Summary
PIK3CA, which encodes the p110α catalytic subunit of phosphatidylinositol 3-kinase α, is frequently mutated in human cancers. Most of these mutations occur at two hot-spots E545K and H1047R located in the helical domain and the kinase domain, respectively. Here, we report that p110α E545K, but not p110α H1047R, gains the ability to associate with IRS1 independent of the p85 regulatory subunit, thereby rewiring this oncogenic signaling pathway. Disruption of the IRS1-p110α E545K interaction destabilizes the p110α protein, reduces AKT phosphorylation, and slows xenograft tumor growth of a cancer cell line expressing p110α E545K. Moreover, a hydrocarbon-stapled peptide that disrupts this interaction inhibits the growth of tumors expressing p110α E545K.
Introduction
PIK3CA is frequently mutated in a variety of human cancers including colorectal cancers (CRCs) (Bachman et al., 2004; Broderick et al., 2004; Campbell et al., 2004; Lee et al., 2005; Levine et al., 2005; Li et al., 2005; Oda et al., 2005; Samuels et al., 2004). PIK3CA encodes the catalytic subunit of phosphatidylinositol 3-kinase α (PI3Kα), which plays a key role in regulating cell proliferation, survival and motility (Liu et al., 2009; Zhao and Vogt, 2008a). PI3Kα consists of a catalytic subunit p110α and one of several regulatory subunits (a major one being p85α). The p110α subunit contains an N-terminal adaptor-binding domain (ABD), a Ras-binding domain (RBD), a C2 domain, a helical domain and a catalytic domain (Amzel et al., 2008). In the basal state, the regulatory p85 subunit stabilizes the catalytic p110α subunit and inhibits its enzymatic activity (Vadas et al., 2011). Upon growth factor stimulation, the SH2 domains of p85 bind to the phospho-tyrosine residues on the receptor protein kinases or adaptor proteins such as insulin receptor substrate 1 (IRS1), thereby activating the lipid kinase activity of PI3Kα (Cantley, 2002). Activated PI3Kα converts phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3). The second messenger PIP3 then activates downstream AKT signaling (Cantley, 2002).
Most p110α mutations occur at two hot spot regions: an acidic cluster (E542, E545 and Q546) in the helical domain and a histidine residue (H1047) in the kinase domain. E545K and H1047R are the two most frequently observed p110α somatic mutations in human cancers. Interestingly, several recent studies indicate that E545K and H1047R mutations exert their oncogenic functions through distinct mechanisms. Zhao and Vogt have shown that the helical domain and the kinase domain mutant proteins could synergistically transform chicken embryonic fibroblasts (Zhao and Vogt, 2008b), suggesting that the two mutations may induce oncogenic transformation through different pathways. Consistently, they further demonstrated that the p110α helical domain mutants require the Ras-binding domain for their transformation activities whereas the oncogenic activity of the H1047R mutant depends on the p85-binding domain. Pang et al. showed that expression of p110α E545K produced a more severe metastatic phenotype than that induced by expressing p110α H1047R in a breast cancer cell line (Pang et al., 2009). The p110α H1047R, but not p110α E545K, is found to enhance HER2-mediated transformation of immortalized mammary epithelial cells (Chakrabarty et al., 2010). Structural analysis indicates that the p110α H1047R mutation alters the interaction between PI3Kα and the cell membrane, thereby activating its kinase activity (Mandelker et al., 2009). It has been suggested that the helical domain mutations activate their enzymatic activities by disrupting the inhibitory effect of the p85 subunits (Huang et al., 2007; Miled et al., 2007). Here we set out to determine how p110α E545K and p110α H1047R differentially activate oncogenic signaling pathways.
Results
The p110α E545K mutant, but not the WT p110α, associates with IRS1
DLD1 is a CRC cell line that carries a wild-type (WT) PIK3CA allele and a PIK3CA E545K allele. DLD1 derivatives carry only the endogenous WT or the mutant PIK3CA allele had been generated previously (Figure 1A) (Samuels et al., 2005). To identify proteins that may differentially bind to WT and E545K mutant p110α, we used recombinant adeno-associated virus (rAAV)-mediated homologous recombination to tag the endogenous WT or mutant p110α with 3xFLAG at their C-termini in DLD1 cells (Figure 1A and Figure S1 A and B) (Zhang et al., 2008). Under serum-starvation conditions, antibodies against FLAG immunoprecipitated a protein of ~ 170 kDa from 3xFLAG-tagged p110α E545K knock-in (KI) cells, but not from the 3xFLAG-WT p110α KI cells (Figure. 1B). Analysis by mass spectrometry identified this protein as IRS1. This IRS1-p110α E545K interaction was validated by immunoprecipitation under serum-starvation conditions in three different settings. In KI DLD1 cells, IRS1 was co-immunoprecipitated with p110α E545K but not the WT p110α (Figure 1C and Figure S1C). Moreover, when IRS1 was immunoprecipitated from various DLD1 derivatives, p110α E545K predominantly associated with IRS1 (Figure 1D and Figure S1D). Furthermore, IRS1 strongly associated with mutant p110α in the mutant only DLD1 cells but its interaction with the WT p110α was barely detectable in the WT only cells (Figure 1D and Figure S1D).
Figure 1. The p110α E545K mutant proteins associate with IRS1.
(A). Schematics of DLD1 genetically engineered cell lines. ABD: adaptor-binding domain; RBD: Ras-binding domain; C2: C2 domain; helical: helical domain; kinase: kinase domain. WT-FLAG: DLD1 cells with the endogenous wild-type p110αy tagged with 3xFLAG. Mutant-FLAG: the endogenous p110α E545K tagged with 3xFLAG. WT only: DLD1 cells with the mutant allele of PIK3CA deleted by homologous recombination. Mutant only: DLD1 cells with the WT allele of PIK3CA deleted as described in (Samuels et al., 2005).
(B). The p110α WT or mutant FLAG tagged cells were grown to full confluence and serum-starved for 24 hr before harvest. Cell lysate from the two cell lines were immunoprecipitated with antibody-conjugated beads against FLAG, resolved on a 4–20% gradient SDS-PAGE gel, and stained with Gelcode Blue Reagent.
(C, D). Lysate from cells of the indicated genotype were immunoprecipitated with antibodies against either FLAG (C) or IRS1 (D). The immunocomplexes were blotted with the indicated antibodies. Arrowheads indicate the p110α E545K and asterisks indicate the WT p110α.
(E). HEK 293 cells were transfeted with a construction expressing FLAG-tagged WT p110α or the indicated tumor-derived p110α mutant together with one expressing MYC-tagged IRS1, cells were grown 2 days post-transfection and then serum-starved for 24 hr. Cell lysates were immunoprecipitated with anti-MYC antibodies and blotted with the indicated antibodies. See also Figure S1.
p110α helical domain mutants, but not the kinase domain mutants, interact with IRS1
In addition to the hot-spot mutations in the helical and kinase domains, cancer-derived mutations also occur in the ABD and C2 domains. We thus proceeded to test whether gain of interaction with IRS1 occurs with other p110α mutations. We constructed a FLAG-tagged p110α expression plasmid and generated frequently observed tumor-derived p110α mutations by site-directed mutagenesis. The p110α WT or mutant plasmids were co-expressed with a MYC-tagged IRS1 construct in HEK 293 cells for immunoprecipitations. As shown in Figure 1E and Figure S1E, in addition to the E545K mutation, other hot-spot p110α mutations in the helical domain, including the E542K, E545A, E545G and Q546K, gain interaction with IRS1 under serum-starvation conditions. In contrast, however, hot-spot mutations in the kinase domain, H1047R, H1047L and G1049R, failed to interact with IRS1. Interestingly, relatively rare mutations including K111N in the ABD domain, N345K in the C2 domain and M1043L in the kinase domain, also interact with IRS1, whereas other rare mutations including R88Q in the ABD domain, C420R mutations in the C2 domain and P539R in the helical domain, failed to interact with IRS1 (Figure 1E).
Interaction between the helical domain mutant p110α and IRS1 does not require p85 or IRS1 phosphorylation
Given that p110α is normally brought to the IRS1 complex through the interaction between the SH2 domain of p85 and phosphorylated tyrosine (pY) residues of IRS1, it was important to determine whether the p110α E545K-IRS1 interaction required p85 and tyrosine phosphorylation of IRS1. As shown in Figure 2A and Fig S2A, p110α E545K, but not the WT p110α, robustly associated with IRS1 under serum-starvation conditions in which tyrosine phosphorylation of IRS1 was undetectable, suggesting that the p110α E545K-IRS1 interaction is independent of IRS1 tyrosine phosphorylation. It appeared that the p110α E545K-IRS1 interaction also did not require p85, as a truncated p110α E545K lacking the ABD domain required for p85 binding (Zhao and Vogt, 2008b) still associated with IRS1 (Figure 2B and Figure S2B). We then proceeded to determine whether p85 proteins were required for the interaction between p110α E545K and IRS1under physiological conditions. This was accomplished by knocking out PIK3R2 (encodes p85β) in the p110α mutant only DLD1 cells using rAAV-mediated homologous recombination (Figure 2C & D and Figure S2C) and knocking down p85α with a siRNA in the p85β knockout (KO) cells (Figure 2E). As shown in Figure 2E and Figure S2D, ablation of p85α and p85β enhanced rather than inhibited the interaction between p110α E545K and IRS1, suggesting that not only p85 proteins were not required but may compete with IRS1 for binding to p110α E545K. However, ablation of both p85 proteins led to destabilization of p110α protein (Figure 2E). Together, our data suggest that p85 proteins are not required for the interaction between p110α E545K and IRS1 but still play a role in the protein complex through stabilization of the mutant p110α. Consistently, p85 proteins were co-immunoprecipitated with p110α E545K (Figure 1C and Figure 2A). Furthermore, gel filtration analysis showed that p85 and p110α E545K formed a protein complex that was larger than that formed by p85 and the WT p110α (Figure S2E). Interestingly, p110α E545K completely co-fractionated with IRS1, while only a portion of p85 proteins co-fractionated with p110α E545K and IRS1 (Figure S2E). This result shows that the p110α E545K mutation changes protein complex formation.
Figure 2. The p110α E545K mutant-IRS1 interaction does not require p85 proteins.
(A). DLD1 cells expressing either the wild-type allele (WT only) or the E545K mutant allele (Mutant only) were serum-starved for 16 hr and then treated with either 1 μg/ml of insulin (I) or pervanadate (V) for 15 min, or left without treatment (−). Cell lysates were immunoprecipitated with anti-IRS1 antibodies and blotted with the indicated antibodies.
(B). Myc-tagged IRS1 plasmid was co-transfected with a FLAG-tagged full-length p110α E545K mutant plasmid (FL), an ABD domain deletion construct of the FLAG-tagged p110α E545K mutant (ΔABD), a FLAG-tagged full-length WT p110α or an ABD domain deletion construct of the FLAG-tagged WT p110α. Cells were grown for 2 days post-transfection and then serum-starved for 24 hr. Cell lysates were immunoprecipitated with anti-FLAG antibodies and blotted with the indicated antibodies.
(C). Schematic of the PIK3R2 knockout strategy is shown.
(D). Cell lysates of the parental p110α E545K mutant only cells and two independently-derived PIK3R2 KO clones were blotted with the indicated antibodies.
(E). p110α E545K mutant only cells of the indicated genotypes were transfected with ether a control siRNA or a siRNA against p85α. Cell lysates were immunoprecipitated with anti-p110α antibodies and blotted with the indicated antibodies.
(F). Purified recombinant His-tagged Myc-IRS1, WT p110α and p110α E545K mutant proteins expressed in Sf9 cells were resolved on SDS-PAGE gels. Asterisk indicates IRS1 proteins and arrow indicates WT and mutant p110α proteins.
(G). The indicated proteins were mixed in vitro and the mixtures were immunoprecipitated with an anti-Myc antibody and blotted with the indicated antibodies. Short: short exposure; Long: long exposure. See also Figure S2.
To test if IRS1 directly binds to p110α E545K, we expressed recombinant IRS1, WT p110α and p110α E545K in Sf9 cell and purified these proteins to homogeneity (Figure 2F). As shown in Figure 2G and Fig S2F, recombinant IRS1 bound directly in vitro with p110α E545K but not WT p110α, providing further evidence that the IRS1-p110α E545K interaction is not mediated by p85 proteins.
Specific regions of IRS1 are required for its interaction with p110α E545K
The altered interaction properties of the mutant forms of p110α prompted us to map the minimal essential regions of IRS1 required for its interaction with p110α E545K mutant protein. For this study, we made a series of 6xHis-tagged IRS1 truncation constructs (Figure 3A and C). The His-tagged IRS1 proteins were first expressed in E. coli and purified with Ni-NTA beads. Then the purified proteins were used to pull down p110α from DLD1 cells expressing either the WT p110α only or the p110α E545K mutant only. With these proteins, we first mapped the mutant p110α interaction domain to the middle part of IRS1 [amino acid (AA) 585–962] (Figure 3B and Figure S3A). This fragment contains three YxxM motifs that mediate the pY-dependent IRS1-p85 protein interaction. We thus constructed an unphosphorylatable IRS1Y3F mutant fragment (Figure 3A). As shown in Figure 3B and Figure S3A, this mutant fragment still bound to p110α E545K, providing further evidence that the protein interaction between IRS1 and p110α E545K mutant is independent of IRS1 phosphorylation. Moreover, we showed that the middle fragment of IRS1 pulled down the p110α E545K, but not the WT counterparts (Figure 3B). Then, we further mapped the interaction domains to two small regions (AA 607–623 and AA 936–962) in the middle part of IRS1 using a combination of 6xHis-tag and GST-fusion constructs (Figure 3C–F and Figure S3 B and C). When expressed in HEK293 cells, a deletion construct of IRS1 (IRS1Δ) devoid of both AA 607–623 and AA 936–962 regions greatly reduced its interaction with p110α E545K under both normal tissue culture and serum-starvation conditions (Figure 3G and H, Figure S3 D to G). In contrast, neither of the single region deletions could interrupt IRS1-mutant p110α interaction (Figure S3 E and F). Importantly, however, IRS1Δ did not affect IRS1-p85 interaction (Figure 3H and Fig, S3D), providing further supporting evidence that IRS1-mutant p110α interaction does not require p85. Finally, similar results were also observed when the IRS1Δ was expressed in p110α E545K 3xFLAG KI DLD1 cells (Figure S3H).
Figure 3. Mapping regions in IRS1 required for its interaction with p110α mutant protein.
(A, C, E, G). Schematic diagrams of IRS1 deletion proteins.
(B). His-tagged IRS1 constructs as shown in (A) were expressed in E. coli and purified using Ni-NTA beads. The beads-bound purified recombinant proteins were incubated with lysates made from either the p110α mutant only or WT only DLD1 cells. The pull-down mixtures were blotted with an anti-p110α antibody.
(D). Recombinant 6xHis tagged IRS1 proteins shown in (C) were incubated with cell lysates made from the p110α mutant only DLD1 cells. The pull-down mixtures were blotted with an anti-p110α antibody.
(F). GST-fusion proteins shown in (E) were expressed in E. coli and purified with Glutathione Sepharose beads. The bead bound proteins were incubated with lysates made from the p110α mutant only DLD1 cells. The pull-down mixtures were blotted with an anti-p110α antibody.
(H). MYC-tagged full-length IRS1 or the IRS1Δ construct was co-transfected with either a WT p110α plasmid or a p110α E545K mutant plasmid and a HA-tagged p85 plasmid into HEK 293 cells. Cells were grown under normal culture conditions for 48 hr and harvested for immunoprecipitation with anti-MYC antibody. Immunocomplexes were blotted with the indicated antibodies. See also Figure S3.
IRS1 stabilizes p110α
Next, we set out to develop a system to test whether disruption of IRS1-mutant p110α interaction affects tumorigenicity. To this end, we first knocked out IRS1, a single exon gene, from DLD1 cells (IRS1 KO) using rAAV-mediated homologous recombination (Figure 4A and B). Two independently-derived IRS1 KO clones were then chosen for further analyses. Interestingly, p110α, but not p85, protein levels were reduced in the IRS1 KO cells in comparison to the parental cells (Figure 4B and C; Figure S4A), suggesting that IRS1Δ stabilizes p110α. It appears that the interaction between IRS1 and p110α E545K is required to stabilize p110α E545K, co-expression of IRS1 with p110α in Sf9 insect cells stabilizes p110α E545K but not the WT p110α (Figure 4D and Figure S4B).
Figure 4. IRS1 stabilizes p110α E545K mutant protein and brings it to cell membrane.
(A). Schematic of IRS1 KO targeting strategy.
(B). Cell lysates from parental and IRS1 KO cells were blotted with antibodies against IRS1, p110α, pan p85 and actin. KO-1 and KO-2 are two independently-derived IRS1 KO clones.
(C). RT-PCR analysis of PIK3CA expression in parental and IRS1 KO cells.
(D). Sf9 insect cells were infected or co-infected with viruses expressing indicated proteins. Cell lysates were blotted with the indicated antibodies.
(E, F). Cell lysates from p110α E545K mutant only cells with IRS1 WT or IRS1 KO were fractionated into cytosolic (Cyto) and cytoplasmic membrane (Mem) fractions then blotted with the indicated antibodies (F). Percentages of membrane-bound p110α are shown in (G). Quantitative measurements are plotted as mean ± SEM. ** p = 0.0006; t test. See also Figure S4.
IRS1-p110α E545K interaction brings the mutant p110α to cytoplasmic membrane
Given that membrane association of p110α is important for its activation, we then determined how IRS1 impacted association of the mutant p110α protein with the membrane. Indeed, more p110α E545K bound to cytoplasmic membrane than the WT proteins (Figure S4C). Importantly, knockout of IRS1 in the p110α E545K mutant only DLD1 cells reduced the amount of membrane-bound p110α mutant proteins (Figure 4E and F), supporting the conclusion that interaction between IRS1 and p110α E545K enhances the association of p110α E545K with the cytoplasmic membrane.
The p110α E545K-IRS1 interaction is required for in vivo tumor growth
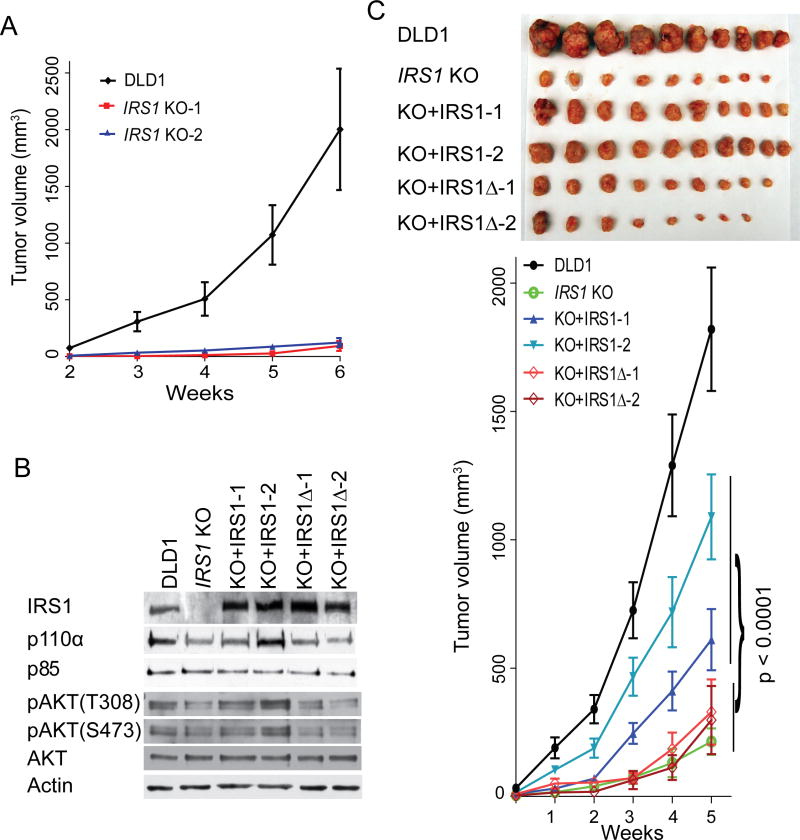
The IRS1 KO cells formed much smaller xenograft tumors in athymic mice (Figure. 5A, although IRS1 KO did not affect clonogenicity and anchorage-independent growth of DLD1 cells under normal tissue culture conditions (data not shown). We reconstituted one of the two IRS1 KO clones with either full-length Myc-IRS1 or Myc-IRS1Δ expression plasmid. Two stable clones expressing each construct were chosen for in-depth analyses. Expression levels of IRS1 in the IRS1Δ reconstituted clones were similar to those of IRS1 reconstituted clones (Figure 5B). Consistent with the hypothesis that IRS1 stabilizes p110α, IRS1, but not IRS1Δ, restored the p110α level to comparable to that observed in parental cells (Figure 5B and Figure S5A). Moreover, AKT phosphorylation levels were also increased in IRS1 reconstituted cells in comparison to that of IRS1 KO and IRS1Δ reconstituted clones under serum starvation (Figure 5B and Figure S5B), suggesting that stabilization of p110α by IRS1 activates down-stream signaling. Importantly, both IRS1 reconstituted clones and IRS1Δ reconstituted clones responded to insulin stimulation (Figure S5C), suggesting that IRS1 does not impair cell signaling pathways activated by growth factors. Notably, Figure 5C show that IRS1 reconstituted clones formed significantly bigger xenograft tumors than IRS1Δ reconstituted clones. Consistently, the IRS1-p110α interaction was also impaired in the xenograft tumors formed by the IRS1Δ reconstituted cells (Figure S5D), which led to reduced phosphorylation of AKT and Foxo1 proteins (Figure S5E). These results suggest that IRS1-p110α E545K interaction is crucial for the mutant p110α to exert its oncogenic functions. Unexpectedly, the full-length IRS1 reconstituted cells formed smaller tumors than the parental cells. We postulate that this difference could be caused by differences in regulating IRS1 between the parental cells and the IRS1 reconstituted cells. The ectopically expressed IRS1 in the IRS1 reconstituted cells may not recapitulate the complex regulation of the endogenous IRS1. In addition, the ectopically expressed IRS1 protein has 3xMyc tag at the N-terminus of the protein, which may not provide full-function equivalent to the endogenous IRS1 proteins.
Figure 5. Disruption of p110α E545K interaction with IRS1 impairs tumorigenicity.
(A). Three million cells of the indicated genotypes were injected subcutaneously into athymic nude mice. Tumor sizes were measured weekly for five week starting 2 weeks after injection.
(B). IRS1 KO DLD1 cells were reconstituted with either full-length IRS1 or IRS1Δ. Cell lysates of two stable clones from each reconstitution were blotted with the indicated antibodies.
(C). Xenograft of the indicated cell lines were established as described in (A). Tumor sizes were measured weekly for five weeks. Mice were then sacrificed and tumors were harvested and photographed. The p value was calculated using ANOVA analysis. All quantitative measurements are plotted as mean ± SEM. See also Figure S5.
A peptide derived from p110α E545K disrupts the p110α helical domain mutant-IRS1 interaction
Because our results indicated that the p110α E545K-IRS1 interaction is required for in vivo tumor growth of the mutant CRC cells, we set out to test whether p110α-derived peptides encompassing the mutation site could be constructed to disrupt this interaction. Based on the crystal structure of p110α (Huang et al., 2007), we first synthesized a 30-AA E545K mutant peptide and its WT counterpart (Figure S6A). These amino acids form two α-helix domains flanking the mutation site. As shown in Figure S6A, the 30-AA mutant peptide inhibited the interaction between p110α E545K and IRS1 when added into cell lysates, whereas the WT peptide failed to cause any inhibition. We then tested if peptides corresponding to only one of the two α-helical motifs could inhibit interactions between the mutant p110α and IRS1. These experiments demonstrated that an 18-AA mutant peptide corresponding to the C-terminal α-helical motif displayed moderate inhibition in cell lysates (Figure S6B), while the N-terminal 21-AA mutant peptide had no effect (Figure S6B).
To improve the potency of the peptide, we converted the aforementioned 18-AA mutant peptide to a stapled macrocyclic derivative as shown in Figure 6A. This approach exploited the well-documented concept that peptide stapling can stabilize the α-helical conformation of a linear peptide and potentially enhance binding affinity toward specific targets (Walensky et al., 2004). Moreover, this approach was taken because stapled peptides are often cell-permeable (Walensky et al., 2004). Strikingly, our stapled mutant peptide almost completely disrupted the IRS1-p110α E545K interaction in cell lysates, but had no effects on p110α-p85 interaction (Figure S6C). In contrast, the corresponding stapled WT p110α peptide did not inhibit IRS1-p110α E545K interactions (Figure S6C). Interestingly, as shown in Figure S6D, the stapled mutant peptide also inhibited the interaction between IRS1 and ΔABD p110α E545K. Thus this result provided further evidence that the protein interaction between IRS1 and p110α E545K mutant is not mediated by p85.
Figure 6. A stapled p110α E545K mutant peptide disrupts the interaction between the mutant p110α and IRS1.
(A). Schematic diagram of linear and stapled WT and mutant peptides derived from p110α.
(B). Linear or stapled WT and mutant peptide were added into culture medium of the p110α E545K 3xFLAG KI cells for 16 hr. Cell lysates were immunoprecipitated with beads conjugated with anti-FLAG antibodies and then blotted with the indicated antibodies.
(C). Vaco481 CRC cells were treated with the stapled mutant peptides at the indicated concentration. Cell lysates were immunoprecipitated with beads conjugated with anti-FLAG antibodies and then blotted with the indicated antibodies.
(D). DLD1 cells expressing only p110α E545K were treated with the indicated stapled peptide. Cell lysates were blotted with the indicated antibodies.
(E). The indicated cancer cell lines were treated or untreated with stapled WT or mutant peptide at 50 μM for 2 hr. Cell lysates were blotted with the indicated antibodies.
(F). The indicated cancer cell lines were treated with or without 50 μM of stapled mutant peptide for 2 hr. Cell lysates were blotted with the indicated antibodies. See also Figure S6.
Importantly, when added to cell culture medium, the stapled mutant peptide effectively disrupted IRS1-p110α E545K interaction in DLD1 cells at 50 μM concentration (Figure 6B and Figure S6E), demonstrating both targeting activity and cell permeability of this stapled peptide. Furthermore, neither the linear mutant peptide nor the stapled WT peptide affected the interaction between p110α E545K and IRS1 when added to cultured DLD1 cells (Figure 6B), providing strong support of the specificity and efficacy of the stapled mutant peptide. Moreover, the p110α E545K stapled peptide also disrupted protein interaction between p110α Q546P and IRS1 in Vaco481 CRC cells (Figure 6C and Figure S6F), suggesting that these helical domain mutant proteins share a common critical protein interaction interface with IRS1. In support of earlier results, it was also notable that the stapled mutant peptide did slightly reduce the protein levels of p110α, but did not affect levels of p85 and IRS1 (Figure 6D and Figure S6G), providing further evidence that the protein interaction between mutant p110α and IRS1 stabilizes p110α. Finally, consistent with the in vitro results, the stapled mutant peptide did not interfere with cellular p110α-p85 interaction or IRS1 phosphorylation (Figure 6B and D).
The p110α E545K mutant peptide reduces AKT phosphorylation in cancer cell lines with p110α helical domain mutations
We next determined the effect of the stapled peptides on the downstream signaling of PI3K. As shown in Figure 6E and Figure S6H, the stapled mutant peptide, but not the WT counterpart, reduced AKT phosphorylation in mutant only DLD1 cells, whereas the mutant peptide did not affect AKT phosphorylation in WT only DLD1 cells. Moreover, treatment of cancer cell lines (DLD1, H460 and MDA-MB361) harboring p110α E545K (Vasudevan et al., 2009) and the Vaco481 cell line harboring p110α Q546P with the stapled mutant peptide resulted in reduced AKT phosphorylation at both T308 and S473 residues (Figure 6F and Figure S6I). Again, in contrast, the stapled mutant peptide had no effect on AKT phosphorylation in cancer cell lines (HCT116, RKO and T47D) harboring p110α H1047R (Figure 6F). Importantly, the stapled mutant peptide has no effect on the kinase activity of recombinant p110α, either alone or in the presence of IRS1, in vitro (Figure S6 J and K). Consistent with previous reports that phospho-IRS1 peptide does not enhance the lipid kinase activity of p110α E545K (Carson et al., 2008; Miled et al., 2007), full-length IRS1 proteins also had no effect on the lipid kinase activity of p110α E545K in vitro (Figure S6 I and K). Thus, the ability of stapled mutant peptide to reduce AKT phosphorylation in cancer cells harboring p110 E545K is not due to direct inhibition of the lipid kinase activity of p110α but by disrupting interaction between the p110α mutant and IRS1. Consistent with the notions that disruption of IRS1-p110α helical domain mutant protein interaction does not perturb AKT activation induced by growth factors, the mutant stapled peptide had no effect on AKT phosphorylation when these cell lines were stimulated by insulin (Figure S6 L and M).
The stapled p110α E545K mutant peptide specifically inhibits in vivo tumor growth of cancer cells with the mutation
Finally, we tested whether the stapled mutant peptide can inhibit xenograft tumor growth of DLD1 cells. Peptides were directly injected into xenograft tumors when the tumors reached ~ 100 to 150 mm3. To better control the experiment, we injected the stapled WT peptide into tumors on the left flanks and the stapled mutant peptide into tumors on the right flanks of the same nude mice (Figure 7A). Tumors of similar size were also injected with equal volume of water as controls. As shown in Figure 7B, the stapled mutant peptide treatment significantly slowed DLD1 xenograft tumor growth in comparison with the control group. In contrast, the stapled WT peptide did not inhibit xenograft tumor growth of DLD1 cells. Moreover, neither the stapled WT nor the stapled mutant peptide inhibited xenograft tumor growth of HCT116 CRC cells that harbor p110α H1047R (Figure 7C and D). In the xenograft tumors, the stapled mutant peptide treatment led to reduced AKT and Foxo1 phosphorylation (Figure S7). Unlike in culture cells where treatment of the stapled mutant peptide reduced AKT phosphorylation at both T308 and S473 residues (Figure 6E and F), peptide treatment in xenograft tumors only decreased AKT phosphorylation at the S473 residue (Figure S7), suggesting that microenvironment in xenograft tumors is more complex than in vitro culture conditions. Consistent with our observation, several studies have indicated that phosphorylation of ATK T308 and S473 are regulated differentially (Hemmings and Restuccia, 2012).
Figure 7. Stapled p110α E545K mutant peptide inhibits xenograft tumor growth of cancer cells harboring the p110α E545K mutation.
(A, B). Five million DLD1 cells were injected subcutaneously and bilaterally into athymic nude mice. Once the xenograft tumors reach 100 to 150 mm3, the left side tumors were injected with the stapled WT peptide and the right side tumors were injected with the stapled mutant peptide daily for 14 days. Ten tumors of similar size in five other athymic nude mice were injected with equal amount of water as controls. Tumor sizes were measured every two days. Representative pictures of mice injected with the peptides are shown in (A) and quantitative measurements are shown in (B).Green arrows indicate tumors treated with the stapled WT peptide and the red arrows indicate tumors treated with the stapled mutant peptide. The p value was calculated using ANOVA analysis.
(C, D). Identical experiments as described above were performed with HCT116 cells. All quantitative measurements are plotted as mean ± SEM. See also Figure S7.
Discussion
Our data demonstrate that the p110α helical domain mutant proteins gain interaction with IRS1 in the absence of growth factor stimulation and that this mutant-specific protein interaction is independent of binding of p85 to phospho-IRS1 (see Figure 8 for our proposed model). Therefore, our study establishes a paradigm in which a tumor-derived mutant protein exerts its oncogenic functions by rewiring an oncogenic signaling pathway. Our observations also have important therapeutic implications. The discovery of frequent mutations of PIK3CA in human cancer provides a strong rationale for inhibition of mutated p110α activities for targeted cancer therapy (Samuels et al., 2005). However, it has remained a challenge to develop p110α isoform-specific inhibitors (Liu et al., 2009). Our data suggest that disruption of the interactions between helical domain p110α mutants and IRS1 may be exploited as a more accessible targeted therapy approach for patients having cancers harboring such mutations. Increasing evidence suggests that protein-protein interactions are druggable targets (White et al., 2008). In fact, small molecules that inhibit MDM2-p53 interaction are being tested in clinical trials to treat cancer patients (Essmann and Schulze-Osthoff, 2011). Further, given that the mutant p110α-IRS1 interaction only exists in tumors harboring these mutations, it is likely that drugs targeting this interaction should have no or minimal toxicity. In this regard, an important future direction is to delineate the protein interaction interface between helical domain p110α mutants and IRS1 by structure analyses in order to provide insights to design chemical compounds that disrupt this mutant-specific protein-protein interaction.
Figure 8. A model of growth factor independent activation of p110α E545K mutant signaling pathway.
(A). When normal cells are stimulated by growth factors, p110α is brought to cell membrane through binding of p85 to phospho-IRS1, whereby converting PIP2 to PIP3.
(B). In cancer cells harboring a p110α helical domain mutation, the p110α mutant directly binds to IRS1, thereby being recruited to cell membrane and converting PIP2 to PIP3.
The current paradigm is that the helical domain mutations weaken p110α’s interactions with the p85 regulatory subunits, alleviate the inhibitory effect of p85, and thus increase the enzymatic activity of p110α (Huang et al., 2007; Miled et al., 2007). In contrast, our results suggest that the weakened p110α-p85 interaction caused by the mutations is not sufficient for the p110α helical domain mutant proteins to exert their oncogenic functions. We provide several pieces of evidence that the p110α E545K-IRS1 interaction plays a critical role in tumorigenesis. Mechanistically, our data suggest that p110α E545K mutant-IRS1 interaction activates the mutant p110α lipid kinase by facilitating the localization of the mutant enzyme complexes from cytosol to plasma membrane and by stabilizing the mutant p110α proteins. We propose that the helical domain mutations of p110α, as well as some of the mutations in the ABD and C2 domains, induce conformational changes that cause p110α to acquire a direct protein interaction with IRS1, which does not require IRS1 tyrosine phosphorylation and p85 proteins. Indeed, a recent study shows that oncogenic mutations of p110α in various domains induce conformation changes of the protein that may affect its structural interactions (Burke et al., 2012).
Lastly, our study may also provide an explanation for observed different phenotypes produced by the helical and kinase domain mutations. Using isogenic breast cancer cell lines, Becker and colleagues demonstrated that expression of p110α helical domain mutants increase sensitivity of cancer cells to chemo-attractants and induce much stronger metastatic phenotypes than the H1047R kinase domain mutant (Pang et al., 2009). These phenotypic differences cannot be simply explained by the increased enzymatic activities of the helical and kinase domain mutations. In fact, Vasudevan et al observed that the basal levels of phospho-AKT are lower in cancer cell lines expressing p110α helical domain mutants compared to the same cell lines expressing the H1047R kinase domain mutant (Vasudevan et al., 2009). Consistently, when complexed with p85 regulatory subunit, p110α H1047R displays higher lipid kinase activity in vitro than p110α E545K (Samuels et al., 2005). We show here that the p110α helical domain mutants, but not the kinase domain mutant, directly associate with IRS1 without growth factor stimulation. In this regard, we suggest that the p110α helical domain mutant-IRS1 complexes may contain additional proteins that do not exist in the p110α kinase domain mutant protein complexes, thereby producing distinct phenotypes. Moreover, we demonstrate that p110α helical domain mutant-IRS1 protein interaction does not require p85, which also provides an explanation to the observation that the helical domain mutants of p110α do not require the binding of p85 to transform chicken embryonic fibroblasts (Zhao and Vogt, 2008b). However, Zhao and Vogt’s study indicates that Ras-binding activity is still required for the helical domain mutants of p110α to exert their transformation activities. It was also reported that interaction between Ras and p110α enhances the substrate binding ability of p110α (Pacold et al., 2000). We postulate that although IRS1 brings p110α helical domain mutant protein to cell membrane, the mutant proteins also require RAS-binding to induce a conformational change to accommodate PIP2.
Experimental Procedures
Tissue culture
Colorectal cancer (CRC) cell lines DLD1, HCT116, Vaco481 and genetically engineered CRC cell lines were maintained in McCoy’s 5A medium containing 10% of fetal bovine serum (FBS). Lung cancer cell line H460 and breast cancer cell line T47D were cultured in RPMI 1640 medium plus 10% of FBS. Breast cancer cell line MDA-MB361 was maintained in Leibovitz's L-15 medium with 20% of FBS. Human embryonic kidney HEK 293 cells were cultured in DMEM medium containing 10% FBS. Mammalian cells were transfected with Lipofectamine in accordance with the manufacturer’s instructions (Invitrogen).
Somatic gene targeting
The targeting vectors were constructed using USER system and targeted cells were generated as described previously (Du et al., 2010). Briefly, vector arms were created by PCR from normal human DNA using HiFi Taq (Invitrogen) and validated by sequencing prior to virus production and infection. The parental DLD1 cells were purchased from ATCC (American Type Culture Collection) and infected with indicated rAAV viruses. Stable G418-resistent clones were then selected for PCR screening as reported. When relevant, targeted clones were genotyped by RT-PCR and sequencing when necessary. Primers for targeting vector construction and PCR screening are listed in supplementary experimental procedures.
Immunoprecipitation and immunoblotting
For Co-immunoprecipitation (Co-IP), cells were harvested, washed with PBS and then lysed in IP buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA (pH 8.0), 0.5% NP40, 1mM PMSF, complete Protease Inhibitor Cocktail tablet (Roche); supplemented with phosphatase inhibitors (1 mM Na3VO4, 20 mM NaF, 0.1 mM β-glycerophosphate, 20 mM sodium pyrophosphate) when necessary]. For immunoblotting (IB) of phospho-specific proteins, unless stated otherwise, cells were lysed in Urea buffer [100 mM NaH2PO4, 10 mM Tris-HCl, 8 M Urea, 1 mM Na3VO4, 20 mM NaF, 0.1 mM β-glycerophosphate, 20 mM sodium pyrophosphate, supplemented with complete Protease Inhibitor Cocktail tablet (Roche), pH 8.0]. Lysates were then cleared by centrifugation (14, 000 rpm, 10 min) and protein concentration in supernatants was determined by the BCA protein assay kit (Pierce, Rockford, IL). Equal amounts of total protein were used for IP or IB as described (Zhang et al., 2007; Zhao et al., 2010).
In vitro binding assay
The 6xHis-MYC-IRS1 baculoviral vector was constructed using the bac-to-bac expression system (Invitrogen). The 6×His-p110α WT and 6×His-p110α E545K baculoviral vectors were kind gifts from Dr. Bert Vogelstein. These recombinant proteins were expressed in Sf9 cell and purified using Ni-NTA agarose beads as described (Yu et al., 2008). For in vitro binding, 100 ng of purified 6×His-MYC-IRS1 proteins were first attached to anti-Myc antibody-bound beads and then incubated with the purified 6×His-p110α WT or 6×His-p110α E545K proteins in 1 ml of IP buffer. The protein-bound beads were washed three times for Western blot analysis.
Pull down assay
His-tagged or GST-tagged truncated IRS1 plasmids were constructed with PCR-based subcloning using USER system. Each construct was transformed into E coli strain BL21. Recombinant protein expression was induced with 0.2 mM IPTG at 16 °C overnight. His-tagged and GST-fusion proteins were purified using either Ni-NTA agarose beads or Glutathione Sepharose 4B beads according to manufactures’ instructions. The purified recombinant proteins were incubated with cell lysates prepared as described above.
Peptide competition assay
For in vitro peptide competition assay, the p110α mutant-FLAG tagged cells were grown to 90% confluency, serum-starved overnight, and lysed as described above. Peptides were then added to the cell lysates, incubated for 30 min at 4 °C and followed by immunoprecipitation assay.
Xenografts
Animal experiments were approved by the Case Western Reserve University Animal Care and Use Committee. As described in (Zhang et al., 2011), 3 million cells were injected subcutaneously into the flanks of 4 to 6-week-old female athymic nude mice. Tumor volume was measured with electronic calipers, and volumes were calculated as length × width2/2.
Peptide treatment of xenograft tumor
Five-week-old male athymic nude mice were injected with 5 million cells subcutaneously and bilaterally. When tumor sizes reached 100–150 mm3, 10 mice with similar size of tumors on both flanks were selected. The left flank tumors were injected with stapled WT p110α peptide, whereas the right flank tumors were injected with stapled p110α E545K mutant peptide. Each tumor was injected with 250 μg of peptide daily for 14 days. Control xeongraft tumors were injected with equal volume of water. The tumor sizes were measured every 2 days.
Supplementary Material
Significance.
PIK3CA is the most frequently mutated oncogene in a variety of human cancers, which opens the door for targeting the mutant enzyme for cancer therapy. However, it remains a challenge to develop p110α isoform-specific inhibitors. Our results show that disruption of the interactions between helical domain p110α mutants and IRS1 may be exploited as a more accessible target for cancer therapy, thereby providing proof-of-principle for treatment of cancer patients harboring such mutations.
Highlights.
p110α E545K helical domain mutant protein directly interacts with IRS1
IRS1-p110α E545K interaction stabilizes p110α E545K and brings it to the membrane
IRS1 mutants that do not interact with p110α E545K reduce oncogenicity
A peptide that disrupts IRS1-p110α E545K interaction inhibits tumor growth in vivo
Acknowledgments
We thank Dr. Bert Vogelstein for providing the PIK3CA knockout cell lines and p110α baculoviral vectors; Drs. Chrys Wesdemiotis and Xiaopeng Li at the University of Akron for the assistance with the mass spectrometric analysis (MALDI-TOF) of the peptides; Dr. Yuxiang Zheng for assistance with in vitro lipid kinase assay. This study was supported by NIH grants R21CA160060, R01CA127590, R01HG004722 and P50CA150964, as well as the National Natural Science Foundation of China (No: 21272094). Weiping Zheng is a Jiangsu provincial specially-appointed professor and a recipient of the Jiangsu provincial “innovation and venture talents” award plan, and the associated financial supports to this study are thus also appreciated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amzel LM, Huang CH, Mandelker D, Lengauer C, Gabelli SB, Vogelstein B. Structural comparisons of class I phosphoinositide 3-kinases. Nat Rev Cancer. 2008;8:665–669. doi: 10.1038/nrc2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA) Proc Natl Acad Sci U S A. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann F, Schulze-Osthoff K. Translational approaches targeting the p53 pathway for anticancer therapy. Br J Pharmacol. 2011 doi: 10.1111/j.1476-5381.2011.01570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, Boyd J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- Li VS, Wong CW, Chan TL, Chan AS, Zhao W, Chu KM, So S, Chen X, Yuen ST, Leung SY. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer. 2005;5:29. doi: 10.1186/1471-2407-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, Kinzler KW, Vogelstein B, Amzel LM. A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci U S A. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- Oda K, Stokoe D, Taketani Y, McCormick F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65:10669–10673. doi: 10.1158/0008-5472.CAN-05-2620. [DOI] [PubMed] [Google Scholar]
- Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, Wu H, Pozzuto M, Goswami S, Condeelis JS, Bresnick AR, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer Res. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Vadas O, Burke JE, Zhang X, Berndt A, Williams RL. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal. 2011;4 doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AW, Westwell AD, Brahemi G. Protein-protein interactions as targets for small-molecule therapeutics in cancer. Expert Rev Mol Med. 2008;10:e8. doi: 10.1017/S1462399408000641. [DOI] [PubMed] [Google Scholar]
- Yu J, Becka S, Zhang P, Zhang X, Brady-Kalnay SM, Wang Z. Tumor-Derived Extracellular Mutations of PTPRT/PTP{rho} Are Defective in Cell Adhesion. Mol Cancer Res. 2008;6:1106–1113. doi: 10.1158/1541-7786.MCR-07-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhao Y, Zhu X, Sedwick D, Zhang X, Wang Z. Cross-talk between Phospho-STAT3 and PLC{gamma}1 Plays a Critical Role in Colorectal Tumorigenesis. Mol Cancer Res. 2011;9:1418–1428. doi: 10.1158/1541-7786.MCR-11-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo A, Yu J, Possemato A, Chen Y, Zheng W, Polakiewicz RD, Kinzler KW, Vogelstein B, Velculescu VE, Wang ZJ. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. PNAS. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo C, Chen Y, Shulha HP, Schnetz MP, LaFramboise T, Bartels CF, Markowitz S, Weng Z, Scacheri PC, Wang Z. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat Methods. 2008;5:163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008a;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008b;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang X, Guda K, Lawrence E, Sun Q, Watanabe T, Iwakura Y, Asano M, Wei L, Yang Z, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci U S A. 2010;107:2592–2597. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.