Abstract
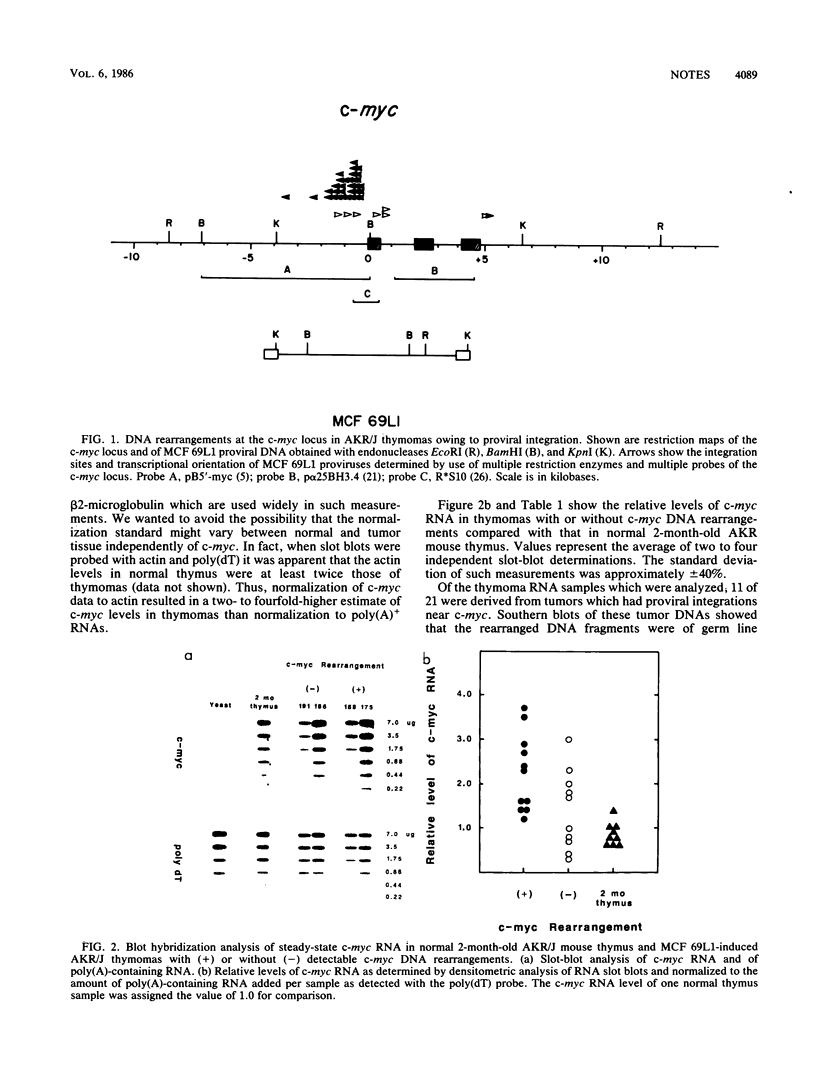
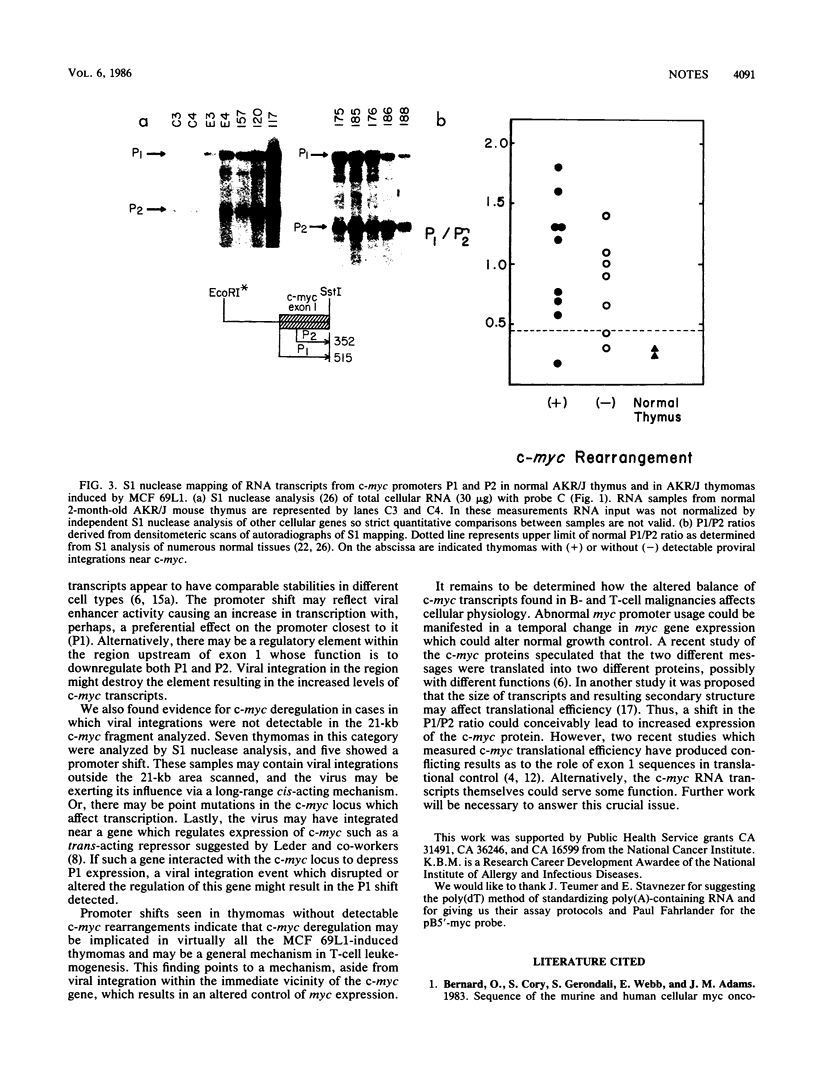
A high frequency (greater than or equal to 65%) of thymomas induced by mink cell focus-forming virus 69L1 in AKR/J mice contain proviral integrations which are clustered 0.7-kilobase upstream of the c-myc oncogene predominantly in the opposite transcriptional orientation. Blot hybridization experiments showed that on the average there was only a twofold elevation of steady-state c-myc RNA in the thymomas as compared with levels in normal AKR/J thymocytes. Such an increase would not appear to be sufficient as a mechanism of oncogene activation in this system. In contrast, S1 nuclease analysis of transcripts initiated from the two known c-myc promoters indicated a strong shift in promoter usage in virtually all thymomas tested. In normal thymus the ratio of transcripts initiated from the proximal promoter P1 to the distal promoter P2 was 0.2 to 0.3. In contrast, most of the thymomas tested (18 of 23) showed an average P1/P2 ratio of 1.2 regardless of whether or not proviral integrations could be detected within a 21-kilobase EcoRI fragment containing the three c-myc exons. We conclude that alterations in P1/P2 ratios are good indicators of c-myc deregulation in thymic lymphomas.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Darveau A., Pelletier J., Sonenberg N. Differential efficiencies of in vitro translation of mouse c-myc transcripts differing in the 5' untranslated region. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2315–2319. doi: 10.1073/pnas.82.8.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrlander P. D., Piechaczyk M., Marcu K. B. Chromatin structure of the murine c-myc locus: implications for the regulation of normal and chromosomally translocated genes. EMBO J. 1985 Dec 1;4(12):3195–3202. doi: 10.1002/j.1460-2075.1985.tb04065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S. R., Eisenman R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984 Nov;4(11):2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keath E. J., Kelekar A., Cole M. D. Transcriptional activation of the translocated c-myc oncogene in mouse plasmacytomas: similar RNA levels in tumor and proliferating normal cells. Cell. 1984 Jun;37(2):521–528. doi: 10.1016/0092-8674(84)90382-9. [DOI] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Li Y., Holland C. A., Hartley J. W., Hopkins N. Viral integration near c-myc in 10-20% of mcf 247-induced AKR lymphomas. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6808–6811. doi: 10.1073/pnas.81.21.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W., Maroney P. A. Translational efficiency of cMyc mRNA in Burkitt lymphoma cells. Mol Cell Biol. 1984 Oct;4(10):2235–2238. doi: 10.1128/mcb.4.10.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell P. V., Fleissner E., Lonial H., Koehne C. F., Reicin A. Early clonality and high-frequency proviral integration into the c-myc locus in AKR leukemias. J Virol. 1985 Aug;55(2):500–503. doi: 10.1128/jvi.55.2.500-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell P. V., Traganos F. Changes in thymocyte proliferation at different stages of viral leukemogenesis in AKR mice. J Immunol. 1986 Jan;136(2):720–727. [PubMed] [Google Scholar]
- O'Donnell P. V., Woller R., Chu A. Stages in development of mink cell focus-inducing (MCF) virus-accelerated leukemia in AKR mice. J Exp Med. 1984 Sep 1;160(3):914–934. doi: 10.1084/jem.160.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saito H., Hayday A. C., Wiman K., Hayward W. S., Tonegawa S. Activation of the c-myc gene by translocation: a model for translational control. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7476–7480. doi: 10.1073/pnas.80.24.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scollay R., Bartlett P., Shortman K. T cell development in the adult murine thymus: changes in the expression of the surface antigens Ly2, L3T4 and B2A2 during development from early precursor cells to emigrants. Immunol Rev. 1984 Dec;82:79–103. doi: 10.1111/j.1600-065x.1984.tb01118.x. [DOI] [PubMed] [Google Scholar]
- Scollay R., Shortman K. Thymocyte subpopulations: an experimental review, including flow cytometric cross-correlations between the major murine thymocyte markers. Thymus. 1983 Sep;5(5-6):245–295. [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Stewart T. A., Bellvé A. R., Leder P. Transcription and promoter usage of the myc gene in normal somatic and spermatogenic cells. Science. 1984 Nov 9;226(4675):707–710. doi: 10.1126/science.6494906. [DOI] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Thompson C. B., Challoner P. B., Neiman P. E., Groudine M. Levels of c-myc oncogene mRNA are invariant throughout the cell cycle. 1985 Mar 28-Apr 3Nature. 314(6009):363–366. doi: 10.1038/314363a0. [DOI] [PubMed] [Google Scholar]
- Yang J. Q., Bauer S. R., Mushinski J. F., Marcu K. B. Chromosome translocations clustered 5' of the murine c-myc gene qualitatively affect promoter usage: implications for the site of normal c-myc regulation. EMBO J. 1985 Jun;4(6):1441–1447. doi: 10.1002/j.1460-2075.1985.tb03800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]





