Abstract
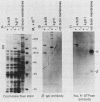
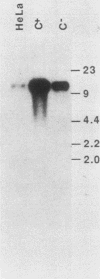
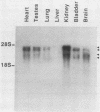
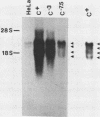
We deduced the complete amino acid sequence of the rat brain Na,K-ATPase beta-subunit from cDNA. The rat brain beta-subunit exhibits a high degree of primary sequence and secondary structural homology with the human and Torpedo beta-subunit polypeptides. Analysis of rat tissue RNA reveals that the beta-subunit gene encodes four separate mRNA species which are expressed in a tissue-specific fashion. In ouabain-resistant HeLa C+ cells, beta-subunit DNA sequences are amplified (approximately 20-fold) and beta-subunit mRNAs are overproduced relative to levels in parental HeLa cells. These results suggest that the beta-subunit plays an important role in Na,K-ATPase structure-function and in the mechanism underlying cellular resistance to the cardiac glycosides.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ash J. F., Fineman R. M., Kalka T., Morgan M., Wire B. Amplification of sodium- and potassium-activated adenosinetriphosphatase in HeLa cells by ouabain step selection. J Cell Biol. 1984 Sep;99(3):971–983. doi: 10.1083/jcb.99.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Emanuel J. R., Garetz S., Schneider J., Ash J. F., Benz E. J., Jr, Levenson R. Amplification of DNA sequences coding for the Na,K-ATPase alpha-subunit in ouabain-resistant C+ cells. Mol Cell Biol. 1986 Jul;6(7):2476–2481. doi: 10.1128/mcb.6.7.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlinger S. Does Na+-K+-atpase have any role in bile secretion? Am J Physiol. 1982 Oct;243(4):G243–G247. doi: 10.1152/ajpgi.1982.243.4.G243. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hall C., Ruoho A. Ouabain-binding-site photoaffinity probes that label both subunits of Na+,K+-ATPase. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4529–4533. doi: 10.1073/pnas.77.8.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert J. J., Schenk D. B., Skelly H., Leffert H. L. Rat hepatic (Na+, K+)-ATPase: alpha-subunit isolation by immunoaffinity chromatography and structural analysis by peptide mapping. Biochemistry. 1986 Jul 15;25(14):4156–4163. doi: 10.1021/bi00362a025. [DOI] [PubMed] [Google Scholar]
- Jørgensen P. L. Mechanism of the Na+, K+ pump. Protein structure and conformations of the pure (Na+ +K+)-ATPase. Biochim Biophys Acta. 1982 Aug 11;694(1):27–68. doi: 10.1016/0304-4157(82)90013-2. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Nojima H., Ohta T., Nagano K. Molecular cloning and sequence analysis of human Na,K-ATPase beta-subunit. Nucleic Acids Res. 1986 Apr 11;14(7):2833–2844. doi: 10.1093/nar/14.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Kyte J. Molecular considerations relevant to the mechanism of active transport. Nature. 1981 Jul 16;292(5820):201–204. doi: 10.1038/292201a0. [DOI] [PubMed] [Google Scholar]
- Noguchi S., Noda M., Takahashi H., Kawakami K., Ohta T., Nagano K., Hirose T., Inayama S., Kawamura M., Numa S. Primary structure of the beta-subunit of Torpedo californica (Na+ + K+)-ATPase deduced from the cDNA sequence. FEBS Lett. 1986 Feb 17;196(2):315–320. doi: 10.1016/0014-5793(86)80270-8. [DOI] [PubMed] [Google Scholar]
- Pauw P. G., Johnson M. D., Moore P., Morgan M., Fineman R. M., Kalka T., Ash J. F. Stable gene amplification and overexpression of sodium- and potassium-activated ATPase in HeLa cells. Mol Cell Biol. 1986 Apr;6(4):1164–1171. doi: 10.1128/mcb.6.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. W., Mercer R. W., Caplan M., Emanuel J. R., Sweadner K. J., Benz E. J., Jr, Levenson R. Molecular cloning of rat brain Na,K-ATPase alpha-subunit cDNA. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6357–6361. doi: 10.1073/pnas.82.18.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweadner K. J., Gilkeson R. C. Two isozymes of the Na,K-ATPase have distinct antigenic determinants. J Biol Chem. 1985 Jul 25;260(15):9016–9022. [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]