Abstract
An unconventional approach for DNA fragmentation was investigated to explore its feasibility as an alternative to the existing DNA fragmentation techniques for next-generation DNA sequencing application. Current methods are based on strong-force liquid shearing or specialized enzymatic treatments. There are shortcomings for these platforms yet to be addressed, including aerosolization of genomic materials, which may result in the cross-contamination and biohazards; the difficulty in multiplexing; and the potential sequence biases. In this proof-of-concept study, we investigated the microwave irradiation as a simple, unbiased, and easy-to-multiplex way to fragment genomic DNA randomly. In addition, heating DNA at high temperature was attempted for the same purpose and for comparison. Adaptive focused acoustic sonication was used as the control. The yield and functionality for the DNA fragments and DNA fragment libraries were analyzed to assess the feasibility and use of the proposed approach. Both microwave irradiation and thermal heating can fragment genomic DNA to the size ranges suitable for next-generation sequencing (NGS) shotgun library preparation. However, both treatments caused severe reduction in PCR amplification efficiency, which led to low production in emulsion PCR (emPCR). The result was improved by amplification prior to emPCR. Further improvements, such as DNA strand repairing, are needed for the method to be applied practically in NGS.
Keywords: next-generation sequencing, genome sequencing, random DNA fragmentation
INTRODUCTION
Since the first introduction of massively parallel DNA sequencing by 454 Life Sciences (Roche, Branford, CT, USA) in 2003, so called next-generation sequencing (NGS) has become a robust tool, dominating genomic discovery research.1–5 This revolutionary technology is greatly accelerating comprehensive understanding of molecular diversification and evolution, human development, gene expression and regulation, genetic variation, and alterations related to the progress of diseases and aging. Moreover, NGS technology has promising perspectives and future impacts on medicine, health care, and preventive and clinical medication.6,7 Nevertheless, there are continuous efforts in technical developments to improve efficiency, consistency, speed, and throughput for upstream nucleic acid sample preparation and to improve downstream data mining and analysis.8,9 Sample preparation has been a major challenge for genome-wide discovery, in part, as a result of the substantially greater technical skill required to process samples compared with previous sequencing technologies, as well as the difficulties encountered in processing multiple samples through these more demanding processes in parallel. Parallel sample processing is desirable, not just for throughput increase but also, for better consistency among samples.
One critical technique for NGS library preparation is the random fragmentation of genomic DNA samples. For genomic DNA fragmentation, several physical approaches are currently used with variable success on different platforms and from laboratory to laboratory. Nebulization was used initially with the Roche 454 genome sequencer and Illumina genome analyzer (Illumina, San Diego, CA, USA) and has become less popular, as acoustic shearing with the Covaris adaptive focused acoustic (AFA) system (Woburn, MA, USA) was applied to generate fragments of tunable size ranges (100 bp–3 kb) for fragment library preparation and aquatic mechanical force shearing with HydroShear (Genomic Solutions, Digilab, Marlborough, MA, USA) to generate large fragments (1.5–8 kb) for jumping library preparation. Various ultrasonic baths, including Bioruptor (Diagenode, Liège, Belgium), Sonifier W-450D (Branson UltrasonicsAmericas, Danbury, CT, USA), and UTR200 (Hielscher, Teltow, Germany), were also shown to be used for DNA fragmentation.10 In addition to these physical shearing approaches, specialized enzymatic methods were developed, which include transposon-mediated fragmentation (Epicentre Biotechnologies, Madison, WI, USA) and time-dependent enzymatic digestion by NEBNext dsDNA Fragmentase (New England Biolabs, Ipswich, MA, USA).
In this study, an approach based on a different principle was proposed and investigated for DNA fragmentation. Microwave irradiation is commonly used as a simple, fast-heating source. One example of application in life science is the rapid preparation of cell lysates by heating with microwaves instead of conventional thermal heating.11,12 It was observed that in addition to heating samples, microwave irradiation accelerated chemical reactions significantly, resulting in drastically shortened reaction time. In peptide synthesis, microwave irradiation can greatly reduce reaction time and facilitate difficult synthesis.13 Microwave irradiation has been used in proteomic research to enhance enzymatic digestion of protein samples to rapidly produce peptide fragments for mass spectroscopic analysis14 and in acceleration of DNA binding and hybridization.15 Application of microwave technology to genomic study has yet to be explored but has the potential to become an effective method to overcome some of the limitations of existing DNA fragmentation methods. Here, we report evaluation of microwave irradiation as a controllable fragmentation procedure that can be used reproducibly and in multiplexing format.
MATERIALS AND METHODS
DNA Samples and Molecular Biology Reagents
Human genomic DNA was purified from a healthy blood sample using Advamax genomic DNA preparations kit (Edge BioSystems, Gaithersburg, MD, USA). Bacterial genomic DNA, purified from Escherichia coli type B cells, strain ATCC 11303, was purchased from Affymetrix (Santa Clara, CA, USA). Bacteriophage λ DNA was purchased from New England Biolabs. Plasmid pUC19 DNA was purified by ion exchange using PurElute IEX Plasmid Maxiprep (Edge BioSystems). Standard molecular biology laboratory chemicals, reagents, and supplies were obtained from commercial vendors.
DNA Fragmentation and Analysis
DNA samples were diluted to the final concentration of 50 μg/ml in 10 mM Tris-Cl, 1 mM EDTA, pH 7.5 (TE buffer) and processed for fragmentation, using different methods and operation parameters described below. The Covaris AFA S2 system was used for ultrasonication shearing of DNA with its 1-kb fragmentation program. Thermal degradation of DNA was conducted by incubating DNA at 95°C in the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) for different durations. Microwave irradiation treatment of DNA was performed using the Discover microwave system with the use of septum-sealed, pressurized sample vials (CEM, Matthews, NC, USA). Microwave energy was tuned by controlling the operation temperature (95°C, 120°C, 145°C, or 170°C) and the duration. The processed and the untreated samples were analyzed by electrophoresis on an agarose gel to determine DNA size distribution. PCR amplification and Sanger sequencing using BigDye v3.1 on 3730xl DNA analyzer (Applied Biosystems) were performed as described previously.16
Genomic DNA Fragment Library Preparation and Roche 454 Pyrosequencing
Genomic DNA fragments were cleaned up and subjected to shotgun library preparation by using GS FLX Titanium rapid ligation (RL) library preparation kit (454 Life Sciences, Roche). The DNA fragment RL libraries were purified and examined by real-time PCR assay, using iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) and primers 454LinkerA and 454LinkerB.17 The RL library for 95°C-incubated DNA and the RL library for microwave-processed DNA were subjected to nick translation and amplification using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) and 454LinkerA and 454LinkerB primers at thermocycling conditions of 72°C for 20 min, 95°C for 5 min, 10 cycles of 95°C for 15 s, 58°C for 15 s, and 72°C for 1 min and then, 72°C for 5 min. After purification, the RL libraries were resolved on a 2% agarose gel. DNA, with the size ranging from 500 to 800 bp, was collected and subjected to DNA gel extraction. Size-selected RL libraries were analyzed for the DNA size distribution by using Agilent 2100 bioanalyzer and Agilent high-sensitivity DNA kit (Agilent Technologies, Santa Clara, CA, USA).
RL library copy number concentration determination, emulsion PCR (emPCR), and NGS pyrosequencing, using the Roche GS FLX Titanium system, were performed by following the manufacturer's protocol. NGS data were analyzed using Roche gsAssembler version 2.5.3 software and Geneious Pro v5.6 software (Biomatters, Auckland, New Zealand).
RESULTS
DNA Fragmentation by Microwave Irradiation and Thermal Incubation
Electromagnetic energy from microwave irradiation was applied on genomic DNA to generate DNA fragments with size and quality suited for NGS. DNA of different types and sizes, including plasmid pUC19, bacteriophage λ, E. coli, and human genomic DNA, was able to be broken to fragments reproducibly. The use of different microwave processing parameters resulted in different patterns of fragment size distribution (Fig. 1A–C). For the CEM Discover microwave system used in this study, microwave energy delivered on samples correlated with the temperature setting. The increase of microwave temperature setting, i.e., the elevation of microwave energy load, clearly led to fragment-size distribution, progressively shifting toward lower molecular weights (Fig. 1B). However, prolonging processing time from minutes to hours at the microwave temperature setting of 95°C, i.e., increasing accumulative microwave energy load, showed no or little effect on enhancing DNA fragmentation (Fig. 1A; 95/microwave, Fig. 1C). As microwave irradiation also generates heat and in consequence, increases temperature of DNA solution to differentiate the microwave irradiation effect from the concurrent thermal effect, DNA was incubated at 95°C in the PCR thermocycler. Heating DNA at 95°C resulted in progressive thermal degradation of DNA (Fig. 1A; 95/thermocycler). DNA was fragmented to 100 bp and shorter after, extensive thermal incubation (Fig. 1D). The evident differences indicate that the electromagnetic energy from microwave irradiation has a distinctive effect on DNA molecules in contrast to the thermal degradation effect. To further prove the observation, a second CEM Discover microwave unit with a forced-air cooling function to remove microwave-generated heat was used to irradiate DNA samples with a power setting of 150 W and a control temperature of 95°C. Interestingly, by using cooling to enhance microwave input, DNA was progressively fragmented at 95°C (Fig. 2). It also enables the convenient use of microwave irradiation for DNA fragmentation at a nonpressurized condition and use of usual sample vessels, such as Eppendorf Safe-Lock microtubes. In addition, no apparent, outstanding DNA band was observed for microwave-treated DNA resolved with agarose gel electrophoresis, implicating a uniform, unbiased DNA fragmentation upon microwave irradiation.
FIGURE 1.
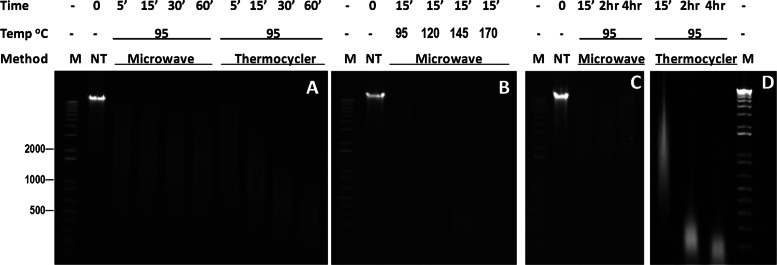
DNA fragmentation with microwave irradiation or direct heating in thermocycler. E. coli genomic DNA solutions were processed using the two methods under the conditions shown above each lane; M, 1 kb plus DNA ladder (Invitrogen); NT, not treated. E-gel agarose electrophoresis (1.2%) was used for the analyses. Two conditions—incubation at 95°C for 5 min and microwave irradiation at 120°C for 15 min—resulted in genomic DNA fragments with size ranges from 500 to 1000 bp.
FIGURE 2.
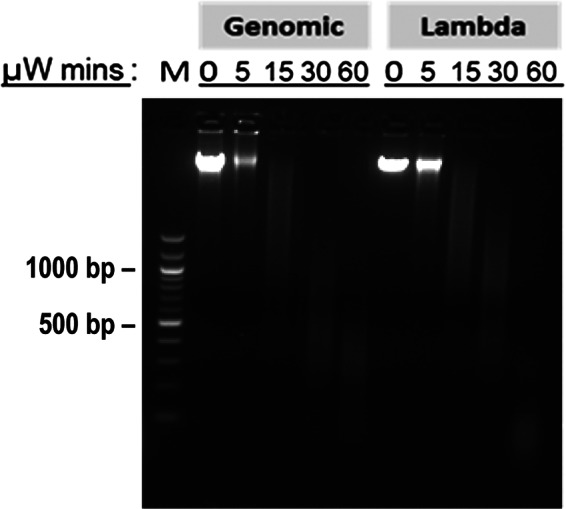
Microwave irradiation of genomic DNA. λ DNA and human genomic DNA in TE were subjected to microwave irradiation (150 W, 95°C) with air cooling for different times, followed by electrophoresis on 2% agarose gel. μW, microwave irradiation; M, 100 bp DNA ladder (New England Biolabs).
Examination of Functionality of the DNA Fragments by PCR and Quantitative PCR (qPCR)
Adverse conditions, such as UV radiation, harsh heating, oxidation, etc., often lead to DNA damages, including apurine/apyrimidine, thymidine dimering, cytosine deamination, and 8-oxo-guanine.18,19 The severe damages may block PCR amplification unless repaired with multiple DNA-modification enzymes, such as PreCR (New England Biolabs). To test sequence integrity of DNA fragmented by microwave, microwave-processed pUC19 samples were amplified by PCR and analyzed by gel electrophoresis. As a control, pUC19 was also sheared using the Covaris AFA S2 system and amplified by PCR. PCR products for 100, 200, and 500 bp fragments of pUC19 were examined for comparison. All amplifications using CEM microwave- or Covaris AFA-processed pUC19 samples as templates were successful and indistinguishable in size and intensity on the agarose gel. Furthermore, all amplified PCR fragments were subjected to Sanger sequencing on Applied Biosystems' 3730xl DNA analyzer. Sequences for all amplicons, totaling 800 bp, are identical to the pUC19 sequence. The results suggest that microwave-irradiated DNA can be amplified, and microwave irradiation does not cause sequence alterations.
Further quantitative comparison was made by SYBR Green real-time PCR using RL libraries as templates and primers corresponding to the RL library adaptors. Three RL libraries for E. coli genomic DNA, processed with Covaris AFA shearing (RL-Sonication), CEM microwave irradiation at 120°C for 15 min (RL-Microwave), or thermocycler heating at 95°C for 5 min (RL-Heating), were first quantified with the 454 RL fluorescence quantitation assay. The assay determines copy number/μl concentration of the RL library by measuring fluorescence-labeled RL adaptors that were ligated to DNA fragments. Gel-size selection was performed for each RL library to collect the fraction in the size range of 500–800 bp (Fig. 3A). For functionality comparison, RL-Sonication was used as standard in a real-time qPCR assay, i.e., RL-Microwave and RL-Heating were quantified against RL-Sonication (arbitrary standard). The effectiveness (efficiency in qPCR) relative to RL-Sonication for RL-Microwave and RL-Heating was assessed by comparing the qPCR assay concentration versus the RL fluorescence assay concentration. The results showed that RL libraries from microwave irradiation and direct thermal heating were not efficient in PCR amplification—only approximately one in 30 as productive in PCR as the control library RL-Sonication (Table 1). In this experiment, microwave irradiation did not show an advantage over direct heating in maintaining DNA structural or chemical stability. One major cause, we speculate, is the heat generated by microwave irradiation and the consequential thermal damage to DNA molecules. Current microwave instruments are designed for reactions at high temperature. It will be interesting to investigate DNA fragmentation by microwave irradiation under low temperature in a microwave system with a robust cooling device, which can quench the heat, or to use nonmicrowave-absorbing solvent instead of water.20 Efficient microwave irradiation under the nonthermal condition will verify explicitly the DNA fragmentation effect of microwave and also enable further study to elucidate the chemical mechanism for electromagnetic energy on DNA macromolecular structures and covalent bonds.
FIGURE 3.
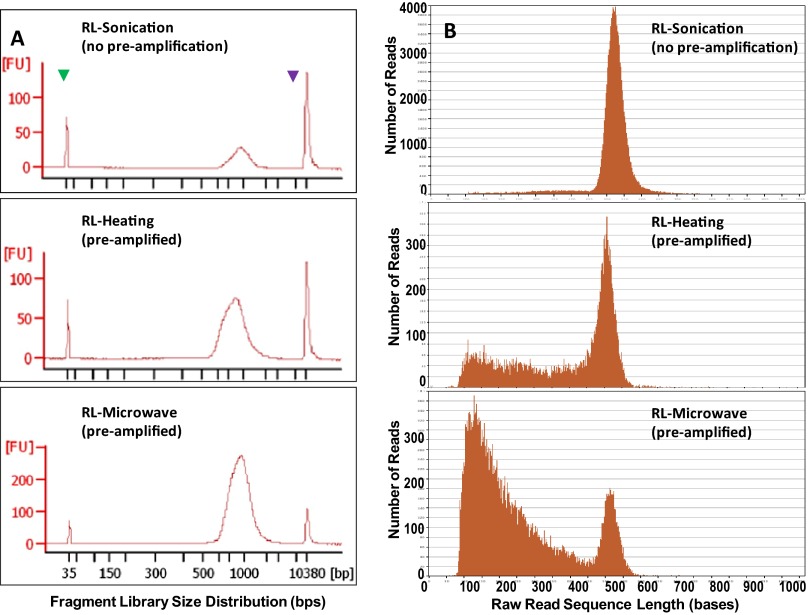
Fragment library-size distributions and Roche 454 sequencing read length graphs. E. coli genomic DNA solutions were processed using Covaris sonication, thermocycler incubation (95°C for 5 min), and microwave irradiation (CEM Discover microwave system; 120°C for 15 min), respectively. (A) Final fragment libraries were analyzed using Agilent bioanalyzer high-sensitivity DNA assay to determine the size distribution. The arrowheads indicate the two internal size markers, 35 bp and 10,380 bp. (B) The fragment libraries were sequenced using the Roche FLX Titanium system. Sequence length distributions for 454 reads were shown for comparison of three DNA fragmentation methods.
Table 1.
Relative PCR Amplification Efficiency
| DNA fragmentation method | Concentration (copies/μl) |
PCR efficiency relative to RL-Sonication | |
|---|---|---|---|
| 454 RL assay | qPCR assay | ||
| Covaris AFA sonication | 5 × 107 | 5 × 107 | 100% |
| Microwave 120°C × 15 min | 5 × 107 | 1.43 × 106 | 2.86% |
| Thermocycler 95°C × 5 min | 5 × 107 | 1.89 × 106 | 3.78% |
E. coli genomic DNA fragment libraries—RL-Sonication, RL-Microwave, and RL-Heating—were first quantified by the 454 RL fluorescence assay and then diluted to 5 × 107and quantified by qPCR using 454 primers and the RL-Sonication library as quantitation standards. Relative PCR efficiency is (qPCR)/(454 RL)%.
RL Library Amplification, emPCR, and Sequencing
emPCR using RL-Microwave and RL-Heating and the purification of Roche 454 pyrosequencing template beads were not successful. The final number of beads produced by following the user's protocol was exceedingly low. Therefore, an additional step prior to emPCR, which was based on Applied Biosystems' SOLiD NGS system's procedure of nick translation and amplification of ligated DNA, was performed to increase the yield of the template beads. The two RL libraries were amplified using primers 454LinkerA and 454LinkerB for 10 PCR thermal cycles, size-selected, quantified by real-time PCR, and used in emPCR. More beads were produced and sequenced in a same 454 run with the beads made for the RL-Sonication, which was size-selected but not amplified. Sequence reads and assembled contig sequences for the three libraries were compared.
All three RL libraries used in emPCR, including RL-Sonication, RL-Microwave, (pre-amplified), and RL-Heating (pre-amplified), were analyzed using the Agilent bioanalyzer. The result shows that the three libraries had almost identical DNA fragment-size distribution (Fig. 3A). However, Roche 454 sequencing read-length distributions for the libraries are very different (Fig. 3B). The control RL-Sonication had an average read length of 515.2 ± 76.2 (mean±sd), with only 6.14% of reads 400 bp or shorter. In contrast, RL-Microwave and RL-Heating had average read lengths of 255.3 ± 141.8 and 385.7 ± 148.7, respectively, and high content of reads 400 bp or shorter at 41.7% and 80.6%, respectively. Sequence data for each RL library were subjected to de novo assembly using Roche gsAssembler software. Sequence comparison for the assembled contigs showed 100% nucleotide identity among three RL-libraries. Moreover, de novo contigs for the control RL-Sonication were used as reference sequence in reference mapping assemblies using Roche gsMapper software for RL-Microwave, RL-Heating, as well as RL-Sonication (re-mapping). There was no nucleotide alteration from the reference in the mapping assembly contigs, and the nucleotide variation for the aligned sequence reads was seemingly random and was likely from the normal sequencing base-calling errors. As we did not remove or destroy the damaged and difficult-to-amplify templates from the RL-Microwave or RL-Heating, the short reads may come from these templates, whereas the long reads may come from pre-amplification products. The speculation is consistent with the difference that the thermocycler incubation was at 95°C for 5 mins, whereas the microwave irradiation was run at 120°C for 15 min—presumably more thermal damages to DNA for RL-Microwave. A potential solution to reduce the short reads is to use DNA modifying enzymes to repair DNA damages prior to PCR pre-amplification or treat PCR pre-amplification products to destroy or digest the damaged templates to small pieces that will be removed in a subsequent size-selection step.
DISCUSSION
In contrast to existing apparatuses that mechanically break DNA, microwave irradiation delivers nonionizing, electromagnetic and thermal energy to solutions of DNA and in so doing, fragments the DNA. The amount of microwave irradiation delivered can be tuned precisely; is distributed uniformly within a large compartment, enabling multiple samples to be processed simultaneously; and readily penetrates through the container into the enclosed solution. Microwave irradiation also effectively relaxes secondary and tertiary structures of macromolecules,14 and in consequence, may enhance more uniform and unbiased DNA cleavage in GC-rich or structural regions. In this study, we showed that microwave irradiation and thermal heating as well can fragment genomic DNA to the size ranges suitable for genomic shotgun library preparation. However, both treatments caused severe reduction in PCR amplification efficiency, which also led to unsuccessful emPCR and low-sequence yield for Roche 454 sequencing applications. The result was improved by a pre-amplification prior to emPCR and nevertheless, needs to be improved further to fulfill the goal of developing an alternative DNA fragmentation approach that has competitive performance and use in NGS.
ACKNOWLEDGMENTS
The study is supported by U.S. National Institutes of Health grant 7R03HG005774 and the U.S. Department of Defense Global Emerging Infections Surveillance (GEIS) program. We thank Mr. Haroun R. Hebron and Drs. Leonard N. Binn, Richard G. Jarman, Stephen J. Thomas, Liang Zhang, and Gnana Rajendran for technical support and critical reading of the manuscript.
DISCLOSURES
The opinions expressed in this work are those of the authors and do not reflect the official policy or position of the U.S. Department of the Army, U.S. Department of Defense, or the U.S. government. We declare that no conflict of interest exists.
REFERENCES
- 1. Loman NJ, Misra RV, Dallman TJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012;30:434–439 [DOI] [PubMed] [Google Scholar]
- 2. Niedringhaus TP, Milanova D, Kerby MB, Snyder MP, Barron AE. Landscape of next-generation sequencing technologies. Anal Chem 2011;83:4327–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 2008;9:387–402 [DOI] [PubMed] [Google Scholar]
- 4. Margulies M, Egholm M, Altman WE. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shendure J, Ji H.Next-generation DNA sequencing. Nat Biotechnol 2008;26:1135–1145 [DOI] [PubMed] [Google Scholar]
- 6. Hutchison CA., III DNA sequencing: bench to bedside and beyond. Nucleic Acids Res 2007;35:6227–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gullapalli RR, Lyons-Weiler M, Petrosko P, Dhir R, Becich MJ, Laframboise WA. Clinical integration of next-generation sequencing technology. Clin Lab Med 2012;32:585–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quail MA, Kozarewa I, Smith F, et al. A large genome center's improvements to the Illumina sequencing system. Nat Methods 2008;5:1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 2012;30:1033–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swanson J. Best practices in GWAS. Genome Technology. New York, NY: GenomeWeb, 2009, http://www.genomeweb.com/node/917734 [Google Scholar]
- 11. Ganzler K, Szinai I, Salgó A.Effective sample preparation method for extracting biologically active compounds from different matrices by a microwave technique. J Chromatogr 1990;520:257–262 [DOI] [PubMed] [Google Scholar]
- 12. Rasmussen JP, Barbez PH, Burgoyne LA, Saint CP. Rapid preparation of cyanobacterial DNA for real-time PCR analysis. Lett Appl Microbiol 2008;46:14–19 [DOI] [PubMed] [Google Scholar]
- 13. Kappe CO, Stadler A. Microwave in Organic and Medical Chemistry. Weinheim, Germany: Wiley-VCH, 2005 [Google Scholar]
- 14. Lill JR, Ingle ES, Liu PS, Pham V, Sandoval WN. Microwave-assisted proteomics. Mass Spectrom Rev 2007;26:657–271 [DOI] [PubMed] [Google Scholar]
- 15. Aslan K, Malyn SN, Bector G, Geddes CD. Microwave-accelerated metal-enhanced fluorescence: an ultra-fast and sensitive DNA sensing platform. Analyst 2007;132:1122–1129 [DOI] [PubMed] [Google Scholar]
- 16. Yang Y, Hebron HR, Hang J.A method for preparing DNA sequencing templates using a DNA-binding microplate. J Biomol Tech 2009;20:165–171 [PMC free article] [PubMed] [Google Scholar]
- 17. Hang J, Forshey BM, Kochel TJ, et al. Random amplification and pyrosequencing for identification of novel viral genome sequences. J Biomol Tech 2012;23:4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bruskov VI, Malakhova LV, Masalimov ZK, Chernikov AV. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res 2002;30:1354–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cadet J, Sage E, Douki T.Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res 2005;571:3–17 [DOI] [PubMed] [Google Scholar]
- 20. Sagripanti JL, Swicord ML, Davis CC. Microwave effects on plasmid DNA. Radiat Res 1987;110:219–231 [PubMed] [Google Scholar]