Abstract
We investigate single-cell reduction of toxic Cr(VI) by the dissimilatory metal-reducing bacterium Shewanella oneidensis MR-1 (MR-1), an important bioremediation process, using Raman spectroscopy and scanning electron microscopy (SEM) combined with energy-dispersive X-ray spectroscopy (EDX). Our experiments indicate that the toxic and highly soluble Cr(VI) can be efficiently reduced to the less toxic and non-soluble Cr2O3 nanoparticles by MR-1. Cr2O3 is observed to emerge as nanoparticles adsorbed on the cell surface and its chemical nature is identified by EDX imaging and Raman spectroscopy. Co-localization of Cr2O3 and cytochromes by EDX imaging and Raman spectroscopy suggests a terminal reductase role for MR-1 surface-exposed cytochromes MtrC and OmcA. Our experiments revealed that the cooperation of surface proteins OmcA and MtrC makes the reduction reaction most efficient, and the sequence of the reducing reactivity of the MR-1 is: wild type > single mutant ΔmtrC or mutant ΔomcA > double mutant (ΔomcA-ΔmtrC). Moreover, our results also suggest that the direct microbial Cr(VI) reduction and Fe(II) (hematite)-mediated Cr(VI) reduction mechanisms may co-exist in the reduction processes.
INTRODUCTION
Hexavalent Chromium [Cr(VI)] contamination of the soil and groundwater is a significant environmental hazard that has attracted considerable attention in the scientific community.1,2 For a human being, the Cr(VI) toxicity can lead to many health impacts such as carcinogenesis and mutagenesis.3 Due to its high aqueous solubility and mobility, Cr(VI) contamination can be proliferated to spread to a vast area through ground water. Therefore, to reduce the highly soluble and toxic Cr(VI) to the less soluble and less toxic Cr(III), in the form of Cr2O3 precipitates, is the primary strategy for the contamination treatment.
In a natural environment, direct microbial reduction or chemical reaction, using Fe2+ or S2− as reductants, are two typical approaches to decrease the Cr(VI) contamination. Microbial reductions have been widely utilized in the bioremediation approaches for the environmental cleanup of the Cr(VI) contaminations in the soil due to being a feasible and promising strategy considering that the bacteria naturally exist and pervade in a wide area even underground and under anaerobic conditions. In recent years, many bacteria have been demonstrated to have the capability of reducing Cr(VI) to Cr(III).4–9 As a dissimilatory metal-reducing bacterium, Shewanella oneidensis MR-1 (MR-1) has also been reported to be capable of reducing Cr(VI), and the reduced product has been shown to occur as nanoparticles on the bacterial cell surface or in the cytoplasm, including our recent report on the reduction activities of outer membrane proteins on reducing Cr(VI).3,6,10–14
Raman spectroscopy is a unique technique, giving the fingerprint information of the molecules (such as proteins and organic molecules), to identify various molecules and probe specific vibrational modes that are sensitive to redox states of the molecules.15,16 Recently, we have applied high resolution AFM-Raman spectroscopy to probe the chemical nature of the cell surface nano domains (i.e., the surface features at nanometer scale) of the MR-1.17 It is demonstrated that the distribution density of the nano domains shows clear differences under aerobic and anaerobic conditions, and the major component of the cell surface domains is identified to be the redox heme proteins. This finding may help to reveal the mechanism of the Cr(VI) reduction by MR-1. Moreover, the surface Heme protein OmcA and MtrC (also known as OmcB) of MR-1 have been proved to play a key role in the reduction of Fe(III), Mn(III/IV) and Cr(VI).10,18–22 In this report, we apply combined surface-enhanced Raman spectroscopy, SEM, and EDX imaging to probe the Cr(VI) reduction mechanism at a single cell level. Our spectroscopic and imaging evidences indicate that: the chemical nature of the reduced nanoparticles is Cr2O3; co-localization of reduced Cr2O3 and MtrC and OmcA, which signifies that the surface proteins OmcA and MtrC are the key components for the Cr(VI) reduction reaction; the cooperation of OmcA and MtrC makes the reduction reaction most efficient, and both direct Cr(VI) reduction by MtrC and OmcA and Fe(II)-mediated Cr(VI) reduction mechanisms, in which Fe(II) is generated by MtrC and OmcA through Fe(III) reduction, are suggested to co-exist in the Cr(VI) reduction process.
EXPERIMENTAL SECTION
Materials and sample preparation
S. oneidensis MR-1 and its cytochrome deleted mutants, ΔmtrC, ΔomcA, and ΔmtrC-ΔomcA, are described in the previous study.10 Wild- type S. oneidensis MR-1 and the various mutants used were routinely cultured at 30°C in dextrose-free tryptic soy broth (TSB, Difco, Lawrence, KS). The Cr(VI) experiments were carried out by using a resting-cell assay. TSB cultures (50 ml) were grown aerobically for 16 hours at 30°C at 100 rpm and harvested by centrifugation at 5,000 × g for 5 minutes. Under these conditions, no growth defect was observed for the mutants used. Cells were washed once in an equal volume of 30 mM sodium bicarbonate buffer (pH 8) at 4°C. Following centrifugation, the cells were re-suspended in the bicarbonate buffer at a density of 2 × 109 cells/ml and purged for 10 minutes with mixed CO2:N2 (80:20) gas. Cr(VI) reduction assays contained 30 mM sodium bicarbonate, pH 8, 0.2 mM K2CrO4 (Sigma, St Louis, MO) and 10 mM sodium lactate that was purged with the mixed CO2:N2 gas and sealed with thick butyl rubber stoppers. Kinetic studies were initiated by adding the purged bacterial cells at a final density of 2 × 108 cells/ml. The same amount of heat-killed wild-type cells was added as a negative control. The reactions were carried out at 30°C with horizontal incubation at 25 rpm. At predetermined time points, cells were harvested. After harvesting by centrifugation, bacterial cells were fixed in 2.5% glutaraldehyde.
For hematite reduction, the cells were prepared in the same way described above. Hematite (11 ± 2 nm; Sample received from Prof. Michael F. Hochella, Center for NanoBioEarth, Department of Geosciences, Virginia Tech) was added at final concentration of 0.1 mM.23 The reduction of hematite was carried out at 30°C with horizontal incubation at 25 rpm.31 At 24 hours, K2CrO4 was added at the final concentration of 0.2 mM. The reductions were carried out under condition described above. At predetermined time points, cells were harvested and fixed in the same way described above.
Surface-enhanced Raman spectroscopy measurements
SERS spectroscopy and imaging were conducted by using an Axiovert 135 inverted scanning confocal microscope, equipped with a 100× and 1.3 NA oil immersion objective (Zeiss FLUAR). A continuous-wave (CW) laser (532 nm, CrystaLaser) was used to pump the sample at 3 µW for SERS and 60 µW for resonance Raman measurements. A beam splitter Z532rdc (Chroma) was used to reflect the excitation light into the microscope objective. Before the scatted light focusing into a monochromator (Triax 550, Jobin Yvon), a band-pass filter HQ580/60M or a long-pass filter HQ545 was positioned before the entrance slit to further reject the Rayleigh light. The Raman spectra were collected by a LN CCD (Princeton Instruments) cooled at about −100 °C with a resolution of 2 cm−1. The setup was carefully calibrated using Mercury lamp and cyclohexane (mode at 801.3 cm−1) before the Raman measurements. For the SERS experiments, we used Ag nanoparticles as substrate and the average size of the nanoparticles is about 50 nm (see Supporting Information for more details.).
SEM and EDX imaging measurements
About 3 µL of the glutaraldehyde-fixed cells was dropped on a clean cover slip to make a dry film. The film was washed carefully with ultrapure water from Millipore. Once the sample dried, an electrically-conductive thin carbon layer of nanometer thickness was coated on the sample surface for a SEM imaging measurement. The SEM imaging was collected by using a FEI-Inspect F scanning electron microscope (FEI, Hillsboro, OR) with a spatial resolution ~ 1 nm. Secondary electrons were probed to get the SEM imaging under a typical acceleration voltage of 20 kV. An EDX system (INCA Penta FET× 3, Oxford Instruments, Abingdon, UK) was attached to the microscope to get the elements (Cr and Fe) imaging.
RESULTS AND DISCUSSION
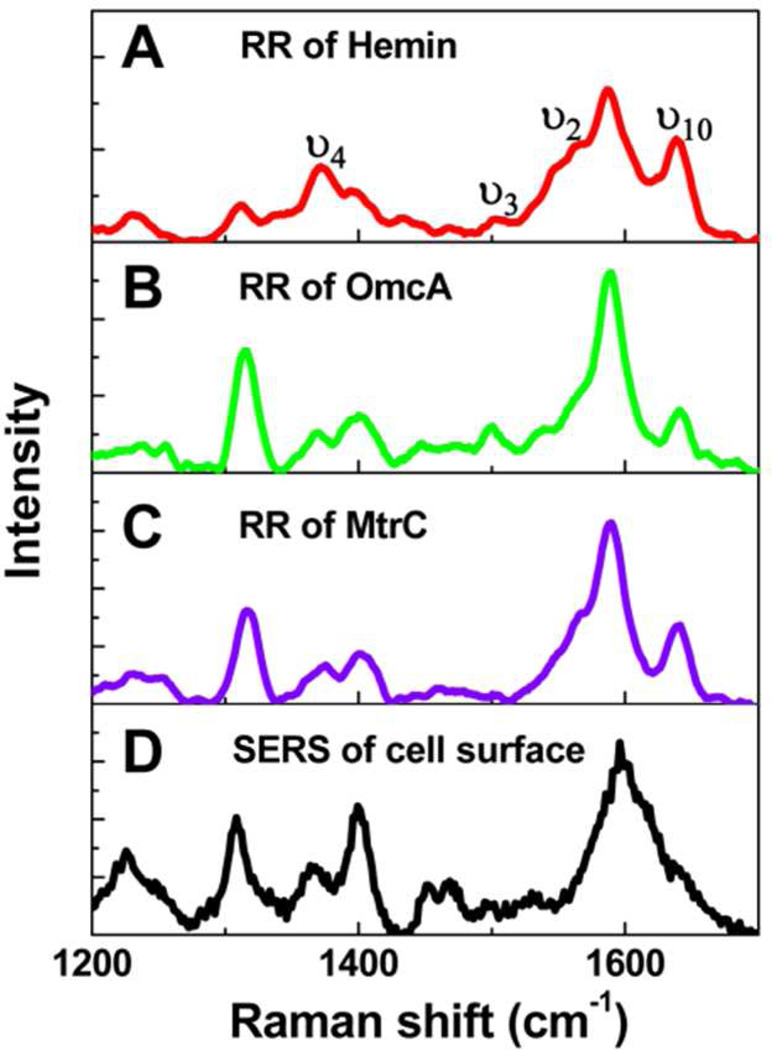
As outer membrane cytochromes (heme proteins), OmcA and MtrC have been suggested to be the major components showing the capability of extracellular respiration at the MR-1 surface.18–22 We have further proved this attribution by our spectroscopic evidences (Figure 1) using resonance Raman (RR) spectroscopy and surface-enhanced Raman spectroscopy (SERS). Figure 1A, 1B, and 1C show the RR spectra of Hemin Chloride (an analogue of heme group in c-type cytochrome), purified OmcA and MtrC. The typical and signature vibrational modes of heme group, such as ν4, ν3, ν2, and ν10 are shown clearly, and the spectral profiles are essentially the same among the examined samples, indicating OmcA and MtrC are intrinsically heme proteins. Moreover, as the oxidation state marker, mode ν4 peaks at 1373 cm−1, implying that all three samples are in their oxidized state, because of having the vibrational frequency in the range of ~1368–1377 cm−1, ferric (FeIII) state.24
Figure 1.
Resonance Raman spectra of Hemin (A), OmcA (B), MtrC (C), and SERS of cell outer surface of wild type MR-1. Although the Raman intensity shows fluctuation, the typical vibrational modes, such as ν4, ν3, ν2, and ν10, are obvious. Based on the similarity of B, C, and D, protein OmcA and MtrC are identified to be the main compositions of the outer membrane of MR-1, which is consistent with the previous reports.18
We have also applied SERS to investigate the chemical nature of the surface proteins of MR-1. As shown in Figure 1D, the Raman spectral profile of the wild type cell surface matches with the RR spectra of OmcA and MtrC, implying that the major components of the cell surface are dominated by c-type cytochrome. These spectroscopic identifications are consistent but beyond our previous measurements.17,18 In addition, we note that the ν4 mode in Figure 1D peaks at 1366 cm−1 unlike the RR of Hemin, OmcA , and MtrC at 1373 cm−1.The observed 7 cm−1 shift of the oxidation state marker is most likely due to the partial charge transfer between the cell surface heme proteins and the Ag substrate used for the SERS measurements.25
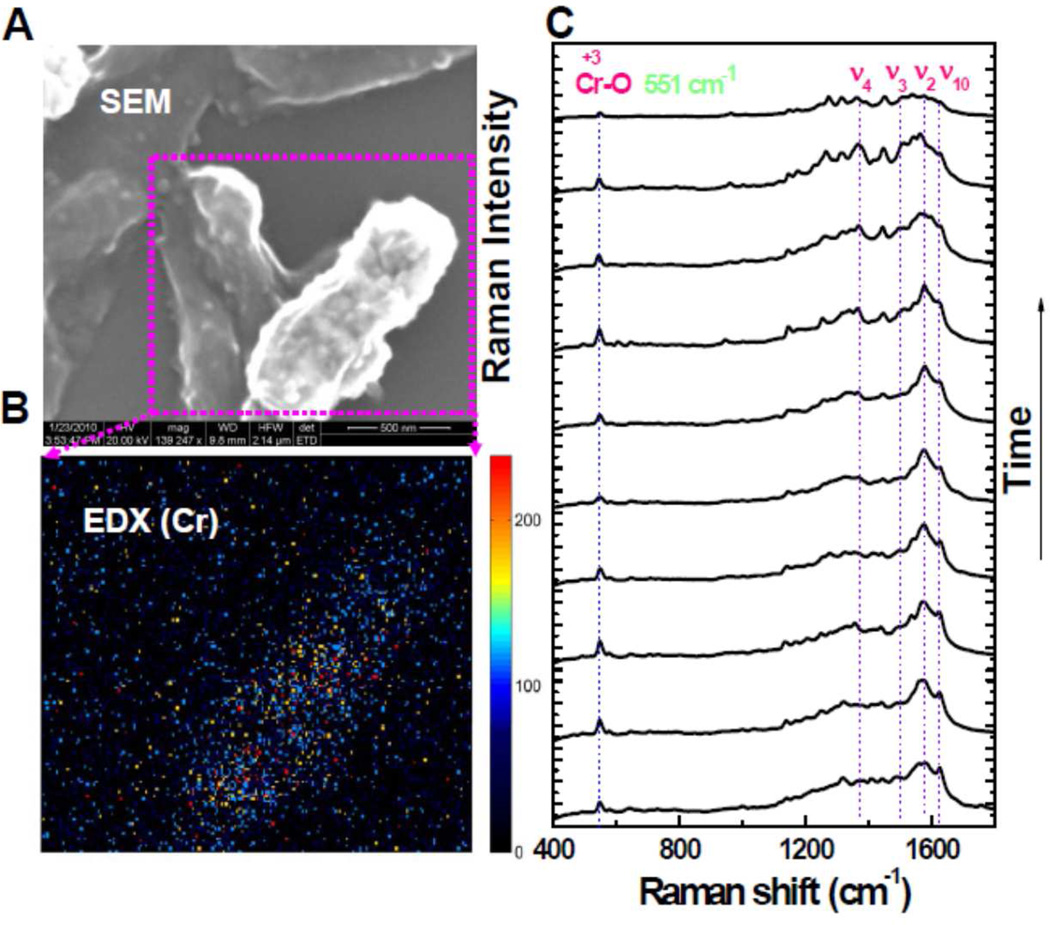
To check the reduction capability of MR-1 cell surface, we performed the biological assay under anaerobic condition by using Cr(VI) in solution. To prove the key roles of the surface proteins, OmcA and MtrC, in the reduction reaction, we have carried out four experiments, under the same measurement conditions and assay protocol,10 on different MR-1 cells: a) wild type, b) ΔmtrC mutant, c) ΔomcA mutant, and d) ΔmtrC-ΔomcA double mutant. We applied SEM, EDX, and SERS to analyze the cell surfaces after Cr(VI) reduction. We have observed rough cell surfaces showing nanoparticles in the case of wild type, ΔmtrC mutant, ΔomcA mutant, but not for the double mutant ΔmtrC-ΔomcA. Because oxygen can be ruled out as an electron acceptor under anaerobic condition, electron transfer from the cell surface to Cr(VI) is the only possible charge transfer pathway. Therefore, the nanoparticles showing on the bacterial MR-1 cell surfaces are attributed to be the reduced insoluble Cr(III) species, i.e., Cr2O3. As a typical case, we show the experimental data of the wild type MR-1 cells in Figure 2. From the SEM image (Figure 2A), nanoparticles can be clearly observed. Figure 2B shows the EDX image of the two adjacent wild type cells in that only one of them is covered with significant number of nanoparticles. Accordingly, high concentrated Cr element is observed on this cell surface. To further probe the chemical nature of the formed nanoparticles, we performed SERS measurements and the results for wild type MR-1 are shown in Figure 2C. Obviously, besides the typical Raman vibrational modes (typical peaks from 1100 to 1650 cm−1) originated from outer membrane c-type cytochromes of MR-1, there is also an additional peak at 551 cm−1, the A1g vibrational mode of Cr(III)-O bond originated from Cr2O3. The other possible compounds, like CrOOH or Cr(OH)3, have different Raman shift for their Cr(III)-O mode. 26,27 Co-localization of cytochromes and Cr(III) suggests that MtrC and OmcA reduce Cr(VI) directly, which is consistent with the terminal reductase roles of these cytochromes in Cr(VI) reduction.10 On the basis of our control experiments, the outer membrane proteins OmcA and MtrC are demonstrated to be the dominant terminal reductases of the Cr(VI) reduction reaction. Combined with the statistical analysis of the density of the nanoparticles on all four samples as well as our recent discoveries,10 we suggest that the reactivity of the reduction reaction changes in the order: wild type > ΔmtrC mutant or ΔomcA mutant > ΔomcA-ΔmtrC double mutant. Our conclusion is consistent with the previous results. 10
Figure 2.
(A) A typical SEM image of MR-1 after the Cr(VI) reduction assay (the scale bar for the SEM image is 500 nm). Nanoparticles emerged on the cell surface after the reduction (an additional SEM image is shown in the Supporting Information to clearly show the nanoparticles on the cell surface.). (B) Nanoparticles are suggested to be Cr by EDX characterization. (C) The chemical composition of the nanoparticles is identified to be the reduced product, Cr(III), i.e., insoluble Cr2O3 by SERS depending on the occurring prominent peak at 551 cm−1, the specific A1g vibrational mode of Cr (III)-O bond, besides the typical peaks originated from the cell surface proteins.
A fundamental understanding of the reduction mechanism of Cr(VI) is critical for the promotion of the bioremediation efficiency. Moreover, a clear and in-depth mechanism can also be used as a reference to study the bioremediation processes of other metal contaminations. Previously, the bioremediation mechanism of Cr(VI) has been suggested to be direct microbial reduction12,14 or Fe (II)-mediated reduction9,28,29 for Shewanella species. Here, we demonstrate that these two mechanisms can co-exist in the Cr(VI) reduction by MR-1.
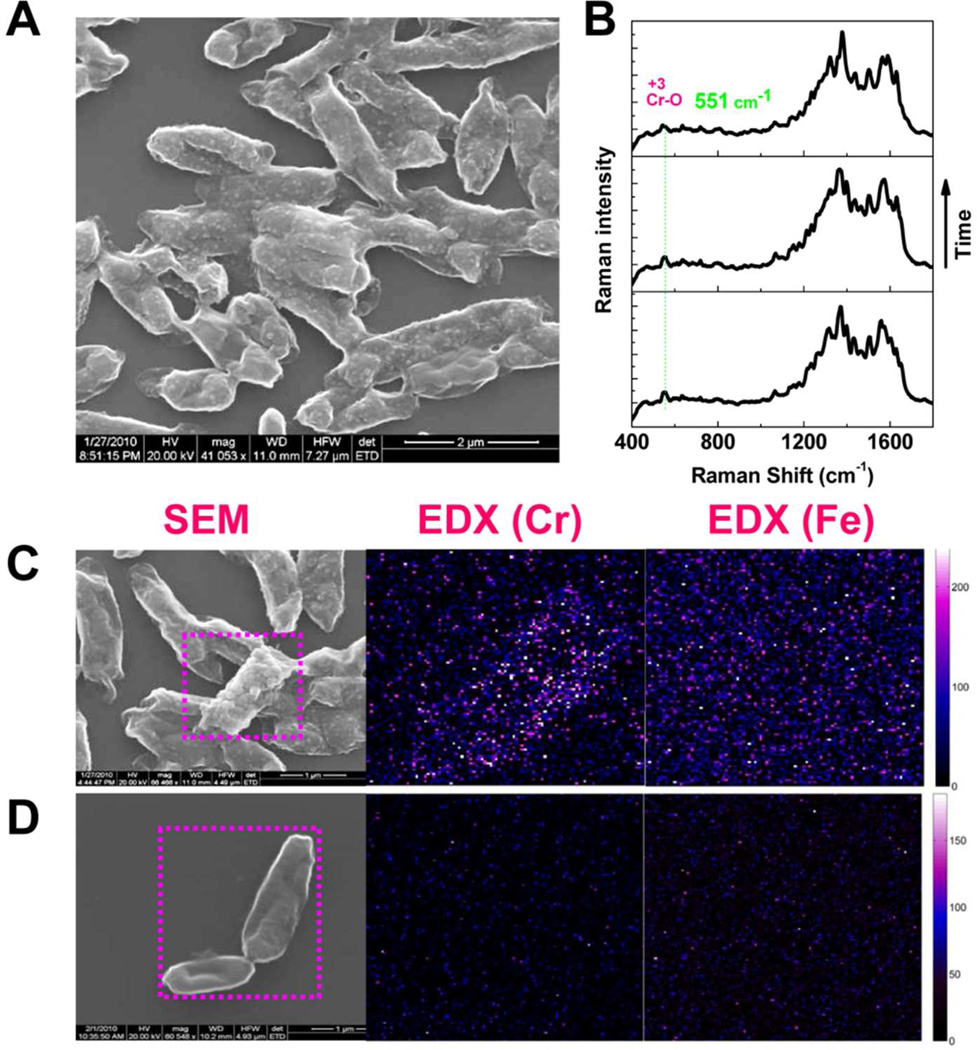
Shewanella species have been observed to be capable of reductively dissolving Fe(III) containing minerals such as ferrihydrite, goethite, and hematite (Fe2O3) in a natural and oxygen limited environment.30 Previously, electron exchange between the MR-1 and hematite nanoparticles has been suggested to occur both direct and indirect mechanisms with the reduction rates altering due to nanoparticle size, shape, and aggregation state.31 Moreover, direct electron exchange between the surface proteins (OmcA and MtrC) of MR-1 and hematite electrodes has been demonstrated by using an electrochemical approach.19 Because hematite is abundant on the earth, if it can mediate the reduction process between Shewanella and Cr(VI), it will help the natural Cr(VI) bioremediation. Therefore, our analytical assay for probing the Cr(VI) reduction mechanism was also performed with additional hematite. Figure 3 shows the imaging and spectroscopic characterizations of the hematite involved Cr(VI) reduction by MR-1 cells. The SEM images (Figure 3A) shows that a significant number of the nanoparticles appeared on the cell surfaces of wild type, ΔomcA mutant and ΔmtrC mutant but not for ΔomcA-ΔmtrC. The additional 551 cm−1 Raman peak (Figure 3B), a signature vibrational signal of Cr2O3, unambiguously identify that the chemical nature of the nanoparticles is Cr2O3, the reduced product of Cr(VI). In addition, we have also applied EDX to analyze the distributions of the elements Cr and Fe on the cell surface. However, for the wild type, ΔomcA mutant and ΔmtrC mutant, we observed Cr but no Fe. Figure 3C shows a typical imaging data of wild type MR-1 after the reduction assay. Under the same background noise level, Cr is clearly observed. For the double mutant ΔomcA-ΔmtrC, most of time we only observe smooth cell surfaces, and there is no concentrated Cr or Fe as shown in Figure 3D. On the basis of these observations, we suggest that most of the Cr2O3 nanoparticles on the cell surfaces are originated from a direct microbial reduction. Our single-cell bioremediation result is consistent with the reported ensemble measurements of a direct microbial reduction mechanism.12,14
Figure 3.
Imaging and spectroscopic characterization of the Cr(VI) reduction with hematite involved. (A) SEM image of the wild type MR-1 after hematite-involved Cr(VI) reduction. The appearance of the nanoparticles is obvious on the cell surface. (B) Using SERS, specific vibrational mode signature signal of Cr(III)-O of Cr2O3 is recorded and used to identify the Cr(III) product redox state. (C) Using EDX, Cr is observed to be concentrated on the cell surface but no Fe is observed for the wild type MR-1. (D) For the double ΔomcA-ΔmtrC as a reducer, cell surface is smooth with less possibility of finding the nanoparticles. There is no concentrated Cr or Fe that can be observed using EDX.
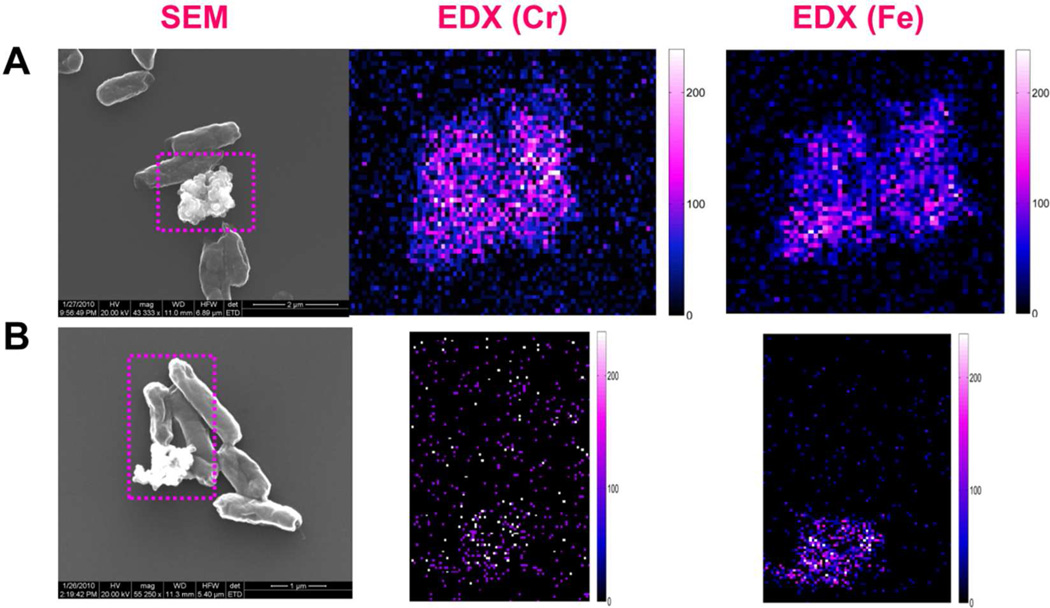
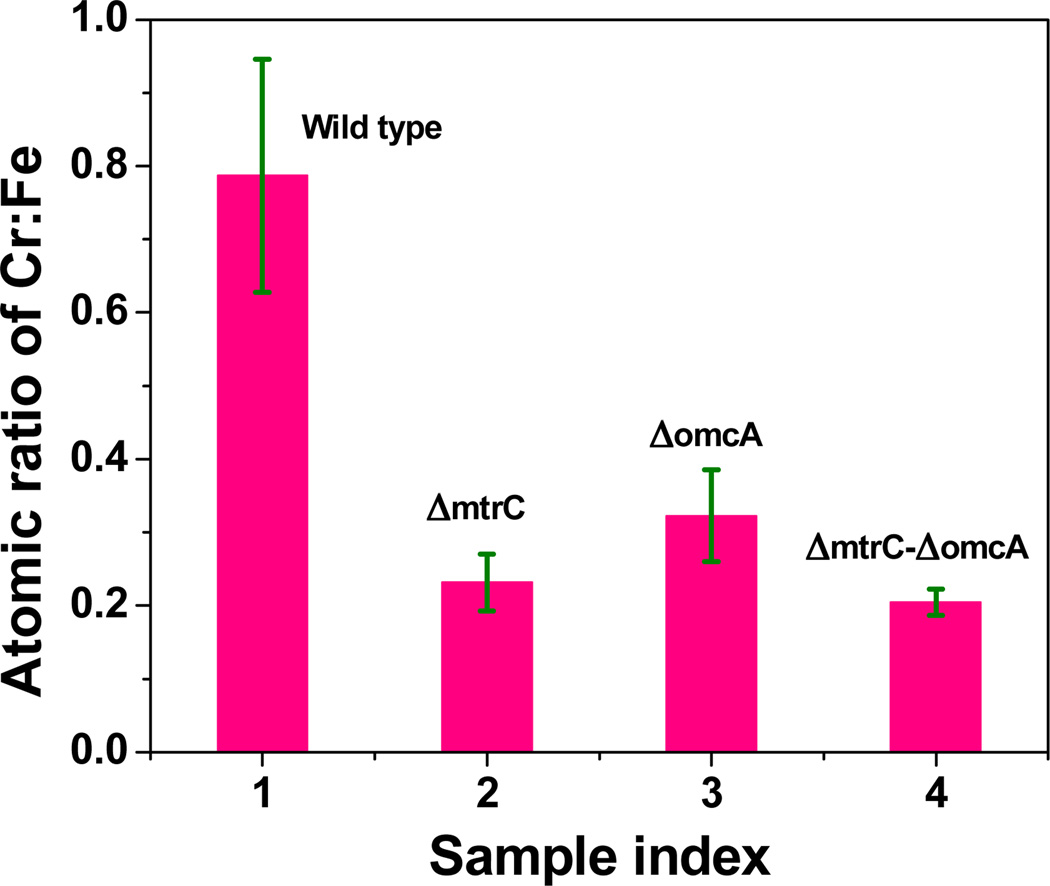
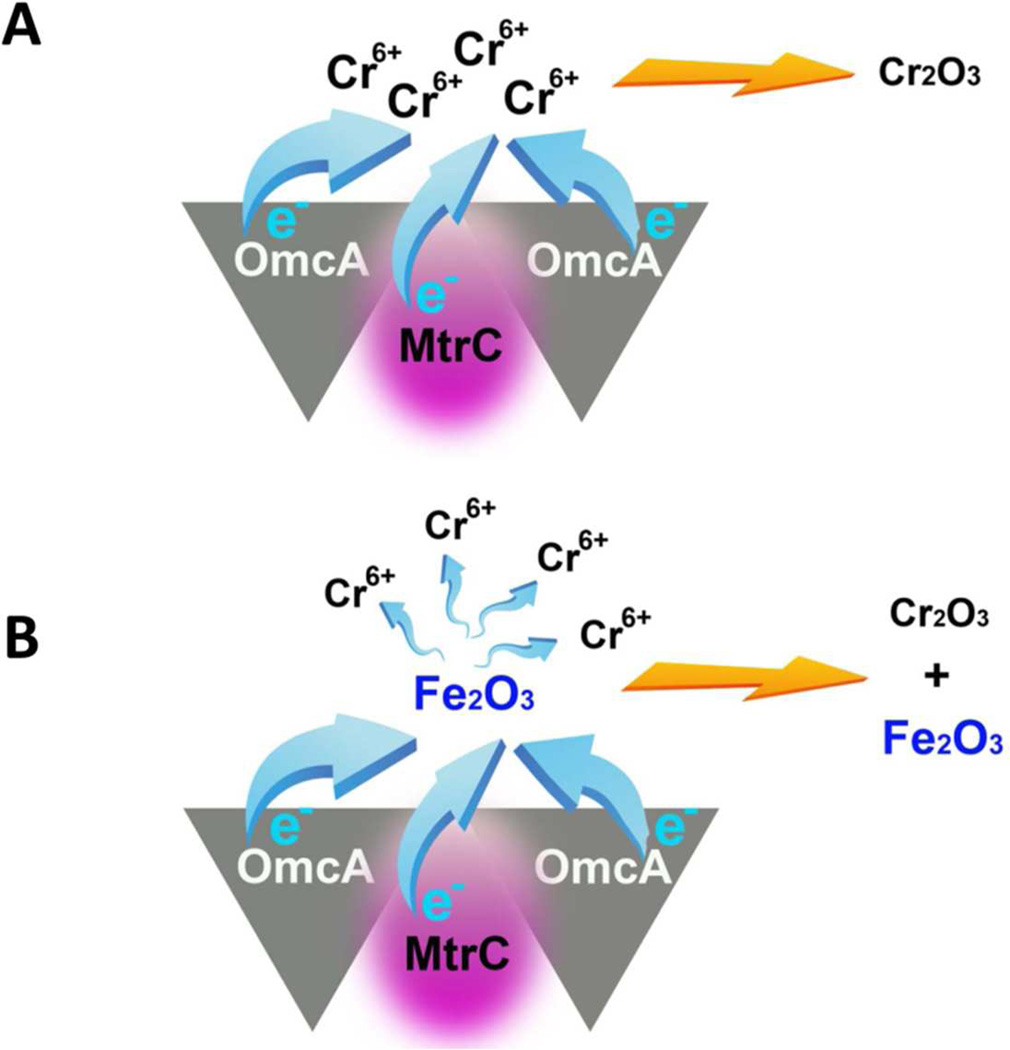
For the hematite involved samples, in addition to the observed nanoparticles on cell surfaces, we also observed large aggregations around the cell surfaces. The SEM images are shown in Figure 4. Using EDX characterization, we found concentrated Cr co-localized with Fe in the aggregations for the samples: wild type, mutant ΔomcA and mutant ΔmtrC. In contrast, for double mutant ΔomcA-ΔmtrC, only low concentrations of Cr were observed. Figures 4A and 4B show the typical SEM and EDX images for the wild type and mutant ΔomcA-ΔmtrC samples. For a quantitative comparison, we measured the amount of Cr and Fe and calculated the Cr : Fe atomic ratios in the aggregations for all four samples with multiple (at least five times) independent sampling by using EDX imaging. The Cr : Fe atomic ratios are calculated to be 0.78 : 1, 0.23 : 1, 0.32 : 1, and 0.2 : 1 for the samples wild type, single mutant ΔmtrC, ΔomcA, and double mutant ΔomcA-ΔmtrC, respectively (Figure 5). The heterogeneous distributions (especially the high concentration for the wild type) of the Cr:Fe atomic ratios imply that the insoluble Cr species (most likely, Cr2O3) in the hematite aggregations is not originated simply from the physical adsorption after the direct microbial reduction. In a direct reduction, the ratios should be close to homogeneity after multiple times of independent sampling. Considering Fe(II) can mediate the electron transfer between MR-1 and Cr(VI),9,28,29 we suggest that hematite could act as an electron shuttle in our reduction assay experiments. In the Cr(VI) reduction process, in addition to the direct microbial reduction (Figure 6A), MR-1 may first reduce hematite, and then the reduced hematite goes on to transfer an electron to the soluble Cr(VI) (Figure 6B). This observation suggests that the direct microbial Cr(VI) reduction and Fe(II)-mediated Cr(VI) reduction mechanisms may co-exist in the bioremediation processes. This proposal is also consistent with the previous reports that suggest several mechanisms may exist in the bacterial metal-reduction process depending on different local environments and conditions.19
Figure 4.
Imaging the chemical compositions of the aggregations after the reduction reaction. (A) wild type MR-1 with Cr(VI) and hematite. (B) Mutant ΔomcA-ΔmtrC with Cr(VI) and hematite.
Figure 5.
Atomic ratios of Cr:Fe in the aggregations of four control experiments. (A) Wild type MR-1 with Cr(VI) and hematite. (B) ΔmtrC mutant with Cr(VI) and hematite. (C) ΔomcA mutant with Cr(VI) and hematite. (D) Mutant ΔomcA-ΔmtrC with Cr(VI) and hematite. For each sample, at least five large aggregations were selected for the measurements.
Figure 6.
Two mechanisms are proposed to co-exist in the in the Cr(VI) reduction process. (A) Direct microbial Cr(VI) reduction. Surface protein OmcA and MtrC form a functional high-affinity complex in vivo and cooperatively serves as electron donor. (B) Fe (II)-mediated Cr(VI) reduction. Hematite acts as electron shuttle between surface proteins and Cr(VI).
To reveal the mechanism of the MR-1 involved bioremediation process, several aspects, including the reduction of the protein species, their localization and identification, and energetics and kinetics of the reduction reactions, still need further specific characterizations. MR-1 genome encodes 42 c-type cytochromes. However, only three of them (OmcA, MtrC, and MtrA) have been demonstrated to involve in the metal reduction.19 Except cytochromes, iron-sulfur proteins and quinones also have been proved to be capable of reducing heavy metals.19,32,33 We, as well as the other groups, have demonstrated that the reduction proteins could localize inner or outer of the cell surface or as external microbial “nanowire” by using Raman spectroscopy and high resolution topographic imaging techniques.14,17,19,34 For the energetics and dynamics of the reduction reaction, both theoretical and experimental approaches have been conducted in recent years.3,10,19 Nevertheless, a real-time and quantitative description of the metal reducing process could include several important parameters such as the electron transfer rate of the direct bacterial reduction reaction, free energy landscape of the reduction reaction, and the working principles of the electron shuttle in an indirect mechanism.
CONCLUSION
We have applied an approach that combines Raman spectroscopic and SEM imaging analyses to characterize the reduction of Cr(VI) to Cr(III) insoluble nanoparticles by c-type cytochromes, MtrC and OmcA, on the bacterial cell surface. Our results demonstrate that (1) MR-1 is capable of reducing the toxic Cr(VI) to low toxic Cr(III) containing Cr2O3 nanoparticle precipitations on the cell surfaces; (2) the cooperation of the surface proteins OmcA and MtrC makes the reduction reaction most efficient, and the sequence of the reducing reactivity of the MR-1 is: wild type > single mutant ΔmtrC or ΔomcA > double mutant ΔomcA-ΔmtrC; (3) the surface proteins such as OmcA and MtrC are the terminal reductases of Cr(VI); and (4) direct microbial Cr(VI) reduction and Fe(II)-mediated Cr(VI) reduction mechanisms may co-exist in the bioremediation process. These conclusions will advance our understanding of the mechanism and the key parameters in the bioremediation process, such as localization and identification of the terminal reductases and mechanical description of the surface chemical reactions. Moreover, these results demonstrate the potential application of SERS in investigating the mechanisms of metal reductions by microbial surfaced proteins.
Supplementary Material
ACKNOWLEDMENTS
This work was supported by NIH NIEHS (5R01ES017070-03).
REFERENCES
- 1.(a) Imam Khasim D, Nanda Kumar NV, Hussain RC. Enviromental Contamination of Chromium in Agricultural and Animal Products Near a Chromate Industry. Bulletin of environmental contamination and toxicology. 1989;43:742–746. doi: 10.1007/BF01701997. [DOI] [PubMed] [Google Scholar]; (b) Lovley DR. Dissimilatory metal reduction. Annu. Rev. Microbiol. 1993;47:263–290. doi: 10.1146/annurev.mi.47.100193.001403. [DOI] [PubMed] [Google Scholar]
- 2.(a) Yeates GW, Orchard VA, Speir TW, Hunt JW, Hermans MCC. Impact of Pasture Contamination by Copper, Chromium, Arsenic Timber Preservative on Soil Biological Activity. Biol. Fert. Soils. 1994;18:200–208. [Google Scholar]; (b) Chen JM, Hao OJ. Microbial chromium (VI) reduction. Crit. Rev.Environ. Sci. Technol. 1998;28:219–251. [Google Scholar]
- 3.Viamajala S, Peyton B, Petersen J. Modeling Chromate Reduction in Shewanella oneidensis MR-1: Development of a Novel Dual-Enzyme Kinetic Model. Biotechnol. Bioeng. 2003;83:790–797. doi: 10.1002/bit.10724. [DOI] [PubMed] [Google Scholar]
- 4.Bencheikh-Latmani R, Obraztsova A, Mackey M, Ellisman M, Tebo B. Toxicity of CRIII) to Shewanella sp. Strain MR-4 during CRVI) Reduction. Environ. Sci. Technol. 2007;41:214–220. doi: 10.1021/es0622655. [DOI] [PubMed] [Google Scholar]
- 5.Bencheikh-Latmani R, Williams S, Haucke L, Criddle C, Wu L, Zhou J, Tebo B. Global Transcriptional Profiling of Shewanella oneidensis MR-1 during CRVI) and U(VI) Reduction. Appl. Environ. Microb. 2005;71:7453–7460. doi: 10.1128/AEM.71.11.7453-7460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daulton T, Little B, Lowe K, Jones-Meehan J. Electron Energy Loss Spectroscopy Techniques for the Study of Microbial Chromium(VI) Reduction. J. Microbiol. Meth. 2002;50:39–54. doi: 10.1016/s0167-7012(02)00013-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y. Environmental microbe-metal interactions: In: Lovley DR, editor. Microbial Reduction of Chromate. Washington, DC: ASM; 2000. pp. 225–235. [Google Scholar]
- 8.Lovley D. Dissimilatory Metal Reduction. Annu. Rev. Microbio. 1993;47:263–290. doi: 10.1146/annurev.mi.47.100193.001403. [DOI] [PubMed] [Google Scholar]
- 9.(a) Wielinga B, Mizuba MM, Hansel CM, Fendorf S. Iron Promoted Reduction of Chromate by Dissimilatory Iron-Reducing Bacteria. Environ. Sci. Technol. 2001;35:522–527. doi: 10.1021/es001457b. [DOI] [PubMed] [Google Scholar]; (b) Fendorf S, Wielinga BW, Hansel CM. Chromium transformations in natural environments: The role of biological and abiological. processes in chromium(VI) reduction. International Geology Review. 2000;42:691–701. [Google Scholar]
- 10.Belchik SM, Kennedy DW, Dohnalkova AC, Wang Y, Sevinc PC, Wu H, Lin Y, Lu HP, Fredrickson JK, Shi L. Extracellular Reduction of Hexavalent Chromium by Cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl. Environ. Microb. 2011;77:4035–4041. doi: 10.1128/AEM.02463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Middleton S, Latmani R, Mackey M, Ellisman M, Tebo BM, Criddle CS. Cometabolism of CRVI) by Shewanella oneidensis MR-1 Produces Cell-Associated Reduced Chromium and Inhibits Growth. Biotechnol. Bioeng. 2003;83:627–637. doi: 10.1002/bit.10725. [DOI] [PubMed] [Google Scholar]
- 12.Myers CR, Carstens BP, Antholine WE, Myers JM. Chromium(VI) Reductase Activity is Associated with the Cytoplasmic Membrane of Anaerobically Grown Shewanella putrefaciens MR-1. J. Appl. Microbiol. 2000;88:98–106. doi: 10.1046/j.1365-2672.2000.00910.x. [DOI] [PubMed] [Google Scholar]
- 13.Neal A, Lowe K, Daulton T, Jones-Meehan J, Little B. Oxidation State of Chromium Associated with Cell Surfaces of Shewanella oneidensis during Chromate Reduction. Appl. Surf. Sci. 2002;202:150–159. [Google Scholar]
- 14.Ravindranath SP, Henne KL, Thompson DK, Irudayaraj J. Raman Chemical Imagining of Chromate Reduction Sites in a Single Bacterium using Intracellularly Grown Gold Nanoislands. ACS nano. 2011;5:4729–4736. doi: 10.1021/nn201105r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Jarvis RM, Goodacre R. Discrimination of Bacteria Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2004;76:40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]; (b) Premasiri WR, Gebregziabher Y, Ziegler LD. On the Difference Between Surface-Enhanced Raman Scattering (SERS) Spectra of Cell Growth Media and Whole Bacterial Cells. Appl. Spec. 2011;65:493–499. doi: 10.1366/10-06173. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Colthup N, Daly L, Wiberley S. Introduction to infrared and Raman spectroscopy. New York: Academic press; 1990. [Google Scholar]
- 16.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Surface-Enhanced Raman Spectroscopy. Annu. Rev. Anal. Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 17.Biju V, Pan D, Gorby YA, Fredrickson J, McLean J, Saffarini D, Lu HP. Combined Spectroscopic and Topographic Characterization of Nanoscale Domains and Their Distrubitions of a Redox Protein on Bacterial Cell Surfaces. Langmuir. 2007;23:1333–1338. doi: 10.1021/la061343z. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Chen B, Wang Z, Elias D, Mayer M, Gorby YA, Ni S, Lower BH, Kennedy DW, Wunschel DS, Mottaz HM, Marshall MJ, Hill EA, Beliaev AS, Zachara JM, Frederickson JK, Squier TC. Isolation of a High-Affinity Functional Protein Complex between OmcA and MtrC: Two Outer Membrane Decaheme c-Type Cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 2006;188:4705–4714. doi: 10.1128/JB.01966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meitl LA, Eggleston C, Colberg PJS, Khare N, Reardon C, Shi L. Electrochemical Interaction of Shewanella oneidensis MR-1 and Its Outer Membrane Cytochromes OmcA and MtrC with Hematite Electrodes. Geochim. Cosmochim. Ac. 2009;73:5292–5307. [Google Scholar]
- 20.Beliaev AS, Saffarini DA, McLaughlin JL, Hunnicutt D. MtrC, an Outer Membrane Decahaem c Cytochrome Required for Metal Reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 2001;39:722–730. doi: 10.1046/j.1365-2958.2001.02257.x. [DOI] [PubMed] [Google Scholar]
- 21.Myers JM, Myers CR. Genetic Complementation of an Outer Membrane Cytochrome omcB Mutant of Shewanella putrefaciens MR-1 Requires omcB Plus Downstream DNA. Appl. Environ. Microb. 2002;68:2781–2793. doi: 10.1128/AEM.68.6.2781-2793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers JM, Myers CR. Role for Outer Membrane Cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in Reduction of Manganese Dioxide. Appl. Environ. Microb. 2001;67:260–269. doi: 10.1128/AEM.67.1.260-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Y, Shi L, Chen B, Mayer MU, Lower BH, Londer Y, Bose S, Hochella MF, Fredrickson JK, Squier TC. High-Affinity Binding and Direct Electron Transfer to Solid Metals by the Shewanella oneidensis MR-1 Outer Membrane c-type Cytochrome OmcA. J. Am. Chem. Soc. 2006;128:13978–13979. doi: 10.1021/ja063526d. [DOI] [PubMed] [Google Scholar]
- 24.Feng M, Tachikawa H. Surface-Enhanced Resonance Raman Spectroscopic Characterization of the Protein Native Structure. J. Am. Chem. Soc. 2008;130:7443–7448. doi: 10.1021/ja8006337. [DOI] [PubMed] [Google Scholar]
- 25.De Groot J, Hester RE. Surface-Enhanced Resonance Raman Spectroscopy of Oxyhemoglobin Adsorbed onto Colloidal Silver. J. Phys. Chem. 1987;91:1693–1696. [Google Scholar]
- 26.Mougin J, Rosman N, Lucazeau G, Galerie A. In situ Raman Monitoring of Chromium Oxide Scale Growth for Stress Determination. J. Raman. Spectrosc. 2001;32:739–744. [Google Scholar]
- 27.(a) Yu T, Shen ZX, He J, Sun WX, Tang SH, Lin JY. Phase control of Chromium Oxide in Selective Microregions by Laser Annealing. J. Appl. Phys. 2003;93:3951–3953. [Google Scholar]; (b) McCreery RL, Packard RT. Raman Monitoring of Dynamic Electrochemical Events. Anal. Chem. 1989;61:775A. [Google Scholar]
- 28.Caccavo F, Jr, Ramsing NB, Costerton JW. Morphological and Metabolic Responses to Starvation by the Dissimilatory Metal-Reducing Bacterium Shewanella alga BrY. Appl. Environ. Microb. 1996;62:4678–4682. doi: 10.1128/aem.62.12.4678-4682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyman JL, Caccavo F, Jr, Cunningham AB, Gerlach R. Metal-Reducing Bacteria Facilitate the Geochemical Elemination of CRVI) from Contaminated Water. Biorem. J. 2002;6:39–55. [Google Scholar]
- 30.Bose S, Hochella Jr MF, Gorby YA, Kennedy DW, McCready DE, Madden AS, Lower BH. Bioreduction of Hematite Nanoparticles by the Dissimilatory Iron Reducing Bacterium Shewanella oneidensis MR-1. Geochim. et Cosmochim. Acta. 2009;73:962–976. [Google Scholar]
- 31.Bose S, Hochella MF, Jr, Gorby YA, Kennedy DW, McCready DE, Madden AS, Lower BH. Geochimica et Cosmochimica Acta. 2009;73:962. [Google Scholar]
- 32.Richardson DJ. Bacterial Resperation: a Flexible Process for a Changing Enviroment. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 33.Giometti C. Microbial proteomics: functional biology of whole organisms. 2006;49:97–111. [Google Scholar]
- 34.Marshall MJ, Beliaev AS, Dohnalkova AC, Kennedy DW, Shi L, Wang Z, Boyanov MI, Lai B, Kemner KM, McLean JS, Reed SB, Culley DE, Bailey VL, Simonson CJ, Saffarini DA, Romine MF, Zachara JM, Fredrickson JK. c-Type Cytochorme-Dependent Formation of U(IV) Nanoparticles by Shewanella oneidensis. PLoS Biol. 2006;4:1324. doi: 10.1371/journal.pbio.0040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.