic ‘Pn3m’ lattice which has a calculated lattice parameter aPn3m = 22.40 nm (Figure 3b, top, black square). The calculated lattice parameters of the two observed cubic phases have a ratio of aIm3m/aPn3m = 1.28, which satisfies the Bonnet ratio of 1.279 for coexisting Im3m and Pn3m lattices. Poly-2 and poly-1 also generated coexisting Pn3m and Im3m phases in these model bacterial membranes (Figure 3b, middle and bottom). Pn3m and Im3m lattices represent two examples of the inverse bicontinuous cubic phases (QII) where two inter-linked but separated water channels weave their way through a single continuous lipid bilayer, leaving the bilayer mid-plane with negative Gaussian curvature (NGC, or equivalently, saddle-splay curvature) at every point (Figure 3c–d). Generation of NGC is topologically necessary for a variety of membrane destabilizing processes including membrane pore formation, micellization, blebbing, and budding. Our observations of QII phases suggest that abiotic SMAMPs may destabilize membranes by generating NGC, as do natural AMPs46. Interestingly, all three SMAMPs generate NGC in the bacterial model membrane, which is consistent with the observed E. coli membrane permeabilization studies above. We next compare the quantity of NGC generated by AMPs and SMAMPs.
Figure 3.
(a) SAXS profiles indicates that exposure to copolymer (starting from top: poly-3, poly-2, poly-1) transits model bacterial membranes (bottom) into inverse cubic phases rich in negative Gaussian curvature topologically necessary for a variety of membrane destabilization processes. (b) Relationships of the measured q positions of the observed scattering peaks show that the copolymer-induced inverse cubic phases are coexisting Pn3m and Im3m lattices. (c) 3D schematic representations of the minimal curvature surface of Pn3m (left) and Im3m (right), respectively. (d) Summary on the average saddle-splay curvature, <K>, induced by copolymers at varying P/L.
The amount of induced NGC tracks with the antimicrobial activity of these SMAMPs. For an induced cubic phase, its average NGC (or average saddle-splay curvature) in the unit cell is <K> = 2πχ/(a2A0), where χ is the Euler characteristic, a is the lattice parameter, and A0 is surface area per unit cell47. From the SAXS profiles (Figure 2), the average NGC in the unit cell of the induced cubic structures enables quantitative comparison between the amount of NGC generated by SMAMPs and that generated by AMPs. Defensins (α-, β-, θ-), a well-characterized family of β-sheet AMPs, generated NGC with <K> of 0.025 – 0.050 nm−246, 48. Compared to defensins, the polymeric SMAMPs clearly show similar curvature generating activity against lipid membranes, giving <K> of 0.008 – 0.042 nm−2 as P/L was varied in the range of 1/210 – 1/55 (Figure 3d), with increasing P/L producing greater <K> for all three SMAMPs. Our results demonstrate that the polydisperse polymer SMAMPs generated similar amounts of saddle-splay curvature as the monodisperse natural AMPs.
Interestingly, the copolymer-induced QII scattering peaks are very sharp and correspond to a single <K> value, as observed with AMPs46. The sharpness of the correlation peaks imply that a large number of unit cells with homogeneous structures are generated by the SMAMPs, which is surprising. Whereas naturally occurring AMPs are generally homogeneous in amino acid sequence and most of them have well-defined secondary structures such as α-helix or β-sheet, SMAMPs are random copolymers which are heterogeneous in terms of chemical structure (random monomer sequence, polydispersity, and tacticity) and conformation (random coli). Gel permeation chromatography (GPC) results confirm that our random copolymers have polydispersity index PDI = 1.52 ~ 1.79 (Table S1). In previous work, we have shown how NGC-inducing peptides generate different quantitative amounts of NGC via changes in peptide sequence49 and length45. Heterogeneous polymeric SMAMPs are therefore intuitively expected to generate heterogeneous structures in lipid membranes. How does an ensemble of heterogeneous copolymers induce highly correlated homogeneous structures in lipid membranes, similar to those induced by homogeneous AMPs? A number of explanations suggest themselves. It is possible that only a subset of the polydisperse ensemble of copolymer chains—those chains with optimal polymer length and monomer sequence for NGC generation—are thereby incorporated into the cubic structures, whereas the rest of polymer chains are excluded from the cubic structures or QII phase forming processes. That the MIC’s of heterogeneous SMAMPs are similar to those of homogeneous natural AMPs argues against this type of explanation. We hypothesize that the conformational flexibility of the lipid membrane itself plays an important role in integrating over the heterogeneity of SMAMP sequences. The distribution of NGC in bicontinuous cubic phases50, 51 implies that different parts of the membrane surface in the induced cubic structures are themselves heterogeneous, and the heterogeneous structures of bicontinuous cubic phases can therefore accommodate copolymers with different lengths, structures, and compositions according to their individual preferred membrane curvature. Moreover, the conformational flexibility of oligomeric SMAMPs can significantly enhance their ability to be incorporated into membranes: flexible acyl-lysyl oligomers efficiently penetrated into membranes composed of lipid A, whereas antimicrobial peptides which adopt rigid secondary structures upon binding a membrane surface and rigid arylamide foldmer mimics failed to do so52. For polymeric SMAMPs which have flexible polymer backbones, their lack of rigid secondary structures may also facilitate their incorporation into the cubic structures induced in lipid membranes. Both of these effects can contribute to the accommodation of heterogeneous polymers chains into an ordered structure with well-defined curvatures.
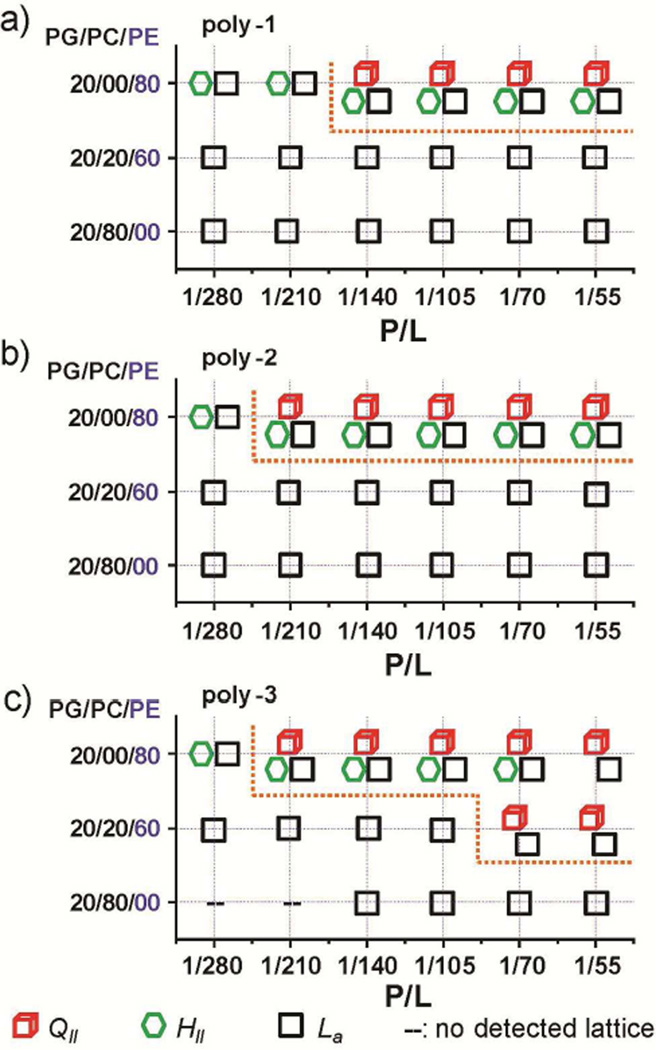
The heterogeneous methacrylate-based SMAMP showed selective activity against bacteria over host cells, as do natural AMPs. The selective activity of AMPs and SMAMPs against bacteria over host cells is commonly ascribed to their preferential electrostatic binding to the anionic bacterial surfaces over the nearly neutral outer leaflets of mammalian membranes1, 53. The electrostatic binding is indeed an important component of activity in that it is a necessary condition for initiating membrane surface association. However, we found that the concentration of negative intrinsic curvature lipids in the target membrane is also an important factor in the cell-selectivity46, 48, 54–60. Bacterial membranes are characterized by anionic lipids such as PG and negative-intrinsic-curvature (NIC) lipids such as PE, both of which are rare in the outer leaflets of mammalian membranes, which are rich in zwitterionic lipids such as phosphocholine (PC). To understand how heterogeneous methacrylate-based SMAMPs result in selective activity, we examined the NGC generating capacity of the SMAMPs for different membrane compositions. To mimic membrane lipid composition of target membranes typically encountered in prokaryotic and eukaroyotic cells, we used ternary membranes composed of DOPG, DOPE, and 1,2-dioleoylsn-glycero-3-phosphocholine (DOPC). Both DOPG and DOPC have zero intrinsic curvature (C0 ~ 0) whereas the zwitterionic DOPE has negative intrinsic curvature (C0 < 0). By varying the ratio of DOPE to DOPC in membranes with fixed DOPG content (20%, a value typical to bacterial membranes), we isolated membrane curvature from membrane surface charge density. Phase diagrams (Figure 4) indicate that induced QII phases from copolymers of different antimicrobial activity profiles require different minimum threshold PE content PE%* in lipid membranes. Both poly-1 (figure 4a) and poly-2 (figure 4b) induced QII phases in PE-rich membranes (DOPG/DOPC/DOPE 20/00/80), but do not do so in membranes with reduced PE% at all tested P/L. In contrast, for poly-3, the induced QII phases persist in PE%-reduced DOPG/DOPC/DOPE = 20/20/60 membranes (Figure 4c). Thus the minimum threshold PE%* values required by copolymer to induce QII phases have the order of poly-1 ~ poly-2 > poly-3. This indicates that, compared to poly-1 and poly-2, poly-3 can induce NGC for membrane destabilization in target membranes of the lower PE concentration. This is consistent with the bacterial membrane permeation studies of Fig 2, where poly-3 caused significantly higher membrane permizabilization in E. coli. In particular, poly-1 also generates significant NGC at PG/PE = 20/80, which is consistent with its inner membrane permeation of E. coli (Figure 2) whose membrane lipids contains PE at ~75% of and PG at ~20%. It is interesting to contrast this phase behavior of random copolymers with that exhibited by monodisperse and structurally homogeneous AMPs. The magainins, which are rich in lysines and rich in aliphatic hydrophobes and are therefore chemically similar to the SMAMPs here, generate NGC over much narrower ranges of P/L ratios61. This difference may also impact the specificity and kinetics of membrane permeabilization, since more lipid diffusion will be necessary for the AMPs to generate the local P/L ratios required for NGC generation.
Figure 4.
Phase diagrams of structural changes in ternary membranes composed of DOPG/DOPC/DOPE with fixed DOPG content (20%, typical to bacterial membranes) but tunable DOPE content, induced by poly-1(a), poly-2 (b), and poly-3(c).
It is informative to compare quantitatively the level of hydrophobicity and cationic charge in the SMAMPs compared to all the known naturally occurring AMPs. For these MA-co-AEMA random copolymers, their hydrophobicity increases (at fixed DP and falkyl) generally led to their activity progresses from non-active, to preferentially active against bacteria, and finally to non-specifically active against both bacteria and eukaryotic cells. Similar trends have been observed for membrane-active antimicrobial peptides (AMPs)62, 63. For AMPs, established scales have been used to quantify peptide hydrophobicity and relate this quantity to peptide activity and selectivity44, 63. For SMAMPs like our methacrylate copolymers, structure-activity relationships have shown a correlation between hemolytic activity and hydrophobicity as quantified by water-octanol partition coefficients, Log P36, 37, 64. Since hydrophobicity scales such as the Wimley-White whole residue octanol scale40 employ similar procedures for determining the hydrophobicity of individual amino acids, in principle, polymers composed of methacrylates can be directly compared to peptides composed of amino acids using a common hydrophobicity scale.
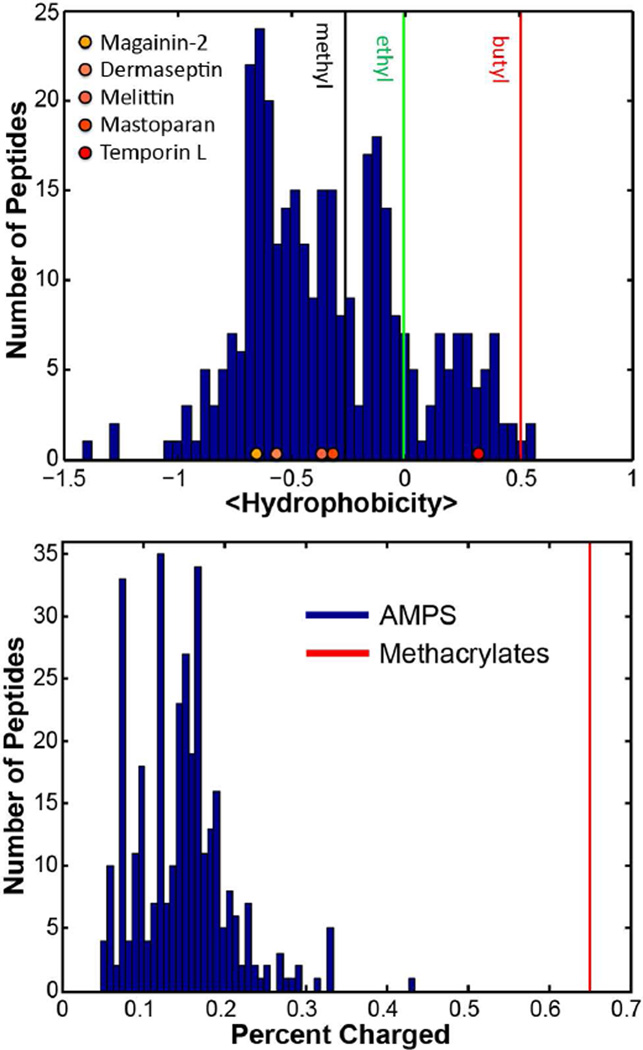
To test the viability of direct comparison between the hydrophobicities of natural AMPs and our methacrylate polymers we translated published Log P values for the methacrylates into free energies of transfer, ΔG, according to the Wimley-White octanol scale (see Methods), and compared the corresponding polymer average hydrophobicity, <Hydrophobicity>, values with those for lysine-rich AMPs (Figure 5a). It should be noted that because the average hydrophobicity, <Hydrophobicity>, is given as a sum of ΔG of each amino acid residue or monomers, <Hydrophobicity> values do not reflect the sequences and conformations of peptide and polymer structures. As expected, methacrylate polymer <Hydrophobicity> noticeably increased with increasing alkyl chain length. The range of <Hydrophobicity> values for these methacrylate copolymers (R = methyl, ethyl, and butyl) overlaps with the broad range exhibited by AMPs. It is interesting to note that the SMAMP random copolymers are generally more hydrophobic than the AMPs. (The published Log P values for each methacrylate differ between studies, which produce differences in <Hydrophobicity> for each polymer, as shown in see Figure S3. However, these trends are still observed when 4 different methacrylate Log P sets are used: There is good overlap between polymers and lysine-rich AMPs, and the polymers are more hydrophobic.) The methyl, ethyl, and butyl versions of the SMAMP are more hydrophobic than 60%, 81%, and 99% of the AMPs respectively. The superimposed <Hydrophobicity> values for common AMPs also display the correlation between hydrophobicity and non-specificity. Both the specifically active magainin-2 and dermaseptin are less hydrophobic than non-specifically active melittin and mastoparan venoms. The short 13AA AMP temporin L is even more hydrophobic, which corresponds well with its hemolytic activity65. These data illustrate that <Hydrophobicity> is a common contributing factor to the activity and specificity of both methacrylate polymers and AMPs.
Figure 5.
Comparison of SMAMPs and AMPs on their <hydrophobicity> and cationic charge percentages.
Comparison of cationicity in the methacrylate polymers and AMPs shows that these copolymers carry substantially more amine groups than lysine-rich AMPs (Fig 5b). With a fixed fAEMA = 65%, the methacrylates are 4× more amine-rich than an average AMP (15%), and at DP = 20, their average +13 charge is much greater than the average number of lysines, NK = 3.75, in an AMP. Therefore, while the activities of these methacrylate copolymers can be tuned by changing their alkyl length of MA without altering the monomer compositions, their overall cationic and hydrophobic properties still deviate from those of natural AMPs. In order to achieve greater fidelity in the mimesis of natural AMPs, existing SMAMPs will need to be tuned to a different range of cationic charge and hydrophobicity. We hypothesize that this shift toward higher hydrophobicity and higher charge in the SMAMPs may be a consequence of the randomness of the monomer sequence and distribution of monomer compositions, compared with AMPs.
One natural question that emerges is why the polydisperse random copolymer-based SMAMPs require simultaneously more cationic charge and more hydrophobicity in order to achieve similar levels of membrane NGC as AMPs? A number of possible explanations can be proposed, but we believe the answer is related at least in part to the fundamental nature of electrostatic and hydrophobic interactions at the nanoscale: A high charge density array of closely-spaced cationic charge does not behave in the same manner as the same number of isolated charges far apart. For example, for a linear charged polymer with an inter-charge distance of less than the Bjerrum length (~7.8Ǻ), Manning condensation will ‘dress’ the polymer with a layer of counterions66, 67. Consequently, there will be a strong thermodynamic driving force for such cationic polymers to bind to anionic membranes of similar charge density, due to the entropy gain of counterion release68. This does not occur if the local charge density of the polymer were significantly lower. Due to their random copolymer nature, only a part of the SMAMPs have local cationic charge densities optimized for membrane binding, while the rest of the cationic charges do not contribute significantly to activity. Likewise, the effective hydrophobicity of two nm-sized hydrophobic residues in close proximity is significantly different than the hydrophobicity of the same two hydrophobic residues far apart. For small hydrophobes (up to ~1nm), the interaction is mainly entropic, whereas for larger hycrophobes, the interaction will have a volume dependent entropic contribution and a surface area dependent enthalpic contribution69. Hydrophobic sequences in AMPs optimized for insertion or binding will be more ‘efficient’, and achieve the same function with few residues compared to random copolymers. Taken together, these considerations imply that by virtue of their specific sequences and the selective clustering that can result, natural AMPs can use their hydrophobic residues and cationic residues more efficiently than a random sequence, and generate the same effects without as many residues.
Conclusions
In summary, we find that SMAMPs permeate bacterial inner membranes in vitro and both SMAMPs and AMPs generate quantitatively similar amounts of membrane NGC. Comparisons between SMAMPs and the chemically similar lysine-containing AMPs indicate that, although the membrane curvatures they generated are similar, the SMAMPs require significantly higher levels of hydrophobicity and cationic charge to achieve the same level of membrane deformation. Such a shift toward higher hydrophobicity and higher charge in the SMAMPs may be a consequence of the cooperative nature of electrostatic interactions and hydrophobic interactions at the nanoscale, combined with the randomness of the methacrylates relative to AMPs.
Supplementary Material
ACKNOWLEDGMENT
The E. coli ML-35 strain was a kind gift from Professor Andrè J. Ouellette at the University of Southern California. GPC characterization was carried out with help of Yuande Long at Chengdu Institute of Organic Chemistry, Chinese Academy of Sciences. Synchrotron SAXS experiments were performed at the Stanford Synchrotron Radiation Laboratory (SSRL, BL4-2), the the Advanced Light Source (beamline 7.3.3), and the Shanghai Synchrotron Radiation Facility (BL16B1). This work was supported in part by the National Natural Science Foundation of China (11074178, 21174138, J1030412) (LY) and Ministry of Education of China (SRF for ROCS 20091001-9-8, SRF for DPHE 20090181120046, FRF for CU WK2060140008) (LY). This work was also supported by NSF CAREER Award DMR-0845592 (KK), NSF DMR1106106 (GW), and NIH UO1 AI082192 (GW).
Funding Sources
Any funds used to support the research of the manuscript should be placed here (per journal style).
Footnotes
ASSOCIATED CONTENT
Supporting Information. GPC traces and results, and additional information on comparison of SMAMPs and AMPs on their <hydrophobicity> and cationic charge percentages. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflict of Interest.
K. Kuroda is a coinventor on a patent application filed by the University of Pennsylvania covering “Antimicrobial Copolymers and Uses Thereof”. The patent application has been licensed to PolyMedix Inc. (Radnor, PA). PolyMedix did not play a role in the design and conduct of this study, in the collection, analysis, or interpretation of the data, or in the preparation, review, or approval of the article.
REFERENCES
- 1.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Hancock REW, Sahl H-G. Nat. Biotech. 2006;24(12):1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 3.Oren Z, Shai Y. Biochemistry. 1997;36(7):1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Mant CT, Farmer SW, Hancock REW, Vasil ML, Hodges RS. J. Biol. Chem. 2005;280(13):12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Won H-S, Jung S-J, Kim HE, Seo M-D, Lee B-J. J. Biol. Chem. 2004;279(15):14784–14791. doi: 10.1074/jbc.M309822200. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Lopez S, Kim H-S, Choi EC, Delgado M, Granja JR, Khasanov A, Kraehenbuehl K, Long G, Weinberger DA, Wilcoxen KM, Ghadiri MR. Nature. 2001;412(6845):452–455. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- 7.Hamuro Y, Schneider JP, DeGrado WF. J. Am. Chem. Soc. 1999;121(51):12200–12201. [Google Scholar]
- 8.Porter EA, Wang X, Lee H-S, Weisblum B, Gellman SH. Nature. 2000;404 doi: 10.1038/35007145. 565-565. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, DeGrado WF. J. Am. Chem. Soc. 2001;123(31):7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]
- 10.Porter EA, Weisblum B, Gellman SH. J. Am. Chem. Soc. 2002;124(25):7324–7330. doi: 10.1021/ja0260871. [DOI] [PubMed] [Google Scholar]
- 11.Patch JA, Barron AE. J. Am. Chem. Soc. 2003;125(40):12092–12093. doi: 10.1021/ja037320d. [DOI] [PubMed] [Google Scholar]
- 12.Tew GN, Liu D, Chen B, Doerksen RJ, Kaplan J, Carroll PJ, Klein ML, DeGrado WF. Proc. Natl. Acad. Sci. USA. 2002;99(8):5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tew GN, Clements D, Tang H, Arnt L, Scott RW. Biochim. Biophys. Acta, Biomembr. 2006;1758(9):1387–1392. doi: 10.1016/j.bbamem.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Choi S, Isaacs A, Clements D, Liu D, Kim H, Scott RW, Winkler JD, DeGrado WF. Proc. Natl. Acad. Sci. U.S.A. 2009;106(17):6968–6973. doi: 10.1073/pnas.0811818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Choi S, Chen B, Doerksen RJ, Clements DJ, Winkler JD, Klein ML, DeGrado WF. Angew. Chem. Int. Ed. 2004;43(9):1158–1162. doi: 10.1002/anie.200352791. [DOI] [PubMed] [Google Scholar]
- 16.Arnt L, Nüsslein K, Tew GN. J. Polym. Sci. A, Polym. Chem. 2004;42(15):3860–3864. [Google Scholar]
- 17.Kuroda K, DeGrado WF. J. Am. Chem. Soc. 2005;127(12):4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 18.Palermo EF, Sovadinova I, Kuroda K. Biomacromolecules. 2009;10(11):3098–3107. doi: 10.1021/bm900784x. [DOI] [PubMed] [Google Scholar]
- 19.Lienkamp K, Madkour AE, Musante A, Nelson CF, Nüsslein K, Tew GN. J. Am. Chem. Soc. 2008;130(30):9836–9843. doi: 10.1021/ja801662y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilker MF, Nüsslein K, Tew GN, Coughlin EB. J. Am. Chem. Soc. 2004;126(48):15870–15875. doi: 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]
- 21.Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. J. Am. Chem. Soc. 2007;129(50):15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 22.Mowery BP, Lindner AH, Weisblum B, Stahl SS, Gellman SH. J. Am. Chem. Soc. 2009;131(28):9735–9745. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]
- 23.Brogden KA. Nat. Rev. Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 24.Shai Y. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1999;1462(1–2):55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki K, Sugishita K-i, Ishibe N, Ueha M, Nakata S, Miyajima K, Epand RM. Biochemistry. 1998;37(34):11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- 26.Huang HW. Biochemistry. 2000;39(29):8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 27.Selsted ME, Ouellette AJ. Nature Immunology. 2005;6(6):551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 28.Ganz T. Nature Reviews Immunology. 2003;3(9):710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda K, Caputo GA, DeGrado WF. Chem.-Eur. J. 2009;15(5):1123–1133. doi: 10.1002/chem.200801523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambhy V, Peterson BR, Sen A. Angew. Chem. Int. Ed. 2008;47(7):1250–1254. doi: 10.1002/anie.200702287. [DOI] [PubMed] [Google Scholar]
- 31.Wei G, Liu X, Yuan L, Ju X-J, Chu L-Y, Yang L. J. Biomater. Sci. Polym. Ed. 2011;22(15):2041–2061. doi: 10.1163/092050610X530982. [DOI] [PubMed] [Google Scholar]
- 32.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. J. Clin. Invest. 1989;84(2):553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skerlavaj B, Romeo D, Gennaro R. Infection and Immunity. 1990;58(11):3724–3730. doi: 10.1128/iai.58.11.3724-3730.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weeks CS, Tanabe H, Cummings JE, Crampton SP, Sheynis T, Jelinek R, Vanderlick TK, Cocco MJ, Ouellette AJ. Journal of Biological Chemistry. 2006;281(39):28932–28942. doi: 10.1074/jbc.M602041200. [DOI] [PubMed] [Google Scholar]
- 35.Falla TJ, Karunaratne DN, Hancock REW. Journal of Biological Chemistry. 1996;271(32):19298–19303. doi: 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- 36.Fujisawa S, Kadoma Y. International Journal of Molecular Sciences. 2012;13(1):758–773. doi: 10.3390/ijms13010758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshii E. Journal of Biomedical Materials Research. 1997;37(4):517–524. doi: 10.1002/(sici)1097-4636(19971215)37:4<517::aid-jbm10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Fujisawa S, Masuhara E. Journal of Biomedical Materials Research. 1981;15(6):787–793. doi: 10.1002/jbm.820150603. [DOI] [PubMed] [Google Scholar]
- 39.Hideji T, Kazuo H. Toxicology Letters. 1982;11(1–2):125–129. [Google Scholar]
- 40.Wimley WC, Creamer TP, White SH. Biochemistry. 1996;35(16):5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 41.White SH, Wimley WC. Annual Review of Biophysics and Biomolecular Structure. 1999;28(1):319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 42.Kim A, Szoka FC., Jr Pharmaceutical Research. 1992;9(4):504–514. doi: 10.1023/a:1015892313856. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Wang G. Nucleic Acids Res. 2004;32(suppl_1):D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GCL. J. Am. Chem. Soc. 2011;133(17):6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra A, Lai GH, Schmidt NW, Sun VZ, Rodriguez AR, Tong R, Tang L, Cheng J, Deming TJ, Kamei DT, Wong GCL. Proc. Natl. Acad. Sci. U.S.A. 2011;108(41):16883–16888. doi: 10.1073/pnas.1108795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GCL. Journal of the American Chemical Society. 2011;133(17):6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harper PE, Gruner SM. The European Physical Journal E: Soft Matter and Biological Physics. 2000;2(3):217–228. [Google Scholar]
- 48.Schmidt NW, Tai KP, Kamdar K, Mishra A, Lai GH, Zhao K, Ouellette AJ, Wong GCL. Journal of Biological Chemistry. 2012;287(26):21866–21872. doi: 10.1074/jbc.M112.358721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao K, Choe U-J, Kamei DT, Wong GCL. Soft Matter. 2012;8(24):6430–6433. doi: 10.1039/C2SM25405K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson DM, Gruner SM, Leibler S. Proceedings of the National Academy of Sciences. 1988;85(15):5364–5368. doi: 10.1073/pnas.85.15.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarz US, Gompper G. Physical Review Letters. 2000;85(7):1472. doi: 10.1103/PhysRevLett.85.1472. [DOI] [PubMed] [Google Scholar]
- 52.Ivankin A, Livne L, Mor A, Caputo GA, DeGrado WF, Meron M, Lin B, Gidalevitz D. Angewandte Chemie International Edition. 2010;49(45):8462–8465. doi: 10.1002/anie.201003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuzaki K. Biochim. Biophys. Acta - Biomembranes. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 54.Yang L, Gordon VD, Mishra A, Som A, Purdy KR, Davis MA, Tew GN, Wong GCL. J. Am. Chem. Soc. 2007;129(40):12141–12147. doi: 10.1021/ja072310o. [DOI] [PubMed] [Google Scholar]
- 55.Yang L, Gordon VD, Trinkle DR, Schmidt NW, Davis MA, DeVries C, Som A, Cronan JE, Jr, Tew GN, Wong GCL. Proc. Natl. Acad. Sci. U.S.A. 2008;105(52):20595–20600. doi: 10.1073/pnas.0806456105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Som A, Tew GN. J. Phys. Chem. B. 2008;112(11):3495–3502. doi: 10.1021/jp077487j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Epand RM, Rotem S, Mor A, Berno B, Epand RF. J. Am. Chem. Soc. 2008;130(43):14346–14352. doi: 10.1021/ja8062327. [DOI] [PubMed] [Google Scholar]
- 58.Epand RF, Umezawa N, Porter EA, Gellman SH, Epand RM. European Journal of Biochemistry. 2003;270(6):1240–1248. doi: 10.1046/j.1432-1033.2003.03484.x. [DOI] [PubMed] [Google Scholar]
- 59.Hristova K, Selsted ME, White SH. Journal of Biological Chemistry. 1997;272(39):24224–24233. doi: 10.1074/jbc.272.39.24224. [DOI] [PubMed] [Google Scholar]
- 60.Epand RF, Raguse TL, Gellman SH, Epand RM. Biochemistry. 2004;43(29):9527–9535. doi: 10.1021/bi049414l. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt NW, Wong GCL. unpublished results [Google Scholar]
- 62.Yeaman MR, Yount NY. Pharmacological Reviews. 2003;55(1):27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 63.Tossi A, Sandri L, Giangaspero A. Peptide Science. 2000;55(1):4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 64.Kuroda K, Caputo GA, DeGrado WF. Chemistry – A European Journal. 2009;15(5):1123–1133. doi: 10.1002/chem.200801523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rinaldi AC, Mangoni ML, Rufo A, Luzi C, Barra D, Zhao H, Kinnunen PKJ, Bozzi A, Di Giulio A, Simmaco M. Biochem. J. 2002;368(1):91–100. doi: 10.1042/BJ20020806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manning GS. The Journal of Chemical Physics. 1969;51(3):924–933. [Google Scholar]
- 67.Manning GS. J. Phys. Chem. B. 2007;111(29):8554–8559. doi: 10.1021/jp0670844. [DOI] [PubMed] [Google Scholar]
- 68.Sanders LK, Xian W, Guaqueta C, Strohman MJ, Vrasich CR, Luijten E, Wong GCL. Proc. Natl. Acad. Sci. USA. 2007;104(41):15994–15999. doi: 10.1073/pnas.0705898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chandler D. Nature. 2005;437(7059):640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.