Abstract
Advanced omics technologies such as deep sequencing and spectral karyotyping are revealing more of cancer heterogeneity at the genetic, genomic, gene expression, epigenetic, proteomic, and metabolomic levels. With this increasing body of emerging data, the task of data analysis becomes critical for mining and modeling to better understand the relevant underlying biological processes. However, the multiple levels of heterogeneity evident within and among populations, healthy and diseased, complicate the mining and interpretation of biological data, especially when dealing with hundreds to tens of thousands of variables. Heterogeneity occurs in many diseases, such as cancers, autism, macular degeneration, and others. In cancer, heterogeneity has hampered the search for validated biomarkers for early detection, and it has complicated the task of finding clonal (driver) and nonclonal (nonexpanded or passenger) aberrations. We show that subtyping of cancer (classification of specimens) should be an a priori step to the identification of early events of cancers. Studying early events in oncogenesis can be done on histologically normal tissues from diseased individuals (HNTDI), since they most likely have been exposed to the same mutagenic insults that caused the cancer in their neighboring tissues. Polarity assessment of HNTDI data variables by using healthy specimens as outgroup(s), followed by the application of parsimony phylogenetic analysis, produces a hierarchical classification of specimens that reveals the early events of the disease ontogeny within its subtypes as shared derived changes (abnormal changes) or synapomorphies in phylogenetic terminology.
Keywords: Cancer, Early events, Heterogeneity, Parsimony, Phylogenetics, Synapomorphies, Systems biology
Cancer remains a particularly challenging disease in terms of prevention and treatment, and our progress in understanding its initiation and progression has been slow and patchy despite the large worldwide investment. While there are many reasons for the slow progress, one issue stands out as a major obstacle to advancement, namely, the various forms of heterogeneity in malignant transformation and progression. Published studies on tumor heterogeneity date back to the early 1950s and continue to this day; however, clinical and pharmaceutical research persistently ignore this crucial problem [Heppner and Miller, 1983; Heng et al., 2009; Michor and Polyak, 2010]. Not surprisingly, many physicians and cancer biologists are frustrated with the lack of progress on circumventing the problem [Couzin-Frankel, 2011]. Heterogeneity has been an underestimated phenomenon in biological systems in general, and an issue that is challenging for scientists who lack training in population biology and evolutionary theory, and for those who view cancer as a static rather than a dynamic disease. There are relatively few areas of bioinformatic research suitable for dealing with heterogeneous data; among the most promising is an analytical paradigm found in the biological field of phylogenetics [Abu-Asab et al., 2008a, b].
Introduction of omics’ high-throughput technologies, such as microarrays, deep sequencing, and mass spectrometry of metabolomics and proteomics has brought to the forefront the challenging problem of analyzing massive heterogeneous data with thousands of variables (also known to computer scientists as high-dimensional data) and increased the awareness about the phenomenon of biological heterogeneity [Clarke et al., 2008]. There are at least 3 levels of intratumoral heterogeneity in tumors; the first being where a tumor has more than one cellular/molecular lineage present despite being of clonal origin. The second level is between tumors within the same individual, such as a primary and its multiple metastases. A third level of heterogeneity is present among tumors of the same histological subtype but from different individuals.
The need for algorithmic solutions to accurately explore heterogeneous data is evident. Without a bioinformatic solution we cannot determine the molecular boundary of diseases, identify biomarkers for early detection, subtype a disease, or stratify patients for treatment options. Heterogeneity is also often the reason for treatment failure, especially when the tumors develop resistance to chemotherapy [Sumer and Gao, 2008; Heng et al., 2010]. Heterogeneity within patient populations likely also contributes to the interindividual variability in both responses to treatment and the distribution of adverse effects [Goldstein et al., 2007].
Early events in cancer initiation remain unknown for many cancers, and there is no concrete agreement on the precise driving events [Pogribny, 2010]. Much of the debate now centers on whether early events are either genetic or epigenetic [Koturbash et al., 2011], involve mitochondrial mutations [Fendt et al., 2011], constitute one ontogenetic pathway or more (e.g. low-vs. high-grade pathways) [Landen et al., 2008; Abu-Asab et al., 2011], and whether early events are similar in every cancer type, persist or are superseded by later events [Ranzani et al., 1995]. Few rigorous approaches or methods have been proposed to identifying early cancer events from omics datasets.
We propose that early events in cancer initiation should be identified in nondiseased rather than neoplastic specimens, specifically in histologically normal tissues from diseased individuals (HNTDI). Such specimens, when taken from tissue that is in proximity to tumor within an organ, often harbor premalignant molecular alterations [Tripathi et al., 2008; Chen et al., 2010; Graham et al., 2010] that differ from healthy controls in gene expression. Ganeshan et al. [2011] suggested that texture analysis of the tissue surrounding a focal breast lesion could be used to identify subgroups of patients with ductal carcinoma in situ who have a lower risk for positive resection. ‘Normal’ tissues proximal/adjacent to the tumor are among the most appropriate for studying early events because they have likely been exposed to the same mutagenic insult(s) that caused the neighboring cancer. We propose that HNTDI data should first be compared with controls via the outgroup comparison in a polarity assessment. Subsequently, data can be processed in a parsimonious phylogenetic analysis. Early events should then be revealed as shared derived changes (abnormal changes) or synapomorphies. Thus, we should be able to classify subtype-related specimens into natural lineages, or clades in phylogenetic terminology, as we describe below.
The Dynamic Nature of Cancer and the Identification of Early Events
Cancer differs from many other diseases in its long time course and the dynamic nature of progression and development: cancer cells continue to change by adapting to selective pressures that often cause dedifferentiation to a more ‘primitive’ neoplastic state. The primitive state here is defined as the lack of differentiation, which makes it difficult to assign some cancer cells to any tissue of origin, a particular problem when the primary tumor’s origin is unknown [Daley, 2008]. Since many cancer therapies are based on knowing the site of origin, this problem has immediate clinical relevance. Often, cancer is described as a multiphasic disease, but it is probably more accurate to describe it as a continuum of change accumulation until the cancerous cells reach their new homeostatic state, often seen in metastatic tumors. Since cancer progression usually drives towards a less differentiated phenotype, cancer could be characterized as a downhill race to dedifferentiation. Superimposed on this continuum are multiple developmental pathways or scenarios that can lead to the same result – a cancer phenotype. Therefore, from its initiation onwards, most tumors will contain evolving lineages (or clones) of cells.
The stochastic nature of events throughout its development is a major source of cancer heterogeneity at many levels [Loeb et al., 2008]. However, not all events in a cancer are driver events; some events are nonexpanded (passenger) and do not occur in all tumors of a cancer type. This mixture of driver and passenger events poses significant biological and bioinformatic challenges, and separating out the driver from passenger events is the key to understanding cancer progression [Bozic et al., 2010]. Despite the many mutations that arise in a tumor, cells survive and produce progenies that are better fit for survival. Thus, from within the mutational chaos there appears to be a selection-for-fitness process that produces successful phenotypes. Early and driver events of cancer initiation and progression could, therefore, be precise and definable because otherwise tumors will collapse by the sheer weight of their mutational load, which rarely takes place.
Precisely defining early events may not easily be ascertained because it is difficult to determine the exact timing of the early event. Since molecular boundaries are ill-defined for most diseases, by attempting to characterize early events as we have described here, data accumulation will eventually help to define disease boundaries and its early events by modeling the disease as a spectrum. The number and sequence of early events may vary across the subtypes of the disease, as we show below. Hence, early events are better viewed as a profile of several variables rather than as single biomarkers.
A bioinformatic paradigm that can accurately decipher or model heterogeneity using multiple high-throughput datasets of each cancer type is clearly needed. Such bioinformatic tools should also include identifying early events of disease initiation and the continuous change that creates multiple ontogenetic pathways and levels of heterogeneity [Abu-Asab et al., 2011]. Datasets from different sources, coupled with objective modeling of various types of cancer by phylogenetics, can produce a ‘Tree of Cancer’. We envision such a cancer tree to be a cladogram that enables us to locate the common changes among all or several types of cancer, which includes shared early events that transcend several cancer types. Here we present the results of analysis of 2 sets of gene-expression breast cancer data to illustrate the possibility of data pooling followed by the mapping of early events as the first steps in building a ‘Tree of Cancer’.
By analyzing these data, we unveil 3 major issues related to cancer gene expression as they occur in microarray data. The first issue is the heterogeneity of gene-expression data. We highlight this characteristic in 2 datasets of breast tissue. The second is the identification of early events in cancer initiation that can be approached by using cancer-adjacent specimens and that appear to be histologically normal but harbor gene-expression abnormality, as revealed by a phylogenetic analysis approach. The third issue, which is related to the first two, deals with the question of whether early events can be biomarkers of early transformation from normal cells into cancer.
Materials and Methods
The data used are from 2 microarray publicly available datasets of breast tissues, GDS3139 [Tripathi et al., 2008] and GDS3716 [Graham et al., 2010], which were downloaded from NCBI’s Gene Expression Omnibus (GEO) DataSets (http://www.ncbi.nlm.nih.gov/gds). GDS3139 is comprised of 29 specimens representing 14 ‘normal’ samples from epithelium adjacent to a breast tumor and 15 samples from patients who underwent a reduction mammoplasty (RM) and so are assumed to be disease free. GDS3716 consists of gene-expression data from 4 sets of histologically normal epithelia breast specimens from 18 RM, 6 prophylactic mastectomy surgeries, 9 biopsies from estrogen receptor-alpha positive breast tumors, and 9 biopsies from estrogen receptor-alpha negative breast tumors. Both datasets were analyzed on GPL96 [HG-U133A] Affymetrix Human Genome U133A Array, which allowed us to combine the 2 datasets after the process of polarity assessment, thus producing a total of 71 specimens.
Data were analyzed by the method described in Abu-Asab et al. [2006]. Briefly, the expression values of each specimen were sorted into either derived (abnormal) or ancestral (normal) groups by comparing the values of the HNTDI specimens against the range of the RM specimens for every gene in the dataset. This process transformed the original data matrix into a qualitative matrix of 0s (ancestral/normal) and 1s (derived/abnormal). This matrix was processed with MIX (the parsimony program in the PHYLIP package), using the Wagner parsimony method as described by Felsenstein [1989].
Shared derived gene expressions (termed synapomorphy[ies] in phylogenetic terminology) for each group of specimens that formed a clade (a branch on the cladogram above a node), and those specific for each specimen, were extracted from the program’s ‘outfile’. The MIX algorithm assigned a number to every cladogram node and listed the synapomorphies of each node. Thus, we could match the lists of synapomorphies with gene identifiers from the microarray experiment.
Results
Phylogenetic Analysis
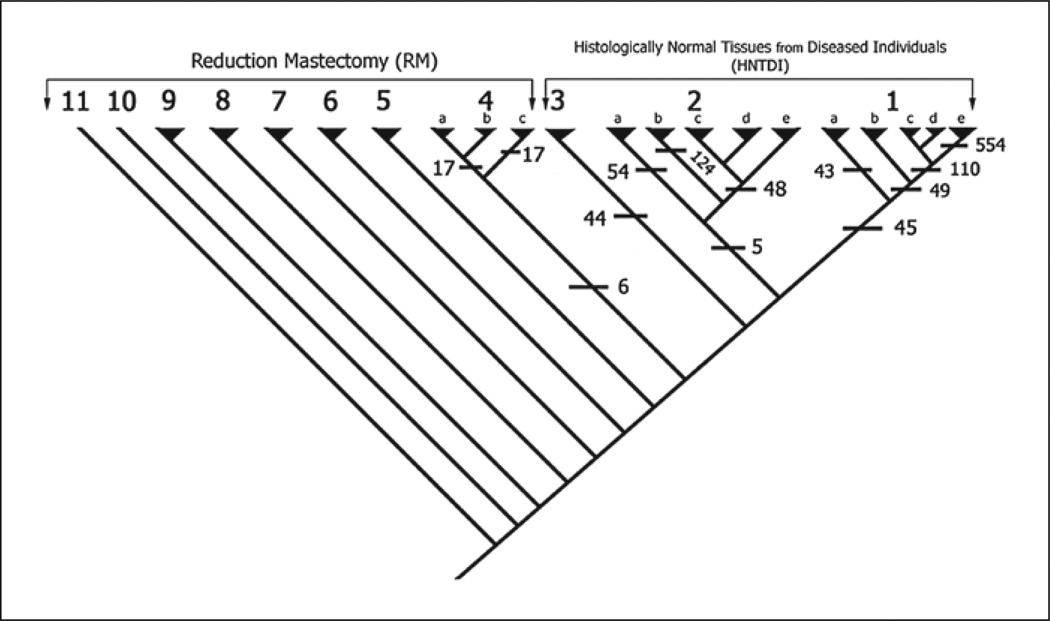
MIX produced 2 similar most parsimonious cladograms with a minor difference in the placement of 2 specimens of the 71 analyzed. Since this difference did not affect either the general topology of the cladograms or the interpretations, we include the first cladogram of the 2 to illustrate our findings (fig. 1).
Fig. 1.
Cladogram showing the classification of all the specimens into clades based on maximum parsimony using MIX of the PHYLIP package. The HNTDI specimens fell into 3 upper clades (1, 2, and 3) each defined by a set of unique synapomorphies (cross bars with numbers), and they do not share any synapomorphies together. The RM specimens fell into 8 clades (4–11) and clade 4 showed 3 subclades. Falling in the closest proximity of the HNTDI specimens, clade 4 may represent a transitional state from healthy towards diseased.
The cladogram classified the 71 specimens into 11 clades (fig. 1), each defined by a number of synapomorphies. Table 1 shows some of the significant clades, their members and their synapomorphies. The base of the cladogram is occupied by 8 clades that belong to the RM group, while the upper 3 clades comprised the HNTDI specimens (clades 1, 2 and 3). The 3 HNTDI clades were arranged in tandem and did not share any synapomorphy (no synapomorphy defined them as a group). Clades 1, 2 and 4 were subdivided into smaller subclades, with each subclade defined by a set of synapomorphies (table 1). Some of these subclades were further subdivided with an increasing number of synapomorphies: clades 1 and 2 were each subdivided into 5 subclades (a–e), where 1a has 43 synapomorphies, and the other 4 subclades (1b–e) share 49 synapomorphies. The more subdivision within the clade the more synapomorphies there are that define these subdivisions. For example, subclade 1e, which occupies a terminal position in its clade, is defined by 554 synapomorphies. Synapomorphies that circumscribe clades and subclades are of greatest interest because they depict the earliest events.
Table 1.
Composition of the clades of interest from the cladogram of figure 1 and their synapomorphies
| Clade 1 |
| 19 Specimens: all from HNTDI |
| GSM512558, GSM512561, GSM512562, GSM512564, GSM512565, GSM512566, GSM512567, GSM512568, GSM512569, GSM512570, GSM512571, GSM512572, GSM512574, GSM512575, GSM512576, GSM512577, GSM512578, GSM512579, GSM512580 |
| 45 Synapomorphies |
| ATP5E, ATP5SL, BLCAP, C19orf10, CALD1, CAPN7, FAM65A, FLOT1, FNDC3B, GANAB, H2AFX, HNRNPA1, HSPD1, IFI35, JUN, KIAA1033, LARP4B, LONP2, LPAL2, MKNK1, MSH6, MYCBP2, NGLY1, ORC2L, PLSCR3, PPCDC, PRDX2, PSMB2, R02172, RAB7A, RARS, RBM16, RBX1, RPIA, SLC25A14, SLC35D2, SNRPB, SWAP70, TARDBP, TEAD4, TIMP1, TRADD, TUBA1C, UBXN7, UQCRC2 |
| Subclade 1a, 43 synapomorphies |
| AKR1C3, ANXA4, ARPC3, BBS9, BLZF1, C3, CTSB, DDR2, EIF3G, ESPL1, FRMD1, GM2A, GPX1, GPX7, HLA-DQA1, IER3, IGF1, LARS2, LGALS3BP, LPAR1, LPCAT1, MED28, MIR936, MSLN, MXRA8, NF1, NLRP1, NPM3, NR4A2, NUDT1, PLTP, PSKH1, QPRT, RPL10P10, RPL28, RPRM, SERTAD2, SRSF1, SSR4, TFAP2A, YTHDF1, ZFP64, ZNF451 |
| Subclades 1b–e, 49 synapom orphies |
| ADAM8, AP2A2, ATP5C1, ATP5C1, AU146983, BAZ1B, BCL7B, CAPZB, CBX6, COMMD4, CSNK1D, CSTB, DDX3X, DNMT1, DUSP8, EDNRB, EPHB4, FRAT2, FYCO1, KLF11, KPNB1, LPPR2, LSM6, MAGEF1, MED27, MRPS31, MTIF2, NAA35, NAP1L1, P4HB, PBXIP1, PDLIM4, PLA2G4B, RAB3GAP1, RARA, RCN3, RNF13, SERINC3, SNRK, SNRNP200, SON, SPTLC2, TMCO6, TMEM159, TMEM2, TMSL3, TNS4, UBR5, ZMYM4 |
| Subclades 1c–e, 110 synapomorphies |
| SEPTIN5, AA719797, ACTB, ADD1, AL109716, ALG6, AMACR, ARFGEF1, ARHGAP11A, ARHGEF7, ARL4A, ARL6IP5, ARNT, ASH1L, ATP6V1B2, BAALC, BAT2, BBS9, BST2, C11orf21, C1orf144, CACNA1A, CBWD7, CD84, CDV3, COL16A1, COMMD3, CSNK1G2, CXCL9, DAZAP1, DECR1, DFNB31, EFNA2, EIF3G, ERP44, F12, FAM125B, FLJ11292, FSHB, FTL, GMEB2, GNAI2, GOLGA1, H49077, HAO1, HIRA, HNRNPL, HNRNPM, ILF3, ITFG1, JMJD6, KCNMB4, KDM5B, KIAA1009, KLHL2, KLRG1, LOC100132247, LOC441899, MACROD1, MAT2A, MON1B, MRAS, MRPL39, MYST2, NDUFS4, NGRN, NLRP1, NSA2, NXT1, PIGC, PPP2R3A, PPP4C, PRDM2, PRUNE, PSMF1, RAD1, RAD23A, RGS5, RHOD, RIOK3, RNF25, RPL28, RPL3, RPLP0, RPLP1, RPP30, RPS16, RPS2, RPS21, RPS3A, RUSC1, SFI1, SFT2D2, SIGLEC6, SIPA1L1, SLC11A2, SNURF, SPTBN1, SRI, STX6, TAF1, THBS1, TRIM32, TRPC1, UBE2Q1, UNC50, WDR1, WDR55, ZNF287, ZNF350 |
| Clade 2 |
| 14 Specimens: all from HNTDI |
| GSM242014, GSM242015, GSM242017, GSM242018, GSM242022, GSM242023, GSM242024, GSM242025, GSM242026, GSM242027, GSM512557, GSM512559, GSM512563, GSM512573 |
| 5 Synapomorphies: |
| CYR61, DUSP1↓, EIF1↓, FOSB, TACSTD2 |
| Subclade 2a, 54 synapomorphies |
| AHNAK, AK000834, AK025360, AKAP13, AL109696, ANKFY1, AP1M2, ARID3B, C1orf116, CCDC88C, CHRNA1, CLCN6, COL4A3, CROCC, DUSP12, DZIP3, EPS8L1, ERLIN2, ERO1LB, FER, FOXO4, GATC, GUSBP3, HOXA5, LHB, MAGEA3, MCOLN1, MXD4, MZF1, PAIP2B, PLGLB1, PRPF39, PRSS22, RB1CC1, RERGL, ROR2, SKAP2, SLC33A1, SLC35A3, SLC6A13, SPATA6, SPRED2, SPTLC1, STK17B, SULF1, TMEM90B, TRMT11, ULK2, WNT2B, ZAK, ZBTB17, ZNF224, ZNF350, ZNF665 |
| Subclade 2b, 124 synapomorphies |
| ADAMTS7, ADRA1B, AGAP2, AK022254, AK024568, AMACR, AP2S1, ARHGDIG, ARHGEF16, ARSJ, ASAH1, ATF4, ATP5H, ATP5L, ATXN7L1, ATXN8OS, AV720803, BAT2, BAX, BGN, BID, BRD2, C7orf28B, CABIN1, CAPZB, CASS4, CCDC88C, CCK, CD58, CDCA4, CDKN2A, CHIT1, CKAP4, COG8, CRCP, CRELD2, CTNNBL1, CYP27B1, CYP2R1, D25272, DGCR8, DRD1, DYSF, EDN2, EHMT2, FGD1, FRS2, FSHB, GALK1, GALNT2, GGCX, GIPR, GLRX3, GNAS, GOLGA2, GPR135, GRHPR, HDLBP, HLA-DOB, HSDL2, HSP90B1, IDH2, IL17B, IL1A, IRAK1, KCNG2, KIAA0319, KIAA1045, KIF1C, KLC1, KLHL20, LANCL2, LMNA, LOC440792, LRCH4, LYZL6, MAP4, MARCO, MDK, MECR, MFNG, NCOR2, NCRNA00260, NFU1, NPHS1, NRIP2, NUP50, OVOL1, P2RY2, PDCD5, PECR, PFDN2, PFN1, PLAUR, PLK3, POLD1, POLR1E, POM121C, PTGIR, PTPLAD1, PUM1, RARG, RFC2, RNF126, RPL22P22, RRP7B, SH3BP2, SLC7A5, SNRPB2, SPIN2A, TGM5, TM9SF1, TMEM132A, TMEM134, TPM4, TSSC1, TUBG1, TXN, UBE2V1P2, UBR4, VEGFA, VENTX, WDR18, Z21967 |
| Subclade 2c–e, 48 synapomorphies |
| AA427737, ADORA1, AF257099, AL137403, BCLAF1, CA3, CCL2, CFLAR, CXCL2, CXCR4, CYLD, DKFZp686O1327, EIF4A1, FKBP8, FRMD1, FUT6, GPR183, GTF2F1, HEY2, IFNGR1, IGHV4-31, KIAA0020, MCL1, MLF1, MYH14, NCLN, NDRG2, NUP88, PDE4B, PNLIPRP2, PPP1R15A, PRNP, PTMA, PTMAP7, PTPN21, RGS1, RNF114, SEC24A, SFRP1, SRF, SRGN, TNFAIP2, TRIM2, UPF3A, WASF3, WSB1, YWHAH, ZFP36 |
| Clade 3 |
| 5 Specimens: all from HNTDI |
| GSM242016, GSM242019, GSM242020, GSM242021, GSM512560 |
| 44 Synapomorphies: |
| ACTR10, ALDH1A3, C14orf147, CAPZA1, CD59, CHMP1B, COPS8, CPD, CSDE1, DCUN1D4, GALNT1, GCOM1, GMFB, HSPH1, KRT10, LEPROTL1, LOC440366, LYST, MCL1, NCRNA00081, NCRNA00120, NUDT4P1, NUPL1, PDS5A, PTP4A1, RAP1A, RBBP7, RBM16, RRAS2, RRP15, SCARB2, SCML1, SET, SLC25A14, SMEK2, SNRNP27, SNRPE, SNX27, SOCS5, YRDC, YWHAZ, ZFP36L1, ZMPSTE24, ZSCAN18 |
| Clade 4 |
| 11 Specimens: all from RM |
| GSM242009, GSM242013, GSM512540, GSM512543, GSM512547, GSM512549, GSM512550, GSM512551, GSM512552, GSM512554, GSM512556 |
| 6 Synapomorphies: |
| DSCAM, DUSP1↑, EIF1↑, HIST1H2BC, JUN, SIK1 |
| Subclades 4a, b, 17 synapomorphies |
| ACAT2, ACBD3, ADAR, ARL4C, BRD7, CAPZB, KATNA1, MRPL39, NEDD8, NSMCE4A, PBX1, PEX19, PTP4A2, SF3A3, SSBP1, TDRD7, WBP4 |
| Subclades 4c, 17 synapomorphies |
| AF198444, AK021633, CITED2, DCUN1D4, DUSP7, FILIP1L, GATM, HIST2H2AA4, HIST2H2AA4, HSPC157, KLF6, NEAT1, PNRC1, REG1A, RHOB, SLC38A2, TOP3A |
In addition to the clades’ synapomorphies, each specimen has its own apomorphies (unique gene-expression aberrations). For example, specimen GSM512567 within subclade 1c has 1,737 apomorphies. These apomorphies are the nonclonal aberrations (passenger) that are specific for this specimen.
The topology of the cladogram has significant biological meaning because it defines the relationships among clades. The arrangement of the HNTDI clades is interpreted as the 3 clades representing 3 independent developmental phenotypes. Thus, the synapomorphies of each HNTDI clade are the early events of a neoplastic phenotype. Also of interest is clade 4, the sister clade of the HNTDI clades. Clade 4 forms a transitional zone from normal RM to cancer-susceptible HNTDI specimens/clades; in this case, clade 4 and its subclades are defined by very small numbers of synapomorphies signifying only slight transformation.
Gene-Expression Heterogeneity
By examining the lists of differentially expressed genes listed by Tripathi et al. [2008] and Graham et al. [2010], no gene from their gene lists had an expression level that was consistently abnormal (derived) across all of the HNTDI specimens when compared with the normal expression range of the RM specimens (fig. 2A). We selected a few genes to illustrate this point. Specimens in figure 2 were arranged according to the HNTDI 3 clades membership to further show the distribution of gene-expression aberration within these clades; it also showed that phylogenetic subtyping of clades identified synapomorphies that have a more consistent expression pattern within each clade than is produced by considering the fold change (fig. 2).
Fig. 2.
Gene-expression heterogeneity in HNTDI. Selected genes from differentially-expressed gene list of Tripathi et al. [2008] and Graham et al. [2010] were not uniformly expressed in all of the HNTDI specimens, while the parsimoniously identified synapomorphies for the 3 clades of HNTDI were more specific to each clade with lesser exceptions than in gene lists of Tripathi et al. [2008] and Graham et al. [2010]. =: No change; −: underexpressed; +: overexpressed.
The patchy (or mosaic) pattern of gene-expression heterogeneity exhibits expression modes within a group of specimens that can be termed the asynchronous and the dichotomously asynchronous [Lyons-Weiler et al., 2004; Abu-Asab et al., 2008b]. The asynchronous mode refers to normal and abnormal gene expressions among the HNTDI specimens of a given gene. The dichotomously asynchronous mode refers to the presence of over- and underexpression of a gene within the HNTDI specimens in relation to the normal range of the RM specimens. Examples of these 2 expression modes can be seen in genes JUN and TIMP1 in figure 2B.
Discussion
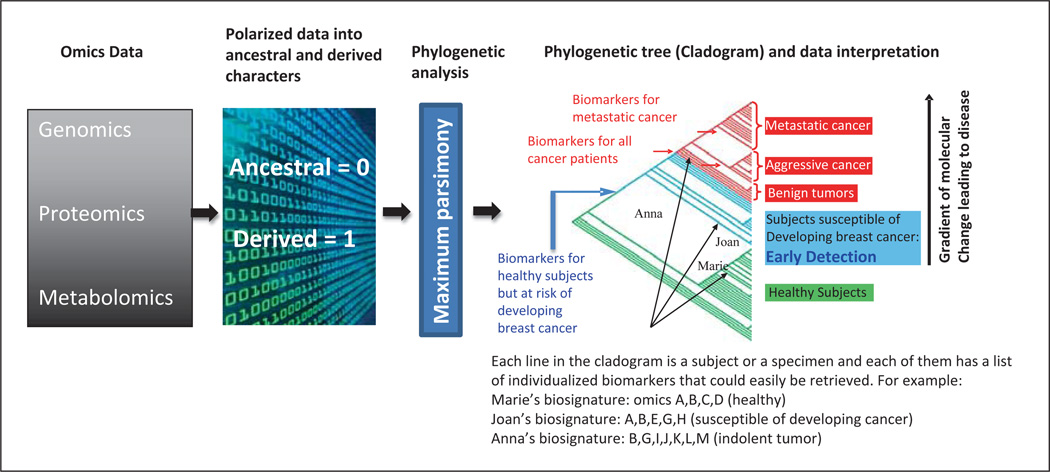
Datasets of gene-expression microarrays in our analysis revealed the extent of existing heterogeneity within the HNTDI breast specimens. This heterogeneity manifests as asynchronous and dichotomously asynchronous gene-expression patterns. However, maximum parsimony remains a most reliable method for analyzing heterogeneous data [Abu-Asab et al., 2008a, b]; the data are modeled onto a cladogram by finding the most parsimonious explanation for data distribution among specimens. Thereafter, the cladogram serves as a multidimensional map that shows the pattern of diversity within the breast cancer by: (i) classifying specimens into groups that share the same gene-expression aberrations, (ii) listing those shared aberrations, and (iii) indicating the direction of change for the entire set of specimens. The cladogram is a hierarchical classification with several levels of branching where each branch is circumscribed by a number of synapomorphies, which are equivalent to clonal changes that may be the driver of the pathological process (fig. 3).
Fig. 3.
Diagram summarizing the application of phylogenetics to omics data (we named it Phylomics) as used in this study. Although Phylomics can be applied to any condition that deviates from its normal/ancestral profile (i.e. mutations, disease onset, response to environmental factors, classification of patients into responders and nonresponders to treatment, etc.), we opted for the example of breast cancer as a disease condition to illustrate its use. In brief, omics data are polarized into ancestral and derived states based on the normal range of each character (i.e. gene, protein or metabolite) using the Universal Parsing Algorithm (UNIPAL) developed by the authors (M.S.A. H.A.). The polarized matrix is then processed using parsimony, a phylogenetic algorithm; the result is a phylogenetic tree, or cladogram, that groups specimens into clades according to their shared derived character states (synapomorphies). This dynamic classification models the disease range on the cladogram from its early stages to its extreme severity. Thus, it plots the specimens with early transformations to the lower end of the disease range on the cladogram, closer to the healthy clade. Red and blue arrows indicate the nodes that delimit the clades for each group sharing common characters; each line within the clade presents a specimen that may have additional characters that are unique to this patient/specimen. For example, Anna shares all the characters that are common among all cancer patients but has additional molecular traits that are unique to her health status.
While microarray data are only one element of a set of events that include epigenetic modifications and genome reorganization, we selected these data because of their widespread usage and public availability. A fold-change analysis of these HNTDI breast specimens has shown that they exhibit signs of early expression transformation of breast cancer [Graham et al., 2010] as well as perturbation of cancer-related pathways that are markers of disease risk, occult disease or the neighboring tissue’s response to an existing tumor [Tripathi et al., 2008].
Due to its dynamic nature, early changes in gene expression in cancer may not be preserved or their signals can be diluted in later stages of cancer progression. Therefore, only tissues showing early signs of transformation are suitable for identifying early events. Using mature cancer specimens – a common practice in cancer research – may not identify biomarkers of early detection. Additionally, the results of our analysis of HNTDI specimens support our earlier conclusion that disease modeling by subtyping (class discovery/classification) should precede exploration of its clonal aberrations or the identification of early events [Abu-Asab et al., 2011]. Both sets of events are different for each subtype of the disease, as we have demonstrated here. The large clades of HNTDI specimens (clades 1, 2, and 3 in fig. 1) are the subtypes that exist in this study collection; their synapomorphies are the early events in breast cancer development (fig. 2).
Our approach and results also call into question the practice of generating differentially expressed genes for a whole set of specimens without a biologically meaningful and discriminatory process. Such lists can be misleading and generate the false impression that they could easily and successfully be used in early detection tests or personalized treatment; this practice has been proven to be unsuccessful [Diamandis, 2010; Buchen, 2011]. The work we describe here offers an alternative approach. The 2 gene lists based on fold-change of differentially expressed genes generated by the 2 previously published studies do not acknowledge the asynchronous nature of the HNTDI gene expression. Other than showing heat maps and qRT-PCR, these lists and descriptions do not mention the asynchronous expressions of differentially expressed genes. Failure to adequately account for heterogeneity is common among microarray studies and has often produced disappointing results. Only by examining the raw data of the 2 studies does the extent of expression heterogeneity of gene expression in the HNTDI specimens become evident.
Do Early Cancer Initiating Events Include Genomic Contributions?
We view cancer initiation and progression as a dynamic continuum of events. Nonetheless, an evident phenotypical distinction can be applied by pathologists to categorize this continuum into 3 phases: histologically normal with minimum change (HNWMC), precancerous and cancerous. The genome theory of cancer evolution asserts a genomic component to cancer initiation in addition to genetic and epigenetic events [Heng et al., 2011a, b]. Genomic instability and its related events are well documented in mature cancers and preneoplastic lesions, but have not yet been shown in HNWMC such as the HNTDI. While precancerous cells are dysplastic with nuclear atypia that cannot always be distinguished from those seen in neoplastic cells [Berman, 2010], HNWMC are not dysplastic and their nuclei cannot be distinguished from normal cells. The implications of declaring early events as only genetic and/or epigenetic affect the conceptual view of cancer and the global search for early detection biomarkers or profiles. However, in the absence of genomic data on HNWMC, it is premature to conclude that early events do not include genomic modifications.
Biomarkers versus Profiles in Early Detection
The general failure of the search for validated early detection biomarkers has exposed the weaknesses of the disease concept within the current biomedical paradigm. The failure may stem partly from the lack of understanding of 2 fundamental characteristics of the disease process. The first is the dynamic nature of disease. Whether one thinks of disease as being either a categorical or a continuous with several phases of events, different events occur at each phase or along the spectrum of the disease (fig. 3). Therefore, markers of early events are most likely different from later events. This necessitates the modeling of each disease by examining as many specimens as possible to establish its spectrum of events from initiation onward. The second fundamental characteristic of the disease process is the heterogeneity that manifests in multiple initiating pathways, each with its own mix of clonal and nonexpanded events.
The dynamic nature of the disease process coupled with heterogeneity of early events (as shown in fig. 2) highlight the inherent weakness of the univariate biomarker concept. Early detection biomarkers remain a hypothesis that needs to be tested [Buchen, 2011]. It seems reasonable to consider that there may not be a reliable single biomarker for early detection. Rather than look for a single or multiple genes, we may be better served looking for a dynamic profile(s) that is capable of classifying each specimen by its closest relationship to known characteristics. The phylogenetic classification of disease specimens could accomplish this goal by modeling disease datasets into a cladogram that can be used as a map of the disease spectrum and for determining the health status of a new specimen by knowing its location on the cladogram.
Conclusions
Heterogeneity in biological data, especially in diseases, may not be easily addressed by reducing high dimensional data to a much smaller number of variables. Heterogeneity is indicative of the many selective processes at work that produce multiple ontogenetic pathways and clonal lineages, and this is likely responsible for variable responses to drugs as well as drug resistance and adverse effects. The linkages among many cellular processes imply that simply selecting differentially expressed genes on an arbitrary basis like fold-change may be suboptimal or produce misleading outcomes.
Separating clonal (driver) from nonexpanded (passenger) aberrations and identifying early events cannot be done without a priori modeling of the data to identify subtypes that reflect the natural classes of a disease. Early events, if they exist as universal clonal aberrations, could be potential biomarkers for early detection. However, as we have shown here, their universality seems unlikely. Early events may be lost in mature cancer specimens and, therefore, the search for early events may be most productive in HNTDI specimens. As shown in our analysis, HNTDI harbors many gene transformations. By comparing HNTDI with normal specimens in a phylogenetic paradigm, it may be possible to identify early events as they relate to the subtypes of the disease.
References
- Abu-Asab MS, Chaouchi M, Amri H. Phyloproteomics: what phylogenetic analysis reveals about serum proteomics. J Proteome Res. 2006;5:2236–2240. doi: 10.1021/pr0504485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab MS, Chaouchi M, Amri H. Evolutionary medicine: a meaningful connection between omics, disease, and treatment. Proteomics Clin Appl. 2008a;2:122–134. doi: 10.1002/prca.200780047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab MS, Chaouchi M, Amri H. Phylogenetic modeling of heterogeneous gene-expression microarray data from cancerous specimens. Omics. 2008b;12:183–199. doi: 10.1089/omi.2008.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab MS, Chaouchi M, Alesci S, Galli S, Laassri M, et al. Biomarkers in the age of omics: time for a systems biology approach. Omics. 2011;15:105–112. doi: 10.1089/omi.2010.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman JJ. Precancer: The Beginning and the End of Cancer. Sudbury: Jones and Bartlett Publishers; 2010. [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchen L. Cancer: Missing the mark. Nature. 2011;471:428–432. doi: 10.1038/471428a. [DOI] [PubMed] [Google Scholar]
- Chen DT, Nasir A, Culhane A, Venkataramu C, Fulp W, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat. 2010;119:335–346. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Ressom HW, Wang A, Xuan J, Liu MC, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008;8:37–49. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin-Frankel J. Personalized medicine. Pushing the envelope in neuroblastoma therapy. Science. 2011;333:1569–1571. doi: 10.1126/science.333.6049.1569. [DOI] [PubMed] [Google Scholar]
- Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:171–174. doi: 10.1101/sqb.2008.73.041. [DOI] [PubMed] [Google Scholar]
- Diamandis EP. Cancer biomarkers: can we turn recent failures into success? J Natl Cancer Inst. 2010;102:1462–1467. doi: 10.1093/jnci/djq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fendt L, Niederstätter H, Huber G, Zelger B, Dünser M, et al. Accumulation of mutations over the entire mitochondrial genome of breast cancer cells obtained by tissue micro-dissection. Breast Cancer Res Treat. 2011;128:327–336. doi: 10.1007/s10549-010-1092-8. [DOI] [PubMed] [Google Scholar]
- Ganeshan B, Strukowska O, Skogen K, Young R, Chatwin C, Miles K. Heterogeneity of focal breast lesions and surrounding tissue assessed by mammographic texture analysis: preliminary evidence of an association with tumor invasion and estrogen receptor status. Front Oncol. 2011;1:33. doi: 10.3389/fonc.2011.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Need AC, Singh R, Sisodiya SM. Potential genetic causes of heterogeneity of treatment effects. Am J Med. 2007;120:S21–S25. doi: 10.1016/j.amjmed.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Graham K, de las Morenas A, Tripathi A, King C, Kavanah M, et al. Gene expression in histologically normal epithelium from breast cancer patients and from cancer-free prophylactic mastectomy patients shares a similar profile. Br J Cancer. 2010;102:1284–1293. doi: 10.1038/sj.bjc.6605576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol. 2009;220:538–547. doi: 10.1002/jcp.21799. [DOI] [PubMed] [Google Scholar]
- Heng HH, Liu G, Stevens JB, Bremer SW, Ye KJ, Ye CJ. Genetic and epigenetic heterogeneity in cancer: the ultimate challenge for drug therapy. Curr Drug Targets. 2010;11:1304–1316. doi: 10.2174/1389450111007011304. [DOI] [PubMed] [Google Scholar]
- Heng HH, Liu G, Stevens JB, Bremer SW, Ye KJ, et al. Decoding the genome beyond sequencing: the new phase of genomic research. Genomics. 2011a;98:242–252. doi: 10.1016/j.ygeno.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Heng HH, Stevens JB, Bremer SW, Liu G, Abdallah BY, Ye CJ. Evolutionary mechanisms and diversity in cancer. Adv Cancer Res. 2011b;112:217–253. doi: 10.1016/B978-0-12-387688-1.00008-9. [DOI] [PubMed] [Google Scholar]
- Heppner GH, Miller BE. Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Rev. 1983;2:5–23. doi: 10.1007/BF00046903. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Beland FA, Pogribny IP. Role of epigenetic events in chemical carcinogenesis – a justification for incorporating epigenetic evaluations in cancer risk assessment. Toxicol Mech Methods. 2011;21:289–297. doi: 10.3109/15376516.2011.557881. [DOI] [PubMed] [Google Scholar]
- Landen CN, Jr, Birrer MJ, Sood AK. Early events in the pathogenesis of epithelial ovarian cancer. J Clin Oncol. 2008;26:995–1005. doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- Lyons-Weiler J, Patel S, Becich MJ, Godfrey TE. Tests for finding complex patterns of differential expression in cancers: towards individualized medicine. BMC Bioinformatics. 2004;5:110. doi: 10.1186/1471-2105-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michor F, Polyak K. The origins and implications of intratumor heterogeneity. Cancer Prev Res (Phila) 2010;3:1361–1364. doi: 10.1158/1940-6207.CAPR-10-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP. Epigenetic events in tumorigenesis: putting the pieces together. Exp Oncol. 2010;32:132–136. [PubMed] [Google Scholar]
- Ranzani GN, Luinetti O, Padovan LS, Calistri D, Renault B, et al. p53 gene mutations and protein nuclear accumulation are early events in intestinal type gastric cancer but late events in diffuse type. Cancer Epidemiol Biomarkers Prev. 1995;4:223–231. [PubMed] [Google Scholar]
- Sumer B, Gao J. Theranostic nanomedicine for cancer. Nanomedicine (Lond) 2008;3:137–140. doi: 10.2217/17435889.3.2.137. [DOI] [PubMed] [Google Scholar]
- Tripathi A, King C, de la Morenas A, Perry VK, Burke B, et al. Gene expression abnormalities in histologically normal breast epithelium of breast cancer patients. Int J Cancer. 2008;122:1557–1566. doi: 10.1002/ijc.23267. [DOI] [PubMed] [Google Scholar]