Abstract
Nosocomial infections are infections acquired by a patient as a result of treatment in a hospital or healthcare service providing center and symptoms occurs within a short period of hospitalization. The study was to determine the antibiotic resistance patterns of Escherichia coli isolated from Kumasi-South, Tafo and Suntreso Hospitals, Kumasi, Ghana. Total of 600 swabs samples from the hospitals were collected between January and June, 2010. The isolates were identified using morphological and biochemical means. A total of 97 E. coli isolates were obtained from the hospitals. Beds in hospital wards had the highest number of E. coli strains (53.6%), followed by floors (20.6%) while drainages had the least isolates (3.1%). Majority of the E. coli isolates (90.7%) exhibited resistance to ampicillin while 6.2 and 3.1% showed intermediate and sensitive respectively. Co-trimoxazole, 78.4% of the isolates were resistant while 9.3 and 12.4% exhibited intermediate and sensitive responses respectively. E. coli isolates (28.6 to 46.4%) were resistant to gentamicin, ciprofloxacin and ceftriaxone while 14.4 to 47.4% gave intermediate responses. Most isolates (80.4%) exhibited multi-drug resistance. There is a need to observe proper personal hygiene, use of effective disinfectants and proper disposal of contaminated/pathogenic materials in these hospitals to control nosocomial infections.
1. Introduction
Nosocomial infections, also called healthcare-associated infections are those infections acquired by a patients as a result of treatment in a hospital, clinic or healthcare service centre. These infections generally appear 48 hours or more after hospital admission or within 30 days after discharge. They occur because of instrumentation, increased use of antimicrobial agents, breaks in aseptic techniques and lack of hand hygiene [1]. At any time, over 1.4 billion people worldwide suffer from infectious complications acquired in hospital [2]. Microorganisms often implicated in these infections include Escherichia coli, Pseudomonas aeruginosa, Klebsiella species, Staphylococcus aureus and Mycobacterium tuberculosis. According to the American National Nosocomial Infections Surveillance, more than 40% of nosocomial infections occurred in parts of Asia, Latin America, and Sub-Saharan Africa [3].
E. coli is facultative anaerobe and can undergo both fermentative and respiratory metabolisms. E. coli is nonspore-forming and beta hemolytic. It usually ferments lactose on MacConkey agar to produce pink colonies with surrounding areas of precipitated bile salts. It also presents with a green sheen on eosin methylene blue agar. E. coli strain will produce indole from tryptophan; it does not produce hydrogen sulfide, urease, and cannot use citrate as sole carbon source [1].
In some hospitals, E. coli ranked first as the most common cause of hospital-acquired infections [4]. E. coli strains were found to be the highest and most frequent among the pathogenic microorganisms isolated from ten teaching hospitals in China [5]. Pathogenic strains of E. coli are responsible for three types of infections in humans; urinary tracts infections, neonatal meningitis, and intestinal diseases [6].
In many West African countries, nosocomial infections are abound but not much study has been done to determine the proportion of infections acquired by patients or health workers from hospital and or healthcare-providing facilities. Hospitals serve a reservoir of various types of microorganisms; some may be multiple resistant to antibiotics [8] and the selective pressure of antimicrobial use in hospitals, therefore makes the environment a repository for these resistant strains [9]. Newman and his colleagues reported on the occurrence of nosocomial infections in Korle-Bu Teaching Hospital in Accra, Ghana [10]. These studies are therefore necessary and need to be conducted in many other parts of the country in order to generate national data on these pathogenic organisms, more especially on their antibiotic resistant patterns in Ghana. This study sought to determine the antibiotic resistance patterns of E. coli isolates from the premises of three hospitals in Kumasi, Ghana.
2. Materials and Methods
The protocols for the study were approved by the individual Hospital's Ethics Committees. The samples were collected from Kumasi South, Tafo and Suntreso Hospitals in Kumasi, Ghana. A total of 600 swabs samples of floors, benches, beds, door handles, and waste water from drainages were collected between January and June, 2010. The swabs were put into sterile test tubes, closed tightly, and labeled appropriately. All the materials including culture media, reference antibiotics, and reagents were purchased from Oxoid, Basingstoke, United Kingdom unless otherwise stated.
2.1. Cultivation and Isolation of Escherichia coli Isolates
The various samples collected were separately inoculated into 10 mL of nutrient broths and incubated at 37°C for 24 hours. Using a sterile platinum loop, each culture was separately streaked onto the surface of MacConkey Agar plates, labeled and incubated at 37°C for 48 hours, and observed for signs of growth and colony appearance.
Colonies that appeared pink on the MacConkey agar plates were removed with sterile inoculating wire and separately streaked onto the surface of eosin methylene blue agar plates. The plates were then incubated at 37°C for 24 hours. Isolated black-colored colonies with metallic sheen were again fished out into nutrient broths and incubated at 37°C for 24 hours. The various subcultures were streaked onto nutrient agar slants, incubated at 37°C for 48 hours, and then kept in the refrigerator at −20°C for further identification and antibiotic sensitivity studies.
2.2. Identification and Confirmation of Escherichia coli Isolates
The E. coli isolates were screened through the various microscopic examination and biochemical reactions to confirm their identities. These included indole, oxidase, and arginine dehydrolase production, citrate utilization, nitrite reduction, fermentation of carbohydrates (such as xylose, maltose, arabinose, glycerol, and starch), methyl red-Voges Proskauer test, and reaction Triple sugar iron agar [11, 12]. All the tests were performed on reference-typed culture of E. coli (ATCC 25922).
2.3. Antibiotic Sensitivity Test
Kirby-Bauer disc diffusion method [13] as recommended by the Clinical and Laboratory Standards Institute [7] was used to determine the in vitro susceptibility of the identified E. coli isolates to gentamicin (GM) 10 μg, ciprofloxacin (CIP) 5 μg, ceftriaxone (CRO) 30 μg, ampicillin (AMP) 10 μg, and cotrimoxazole (SXT) (trimethoprim-sulphamethoxazole) 25 μg. A standardized suspension of the isolated E. coli was prepared by inoculating a colony into 10 mL peptone water and incubated at 37°C for 24 hours. It was then diluted to 0.5 MacFarland turbidity standards. A sterile swab was dipped into the standardized inoculum and used to inoculate evenly the surface of already prepared Mueller-Hinton agar (Oxoid Basingstoke, UK). The agar was left for 15 minutes for the surface moisture to dry. A multichannel disc dispenser (Oxoid, Basingstoke, UK) was used to deposit the antibiotics discs onto the surface of the inoculated medium. The plate was then incubated at 37°C for 18 hours. The zones of growth inhibition were recorded. The method was replicated three times and the mean zones of inhibition compared with figures (Table 1) provided by the Clinical and Laboratories Standards Institute [13]. Escherichia coli ATCC 25922 was used as control.
Table 1.
Acceptable susceptible zone of inhibition values for antibiotics used based on *CLSI.
| Antibiotic | Resistant | Intermediate | Susceptible |
|---|---|---|---|
| Ciprofloxacin (5 μg) | ≤15 | 16–20 | ≥21 |
| Ampicillin (10 μg) | ≤13 | 14–16 | ≥17 |
| Gentamicin (10 μg) | ≤12 | 13-14 | ≥15 |
| Ceftriaxone (30 μg) | ≤13 | 14–20 | ≥21 |
| Co-trimoxazole (25 μg) | ≤10 | 11–15 | ≥16 |
*CLSI: Clinical and Laboratory Standards Institute [7].
3. Results and Discussion
A total of 150 (lactose fermenter) isolates recovered on MacConkey agar (Oxoid, Basingstoke, UK) were suspected to be E. coli. These were screened through the various microscopic examination and biochemical reactions. E. coli isolates were identified from the various locations (benches, floor, door handles and drainages, male, female, and pediatrics wards) in the three hospitals (Figure 1). A total of 97 isolates from the three hospitals were confirmed as E. coli.
Figure 1.
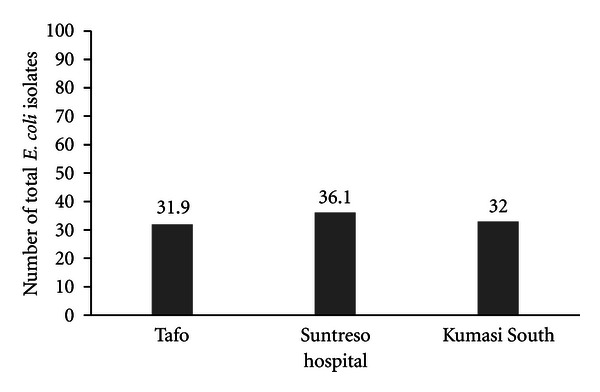
Distribution of E. coli isolates within the three hospitals: Kumasi South, Tafo and Suntreso hospitals, Kumasi, Ghana.
Jarvis and Martone [14] reported that E. coli as the most common nosocomial pathogen in some hospitals in the United States. Also, E. coli has been reported to be among the most frequent isolates in hospitals in Ethiopia [15].
Among the three hospitals from which the samples or swabs were taken, E. coli isolates were widely distributed in various locations throughout the three hospitals for which samples were analyzed (Table 2).
Table 2.
Number of samples containing E. coli isolates in the swabs/samples taken from the three hospitals.
| Tafo hospital (n) | Kumasi South hospital (n) | Suntreso hospital (n) | |
|---|---|---|---|
| Beds | 17 (90) | 17 (90) | 18 (90) |
| Floors | 7 (30) | 3 (30) | 10 (30) |
| Benches | 5 (30) | 6 (30) | 6 (30) |
| Door handles | 0 (30) | 5 (30) | 0 (30) |
| Drainages | 0 (20) | 3 (20) | 0 (20) |
Key: n: total number of swabs/samples taken from the hospital.
About 90% of the E. coli isolates exhibited resistance to ampicillin while 6.2 and 3.1%, respectively, showed intermediate and sensitive. For cotrimoxazole (trimethoprim-sulphamethoxazole), 78.4% of the isolates were resistant while 9.3 and 12.3% intermediate and sensitive responses. Between 26.8 to 46.4% of the E. coli isolates also showed resistance to gentamicin, ciprofloxacin, and ceftriaxone, while 14.4 to 47.4% gave intermediate responses. Ceftriaxone, ciprofloxacin, and gentamicin sensitive isolates were also in the range of 23.7 to 39.2% (Figure 2).
Figure 2.
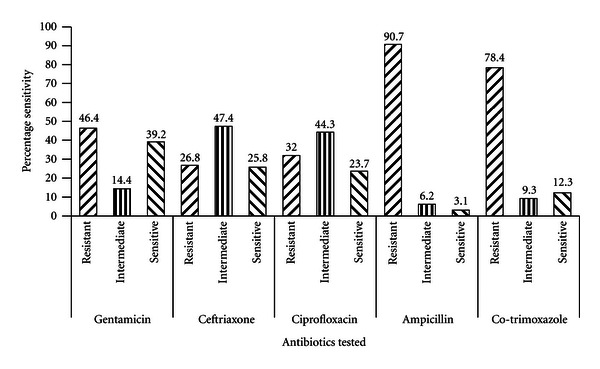
Antibiotic sensitivity patterns of E. coli isolates to reference antibiotics determined by Kirby-Bauer agar disc diffusion method and the mean zones of inhibition compared with the values provided by the Clinical and Laboratory Standards Institute [7].
The majority of the gentamicin sensitive E. coli isolates (28.9%) was isolated from the male wards followed by floor samples (21.1%) as shown in Table 3. None of the drainage samples were resistant to gentamicin, while 20% each from the floor and female wards proved resistant. 26.9% isolates from bench samples exhibited intermediate response to gentamicin. Out of the total, 46 E. coli isolates exhibited intermediate response to ceftriaxone, 30% were from the male wards, 21.4% from floor and 2.2% from drainage samples. Most of the E. coli resistant isolates (26.9%) were from the benches and 19.2% from male wards while no resistant strains were recovered from door handles (Table 3).
Table 3.
Sources of E. coli isolates in relation to antibiotic sensitivity patterns as compared to with values provided by the Clinical and Laboratories Standards Institute [7].
| Type of reference antibiotics and resistance patterns of E. coli isolates | Source of sample | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Floors | Drainages | Door handles | Benches | Female wards | Male wards | Pediatric wards | |||
| Gentamicin | |||||||||
| Intermediate | Count (%) | 3 (21.4) | 1 (7.1) | 0 (0) | 4 (28.6) | 3 (21.4) | 1 (7.1) | 2 (14.3) | 14 (100) |
| Resistant | Count (%) | 9 (20.0) | 0 (0) | 1 (2.2) | 6 (13.3) | 9 (20.0) | 11 (24.4) | 9 (20.0) | 45 (100) |
| Sensitive | Count (%) | 8 (21.1) | 2 (5.3) | 1 (2.6) | 7 (18.4) | 6 (15.8) | 11 (28.9) | 3 (7.9) | 38 (100) |
| Ceftriaxone | |||||||||
| Intermediate | Count (%) | 12 (26.1) | 1 (2.2) | 2 (4.3) | 4 (8.7) | 8 (17.4) | 14 (30.4) | 5 (10.9) | 46 (100) |
| Resistant | Count (%) | 3 (11.5) | 2 (7.7) | 0 (0) | 7 (26.9) | 4 (15.4) | 5 (19.2) | 5 (19.2) | 26 (100) |
| Sensitive | Count (%) | 5 (20.0) | 0 (0) | 0 (0) | 6 (24.0) | 6 (24.0) | 4 (16.0) | 4 (16.0) | 25 (100) |
| Ciprofloxacin | |||||||||
| Intermediate | Count (%) | 8 (18.6) | 1 (2.3) | 0 (0) | 10 (23.3) | 5 (11.6) | 15 (34.9) | 4 (9.3) | 43 (100) |
| Resistant | Count (%) | 9 (29.0) | 1 (3.2) | 1 (3.2) | 4 (12.9) | 6 (19.4) | 4 (12.9) | 6 (19.4) | 31 (100) |
| Sensitive | Count (%) | 3 (13.0) | 1 (4.3) | 1 (4.3) | 3 (13.0) | 7 (30.4) | 4 (17.4) | 4 (17.4) | 23 (100) |
| Ampicillin | |||||||||
| Intermediate | Count (%) | 0 (0) | 1 (16.7) | 0 (0) | 0 (0) | 1 (16.7) | 2 (33.3) | 2 (33.3) | 6 (100) |
| Resistant | Count (%) | 18 (20.5) | 2 (2.3) | 2 (2.3) | 17 (19.3) | 17 (19.3) | 20 (22.7) | 12 (13.6) | 88 (100) |
| Sensitive | Count (%) | 2 (66.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (33.3) | 0 (0) | 3 (100) |
| Co-trimoxazole | |||||||||
| Intermediate | Count (%) | 2 (22.2) | 1 (11.1) | 0 (0) | 1 (11.1) | 2 (22.2) | 2 (22.2) | 1 (11.1) | 9 (100) |
| Resistant | Count (%) | 14 (18.4) | 1 (1.3) | 2 (2.6) | 15 (19.7) | 14 (18.4) | 17 (22.4) | 13 (17.1) | 76 (100) |
| Sensitive | Count (%) | 4 (33.3) | 1 (8.3) | 0 (0) | 1 (8.3) | 2 (16.7) | 4 (33.3) | 0 (0) | 12 (100) |
Ciprofloxacin-resistant E. coli isolates were recovered from floor samples (29%) followed by the samples from pediatric wards (19.4%). E. coli isolates which exhibited intermediate response to ciprofloxacin (30%) were in the samples/swabs from the male ward and none from door handles. Majority of the E. coli sensitive isolates were from female wards (30.4%) followed by male and pediatric wards samples (17.4%). Floors and benches samples showed equal ciprofloxacin sensitive E. coli strains of 13%. The distributions of ampicillin resistant E. coli isolates were 22.7, 20.5, and 19.3% for male wards, floors, and benches, respectively (Table 3). Many of the isolates obtained were found to be resistant to more than two different classes of the reference antibiotics (Table 4).
Table 4.
Multiple-drug resistant (MDR)* pattern of E. coli isolates determined by Kirby-Bauer agar disc diffusion method and mean zones of inhibition compared with the values provided by the Clinical and Laboratory Standards Institute [7].
| Hospital | Total No. of MDR E. coli | Total no. of E. coli isolates |
Percent of MDR |
|---|---|---|---|
| Tafo | 27 | 30 | 90 |
| Suntreso | 28 | 34 | 82.4 |
| Kumasi-South | 23 | 33 | 69.7 |
|
| |||
| Total | 78 | 97 | 80.4 |
*Multiple drug resistance (MDR) is defined as resistance of organism (bacteria) to at least three different antibiotics.
Majority of the E. coli isolates (53.6%) were isolated from the hospital beddings while about 21% were from floor samples (Table 2). Most of the E. coli isolates (90 to 78%) were resistant to ampicillin and cotrimoxazole, respectively (Figure 1). The high occurrence of E. coli isolates in these samples could be attributed to poor hygienic conditions in these hospitals and the overcrowding in these hospitals due to inadequate number of health care facilities in the region.
A total of 46.4, 32.0, and 26.8% of the E. coli isolates exhibited resistance to gentamicin, ciprofloxacin, and ceftriaxone, respectively, and these were similar to what was reported by Yismaw and his colleagues [16]. Yismaw et al. [16] also reported a similar resistance pattern of E. coli to gentamicin (47%), ciprofloxacin (33%), and ceftriaxone (26%). And these high levels of antibiotic resistance have been attributed to widespread abuse of these antibiotics [15]. Out of 97 E. coli isolates, 78 isolates or 80.4% exhibited multiple drug resistance [17] to at least three different classes of the reference antibiotics used. These high numbers of resistant E. coli isolates in the hospitals are potential reservoirs of resistant genes which can easily be transferred to other pathogens. Hence, there is a need to observe proper hygiene, use of effective disinfectants, and monitor the administration and prescription of antibiotics in hospitals.
4. Conclusion
Most of the E. coli isolates (80.4%) exhibited multiple drug resistance and measures such as observation of proper personal hygiene by health staff and patients, use of effective disinfectants in reducing the possible pathogenic organisms in these hospitals, and so forth. These findings have therefore showed the need for the hospital management to be concerned about the potential of hospitalized patients becoming infected with some nosocomial infections, especially resistant strains of E. coli.
Acknowledgments
The authors would like to thank the management and laboratory staff of Kumasi South, North Suntreso and Tafo hospitals, Kumasi, Ghana for their assistance and cooperation during the study. They are also grateful to the United States Agency for International Development (USAID) and Government of Liberia for the financial support to D. F. George.
References
- 1.Mahon CR, Lehman DC, Manuselis G. Textbook of Diagnostic. 3rd edition. St. Louis, Mo, USA: Saunders; 2007. [Google Scholar]
- 2.Tikhomirov E. WHO programme for the control of hospital infections. Chemioterapia. 1987;6(3):148–151. [PubMed] [Google Scholar]
- 3.National Nosocomial Infections Surveillance (NNIS) National Nosocomial Infections Surveillance (NNIS). System Report, data summary from January 1992 through June 2004. American Journal of Infection Control. 2004;32:470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 4.Berkley JA, Lowe BS, Mwangi I, et al. Bacteremia among children admitted to a rural hospital in Kenya. The New England Journal of Medicine. 2005;352(1):39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Chen M, Ni Y, et al. Antimicrobial resistance among clinical isolates from the Chinese Meropenem Surveillance Study (CMSS), 2003–2008. International Journal of Antimicrobial Agents. 2010;35(3):227–234. doi: 10.1016/j.ijantimicag.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Todar K. Pseudomonas aeruginosa. Todar’s Online Textbook of Bacteriology, pp 1. 2008, http://textbookofbacteriology.net/pseudomonas.html.
- 7.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, Seventeenth Informational Supplement. Wayne, Pa, USA: 2010. M100-S17.CLSI. [Google Scholar]
- 8.Haley RW, Cushion NB, Tenover FC, et al. Eradication of endemic methicillin-resistant Staphylococcus aureus infections from a neonatal intensive care unit. Journal of Infectious Diseases. 1995;171(3):614–624. doi: 10.1093/infdis/171.3.614. [DOI] [PubMed] [Google Scholar]
- 9.Newman MJ. Neonatal intensive care unit: reservoirs of nosocomial pathogens. West African Journal of Medicine. 2002;21(4):310–312. doi: 10.4314/wajm.v21i4.28007. [DOI] [PubMed] [Google Scholar]
- 10.Newman MJ. Nosocomial and community acquired infections in Korle Bu Teaching Hospital, Accra. West African Journal of Medicine. 2009;28(5):300–303. doi: 10.4314/wajm.v28i5.55005. [DOI] [PubMed] [Google Scholar]
- 11.Harley J, Prescott L. Laboratory Exercises in Microbiology. Dubuque, La, USA: Wm. C. Brown Pubishers; 1990. [Google Scholar]
- 12.Forbes BA, Sahm DF, Weissfeld AS. Bailey and Scott’s Diagnostic Microbiology. 12th edition. St. Louis, Mo, USA: Mosby Company; 2007. [Google Scholar]
- 13.Gloria A, Cheryl B, John E, et al. Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World. Atlanta, Ga, USA: Centers for Disease Control and Preventionand World Health Organization, Department of Communicable Disease Surveillance and Response; 2003. [Google Scholar]
- 14.Jarvis WR, Martone WJ. Predominant pathogens in hospital infections. Journal of Antimicrobial Chemotherapy. 1992;29:19–24. doi: 10.1093/jac/29.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 15.Namboodiri SS, Opintan JA, Lijek RS, Newman MJ, Okeke IN. Quinolone resistance in Escherichia coli from Accra, Ghana. BMC Microbiology. 2011;11, article 44 doi: 10.1186/1471-2180-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yismaw G, Abay S, Asrat D, Yifru S, Kassu A. Bacteriological profile and resistant pattern of clinical isolates from pediatric patients, Gondar University Teaching Hospital, Gondar, Northwest Ethiopia. Ethiopian Medical Journal. 2010;48(4):293–300. [PubMed] [Google Scholar]
- 17.Obritsch MD, Fish DN, MacLaren R, Jung R. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy. 2005;25(10):1353–1364. doi: 10.1592/phco.2005.25.10.1353. [DOI] [PubMed] [Google Scholar]


