Abstract
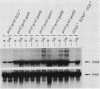
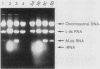
GCN4 encodes a positive regulator of multiple unlinked genes encoding amino acid biosynthetic enzymes in Saccharomyces cerevisiae. Expression of GCN4 is coupled to amino acid availability by a control mechanism involving GCD1 as a negative effector and GCN1, GCN2, and GCN3 as positive effectors of GCN4 expression. We used reversion of a gcn2 gcn3 double mutation to isolate new alleles of GCD1 and mutations in four additional GCD genes which we designate GCD10, GCD11, GCD12, and GCD13. All of the mutations lead to constitutive derepression of HIS4 transcription in the absence of the GCN2+ and GCN3+ alleles. By contrast, the gcd mutations require the wild-type GCN4 allele for their derepressing effect, suggesting that each acts by influencing the level of GCN4 activity in the cell. Consistent with this interpretation, mutations in each GCD gene lead to constitutive derepression of a GCN4::lacZ gene fusion. Thus, at least five gene products are required to maintain the normal repressed level of GCN4 expression in nonstarvation conditions. Interestingly, the gcd mutations are pleiotropic and also affect growth rate in nonstarvation conditions. In addition, certain alleles lead to a loss of M double-stranded RNA required for the killer phenotype. This pleiotropy suggests that the GCD gene products contribute to an essential cellular function, in addition to, or in conjunction with, their role in GCN4 regulation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball S. G., Tirtiaux C., Wickner R. B. Genetic Control of L-a and L-(Bc) Dsrna Copy Number in Killer Systems of SACCHAROMYCES CEREVISIAE. Genetics. 1984 Jun;107(2):199–217. doi: 10.1093/genetics/107.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S. S., Theodorakis N. G., Morimoto R. I. Heat shock-induced translational control of HSP70 and globin synthesis in chicken reticulocytes. Mol Cell Biol. 1984 Nov;4(11):2437–2448. doi: 10.1128/mcb.4.11.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Chattoo B. B., Sherman F., Azubalis D. A., Fjellstedt T. A., Mehnert D., Ogur M. Selection of lys2 Mutants of the Yeast SACCHAROMYCES CEREVISIAE by the Utilization of alpha-AMINOADIPATE. Genetics. 1979 Sep;93(1):51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A. Curing of a killer factor in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2846–2849. doi: 10.1073/pnas.69.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Fink G. R. Electron microscopic heteroduplex analysis of "killer" double-stranded RNA species from yeast. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4224–4228. doi: 10.1073/pnas.75.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S. A., Donahue T. F., Culbertson M. R. Selection of spontaneous mutants by inositol starvation in yeast. Mol Gen Genet. 1975 Dec 30;143(1):5–11. doi: 10.1007/BF00269415. [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr, Rosbash M., Kaback D. B. Isolation of yeast histone genes H2A and H2B. Cell. 1979 Dec;18(4):1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopotowski T., Wiater A. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Arch Biochem Biophys. 1965 Dec;112(3):562–566. doi: 10.1016/0003-9861(65)90096-2. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie S. L., Henikoff S., Meselson M. Localization of RNA from heat-induced polysomes at puff sites in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1117–1121. doi: 10.1073/pnas.72.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Delforge J. Role of transfer ribonucleic acids in the regulation of several biosyntheses in Saccharomyces cerevisiae. Eur J Biochem. 1976 Aug 16;67(2):335–339. doi: 10.1111/j.1432-1033.1976.tb10696.x. [DOI] [PubMed] [Google Scholar]
- Miozzari G., Niederberger P., Hütter R. Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway. J Bacteriol. 1978 Apr;134(1):48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Botstein D. Structure and function of the yeast URA3 gene. Differentially regulated expression of hybrid beta-galactosidase from overlapping coding sequences in yeast. J Mol Biol. 1983 Nov 15;170(4):883–904. doi: 10.1016/s0022-2836(83)80193-4. [DOI] [PubMed] [Google Scholar]
- Rose M., Casadaban M. J., Botstein D. Yeast genes fused to beta-galactosidase in Escherichia coli can be expressed normally in yeast. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2460–2464. doi: 10.1073/pnas.78.4.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. J., Rose M., Botstein D., Fink G. R. Regulation of HIS4-lacZ fusions in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1212–1219. doi: 10.1128/mcb.2.10.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Regulatory sites for his3 gene expression in yeast. Nature. 1982 Nov 18;300(5889):285–286. doi: 10.1038/300284a0. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Oshima T., Araki H., Harashima S., Oshima Y. Mating type control in Saccharomyces cerevisiae: a frameshift mutation at the common DNA sequence, X, of the HML alpha locus. Mol Cell Biol. 1984 Jan;4(1):203–211. doi: 10.1128/mcb.4.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Ridley S. P., Fried H. M., Ball S. G. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]