Abstract
Endolysins are bacteriophage enzymes that lyse their bacterial host for phage progeny release. They commonly contain an N-terminal catalytic domain that hydrolyzes bacterial peptidoglycan (PG) and a C-terminal cell wall-binding domain (CBD) that confers enzyme localization to the PG substrate. Two endolysins, phage lysin L (PlyL) and phage lysin G (PlyG), are specific for Bacillus anthracis. To date, the cell wall ligands for their C-terminal CBD have not been identified. We recently described structures for a number of secondary cell wall polysaccharides (SCWPs) from B. anthracis and B. cereus strains. They are covalently bound to the PG and are comprised of a -ManNAc-GlcNAc-HexNAc- backbone with various galactosyl or glucosyl substitutions. Surface plasmon resonance (SPR) showed that the endolysins PlyL and PlyG bind to the SCWP from B. anthracis (SCWPBa) with high affinity (i.e. in the μM range with dissociation constants ranging from 0.81 × 10−6 to 7.51 × 10−6 M). In addition, the PlyL and PlyG SCWPBa binding sites reside with their C-terminal domains. The dissociation constants for the interactions of these endolysins and their derived C-terminal domains with the SCWPBa were in the range reported for other protein–carbohydrate interactions. Our findings show that the SCWPBa is the ligand that confers PlyL and PlyG lysin binding and localization to the PG. PlyL and PlyG also bound the SCWP from B. cereus G9241 with comparable affinities to SCWPBa. No detectable binding was found to the SCWPs from B. cereus ATCC (American Type Culture Collection) 10987 and ATCC 14579, thus demonstrating specificity of lysin binding to SCWPs.
Keywords: Bacillus anthracis, bacteriophage, endolysin, polysaccharide, secondary cell wall polymer
Introduction
Bacillus anthracis is the etiologic agent of anthrax, a disease primarily of herbivores, although humans can also be infected. Regardless of B. anthracis' route of entry (cutaneous, gastrointestinal or inhalation), systemic anthrax has a high case-fatality ratio. Bacillus anthracis has long been recognized as a potential biological weapon (Hilleman 2002; Baillie 2005). In particular, since the anthrax bioterrorism events in 2001, there has been a renewed interest in effective diagnostic tools and medical countermeasures (Russell 2007).
Endolysins are bacteriophage-encoded enzymes capable of hydrolyzing the peptidoglycan (PG) of the cell wall murein layer. Because of this activity, endolysins have also been termed phage lysozymes, lysins or muralytic or mureolytic enzymes. They are considered potent antibacterials and a potential alternative to antibiotics for the control of pathogenic bacteria, including B. anthracis and B. cereus. Endolysins are expressed intracellularly in phage-infected bacterial cells at the end of the phage lytic cycle during multiplication to release the phage progeny. Endolysins are characterized by their ability to target and cleave bonds of the PG thereby degrading the rigid PG murein layer, which leads to cell lysis and, in turn, to the release of the newly assembled virions (for review, see Loessner 2005).
Two well-characterized endolysins capable of lysing B. anthracis cells are phage lysin G (PlyG) and phage lysin L (PlyL) (Schuch et al. 2002; Low et al. 2005). The PlyG protein was initially identified in the γ-phage (Schuch et al. 2002). This phage, at first isolated as a variant of the original Bacillus strain W bacteriophage, was described as B. anthracis-specific (McCloy 1951; Brown and Cherry 1955). The PlyL protein, on the other hand, was isolated from a lysogenic copy of the λ Ba02-prophage integrated into the B. anthracis genome (Low et al. 2005). Investigations into the activity spectrum of these phages showed that the lytic activity of both enzymes is not strictly specific for the lysis of B. anthracis cells, since certain B. cereus strains closely related to B. anthracis are also lysed (e.g. strain B. cereus ATCC 4342 by PlyG; Schuch et al. 2002; Low et al. 2005, 2011), but these strains are exceptions. Of applied interest is the fact that when added to cells extracellularly these endolysins retain their lytic activity, even in the presence of capsular S-layer proteins, showing that the PG substrate is accessible to the endolysins to some degree even in the presence of S-layer proteins (Schuch et al. 2002; Fischetti 2010; Low et al. 2011).
The endolysin molecular architecture is dictated by two basic functions: substrate recognition and hydrolysis (Loessner 2005). As with other endolysins, these two functions are reflected in the modular architecture of PlyL and PlyG.
The C-terminal domain of these proteins contain a cell wall-binding domain (CBD) conferring specificity for a cell wall component; their CBDs are not of a well-characterized type and their receptor ligands have not yet been described. In the N-terminal domain of these proteins, they harbor the catalytic domain for the hydrolysis of PG (Schuch et al. 2002; Low et al. 2005, 2011; Fujinami et al. 2007; Kikkawa et al. 2007, 2008). The related functions of these enzyme domains are reflected in the homology of their amino acid sequences, which show 60% identity in the C-terminal region and 93% in the N-terminal region of the protein (Low et al. 2005). The less pronounced homology of the C-terminal domains could indicate that the C-terminal CBDs of both enzymes bind to a related structure in the B. anthracis cell wall, but with somewhat different binding properties. On the other hand, the high homology of the N-terminal regions corroborates the N-acetylmuramoyl-l-alanine amidase activity suggested for both catalytic domains (Schuch et al. 2002; Low et al. 2005; Kikkawa et al. 2008).
Classical lysins require the CBD to bind to a cell wall component and orient the catalytic domain of the enzyme to the PG. In this regard, PlyG and PlyL may show a difference. It has been reported that PlyG behaves as a classical PG-degrading enzyme with its CBD binding to the cell wall ligand, a precondition for reactivity (Schuch et al. 2002, Kikkawa et al. 2008). However, somewhat similar to what had previously been observed for a B. anthracis-encoded PG hydrolyzing autolysin (Mesnage and Fouet 2002), a recent paper reports lytic activity and non-classical behavior for the catalytic domain of PlyG, i.e. lytic activity in the absence of the CBD (Low et al. 2011). This contradiction to the older PlyG findings is as yet unexplained. For PlyL, non-classical behavior has also been reported with the isolated N-terminal region of PlyL retaining PG-degrading activity without an attached CBD (Low et al. 2005, 2011). It has been reported for both PlyL and PlyG that the removal of the C-terminal domain of the full-length proteins decreases the reactivity for B. anthracis strains and also broadens the reactivity to more Bacillus species (Low et al. 2005, 2011). Hence, either for efficient lysis or cell lysis specificity of host cells, both PlyG and PlyL require their CBD to bind to a cell wall ligand. To date, this cell wall ligand in the B. anthracis and B. cereus strains has not been identified.
Interest in B. anthracis carbohydrates has increased, since the discovery by Mesnage et al. (1999) that S-layer cell surface proteins contain an S-layer homology (SLH) domain that non-covalently binds to a B. anthracis cell wall carbohydrate and this way anchors the proteins to the bacterial vegetative cell wall (Mesnage et al. 1999, 2000; Fouet 2009; Kern and Schneewind 2010). This carbohydrate, categorized as a non-classical, secondary cell wall polysaccharide (SCWP; Schäffer and Messner 2005), is thought to be covalently attached to the PG through a phosphate group at the reducing end of the polysaccharide (Leoff, Saile, et al. 2008; Kern et al. 2010; Schneewind and Messiakas 2012) and can be released from the cell wall by treatment with cold aqueous hydrofluoric acid. We recently used this procedure to isolate and determine the structures of the SCWPs from B. anthracis Sterne strains 34F2 and 7702 (both strains have a S-layer, but lack virulence plasmid pXO2 coding for the poly-γ-glutamate capsule), from B. anthracis Pasteur (this strain has a S-layer, but lacks virulence plasmid pXO1), and the fully equipped B. anthracis Ames strain (Choudhury et al. 2006), as well as from two B. cereus strains closely related to B. anthracis (i.e. B. cereus G9241, isolated from a case of severe pneumonia in humans, and B. cereus ATCC 10987, isolated from dairy products; Leoff, Choudhury, et al. 2008; Forsberg et al. 2011, 2012). In addition, recently Candela et al. (2011) have reported the SCWP structure from the more distantly related type strain, B. cereus ATCC 14579.
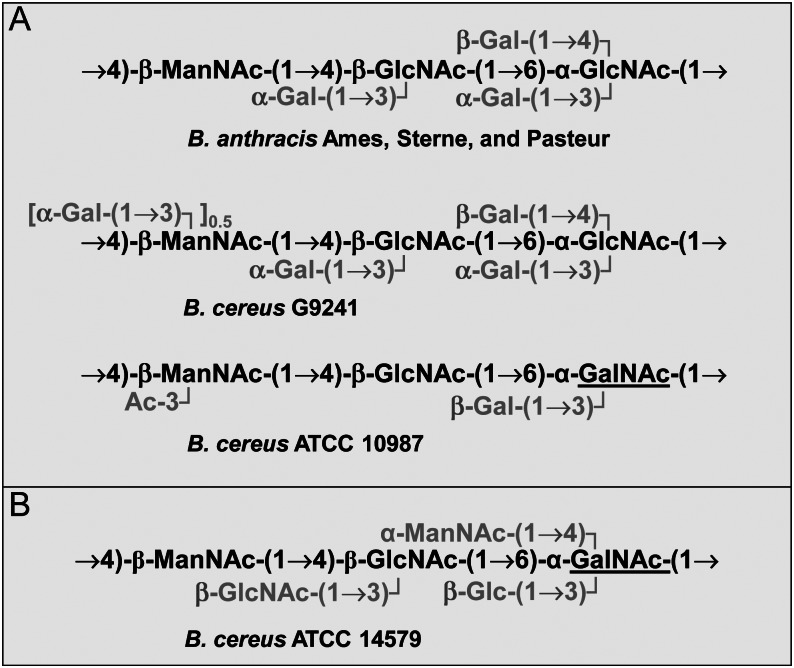
The structures of the SCWPs found in the vegetative cell walls of these strains are shown in Figure 1 (compiled from Choudhury et al. 2006; Leoff, Choudhury, et al. 2008; Candela et al. 2011; Forsberg et al. 2011). In B. anthracis strains examined to date, the SCWPs (SCWPBa) have the same structure comprised of an amino sugar backbone of →4)-β-d-ManpNAc-(1→4)-β-d-GlcpNAc-(1→6)-α-d-GlcpNAc-(1→ in which the α-GlcNAc residue was substituted with α-Gal and β-Gal at O3 and O4, respectively, and the β-GlcNAc substituted with α-Gal at O3 (Choudhury et al. 2006). This SCWPBa structural theme is conserved in strain B. cereus G9241 SCWP (SCWPBcG9241), but contains an additional α-Gal substitution on ∼50% of the β-ManpNAc residues at O3 (Forsberg et al. 2011). In comparison, the SCWP from the closely related strain B. cereus ATCC 10987 (SCWP10987) consists of a →4)-β-d-ManpNAc-(1→4)-β-d-GlcpNAc-(1→6)-α-GalpNAc-(1→ backbone in which the α-GalNAc is substituted at O3 with a β-Gal residue and the β-ManNAc is acetylated at O3 (Leoff, Choudhury, et al. 2008). From these different SCWP structures identified so far (Figure 1), a common structural theme has emerged consisting of a -ManNAc-GlcNAc-HexNAc- backbone that is substituted with terminal galactosyl (Gal) or glucosyl (Glc) residues or non-carbohydrate substituents such as acetyl, amino or pyruvyl groups (Figure 1A; Choudhury et al. 2006; Leoff, Saile, et al. 2008; Forsberg et al. 2011, 2012). The SCWP from the type strain B. cereus ATCC 14579 (SCWP14579) presents a significantly different structural motif in which the -ManNAc-GlcNAc-HexNAc- backbone is substituted with terminal GlcNAc, ManNAc and Glc residues (Figure 1B; Candela et al. 2011). Recent studies have provided sufficient evidence that the SCWPs, compared with other bacterial surface carbohydrates such as exopolysaccharides, are relatively small polysaccharides (their molecular weights ranging from ∼10,000 to 20,000 Da depending on the strain) that display rather minimal size heterogeneity during size exclusion chromatography (Mesnage et al. 2000; Forsberg et al. 2011).
Fig. 1.
(A) The SCWPs from B. anthracis and the indicated B. cereus strains with a related glycosyl backbone structure. The glycosyl backbones of these SCWPs all consist of amino sugar shown in black, which are decorated by glycosyl or acetyl residues shown in gray. The GalNAc of the B. cereus ATCC 10987 SCWP backbone that is different from the B. anthracis SCWP is underlined. Strain B. cereus G9241 is a clinical isolate from a case of severe pneumonia, and B. cereus strain ATCC 10987 does not cause severe or fatal disease. (B) The SCWP of the type strain B. cereus ATCC 14579 differs significantly. Its trisaccharide backbone is the same as that for B. cereus ATCC 10987; however, the terminal glycosyl residues also include aminoglycosyl residues and Glc instead of Gal or acetyl substituents (adopted from Leoff, Saile, et al. 2008; Candela et al. 2011).
The attachment of these SCWPs to PG, together with the presence of their strain-specific as well as more general species-specific structural features, lead us to hypothesize that these polysaccharides could function as ligands for endolysin binding in the B. anthracis and B. cereus cell walls. Therefore, we used SPR (BIAcore, Piscataway, NJ, USA) in conjunction with full-length PlyL and PlyG proteins and their C- and N-terminal domains to investigate the binding affinities of these endolysins to SCWPs. The SCWPs from the different B. anthracis and B. cereus strains that went into this study were chosen because they are structurally related (Figure 1) and were used to test whether identical SCWPs (as in the case of the SCWPs from B. anthracis Sterne 34F2 and 7702) display similar binding properties. We show here that the full-length PlyL and PlyG and their C-terminal domains bind to the SCWPBa from B. anthracis but not to the SCWPs from the closely related B. cereus ATCC 10987 and the more distantly related type strain, B. cereus ATCC 14579. Interestingly, PlyG also bound to the SCWP from B. cereus G9241, an isolate from a case of severe pneumonia. Our findings support the model that the SCWPBa is the ligand for the CBD of PlyL and PlyG and that this binding localizes and orients PlyL and PlyG, so they can carry out their lytic activity on the PG. This binding also accounts for the specificity of these endolysins.
Results
Lysin PlyL C-terminal domain binds to B. anthracis cells
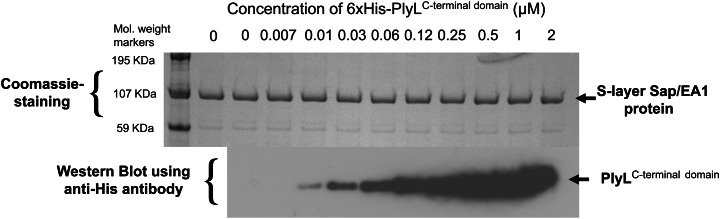
Previous work showed PlyG's and PlyL's modular protein architecture is comprised of a catalytic N-terminal domain and a C-terminal CBD (Low et al. 2005; Kikkawa et al. 2008) and directly binds the PlyG C-terminal domain to B. anthracis cells (Fujinami et al. 2007). Binding of PlyL CBD to B. anthracis cells has only recently and as yet rather selectively been addressed (Low et al. 2011). To confirm and extend the observations on the binding ability of the PlyL C-terminal domain to whole B. anthracis cells, we performed a pull-down experiment with strain B. anthracis 34F2 cells pre-exposed to increasing amounts of the PlyL C-terminal protein domain. As shown in Figure 2, gels loaded with pulled-down B. anthracis cells contained bound PlyL C-terminal domains. The amount of the C-terminal domain that was pulled down was dependent on the amount of the PlyL C-terminal domain used for pre-exposure and reached a maximum signal at ∼0.5–1.0 μM. This shows that the PlyL C-terminal domain binds to a ligand in the B. anthracis cell wall. The finding indicates that under the chosen experimental conditions either the number of binding sites on the cell surface is limited or the amounts of PlyL bound to the nitrocellulose sheet is saturable.
Fig. 2.
Pull-down assay of the endolysin PlyL C-terminal domain using live B. anthracis Sterne 34F2 cells. The top panel shows a Coomassie-stained PAGE gel of the samples analyzed. Equal loading of B. anthracis cells was confirmed making use of the prominent ∼100 kDa band of the S-layer proteins Sap and EA1 (at 0 μM PlyL C-terminal domain, the equal number of B. anthracis cells was analyzed without PlyL C-terminal domain pretreatment). The western blot at the bottom of the figure shows the increasing amounts of the pulled-down PlyL C-terminal domain bound to B. anthracis cells after pretreatment with increasing concentrations of the PlyL C-terminal domain. For that, tubes containing a cell suspension/endolysin protein mixture were placed on a rocking platform and incubated for 10 min. After incubation, the cells were pelleted and resuspended in phosphate-buffered saline, washed and mixed with an equal volume of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) sample loading buffer, boiled and analyzed on SDS–PAGE gels. For western blots, the anti-His antibody and the Goat horseradish peroxidase conjugated anti-mouse antibody were used as the primary and the secondary antibodies, respectively (for more details, see Experimental).
Ply binding studies
We used SPR to investigate SCWP–Ply interactions as it is a rapid and sensitive method to evaluate molecular affinities; the method is based solely on mass changes occurring during the interactions, allowing for the assessment of the interactions in real time and without the involvement of external labels, e.g. flourophores, which can have modifying effects on the molecular interactions. This may be particularly relevant to this study, as the method avoids the increase in sample heterogeneity in the already size heterogenous polysaccharide analyte samples. The full-length proteins as well as the C- and N-terminal domains of the PlyL and PlyG proteins were immobilized onto N-hydroxysuccinimide (NHS)-activated CM5 sensor chips and titrated with increasing amounts of SCWP isolated from different B. cereus group strains. Bulk refractions instigated by sample injections to the running buffer and non-specific binding were ameliorated with the help of a control cell pretreated with ethanolamine. Sensorgrams were evaluated using double referencing, global fitting and interaction models as indicated in figures and tables.
PlyL and PlyG bind to SCWPBa from strains B. anthracis 34F2 and 7702
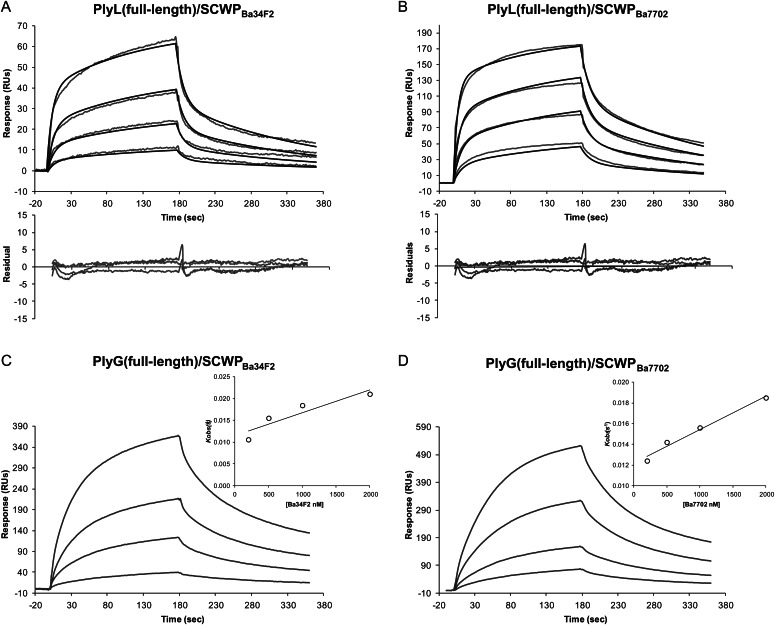
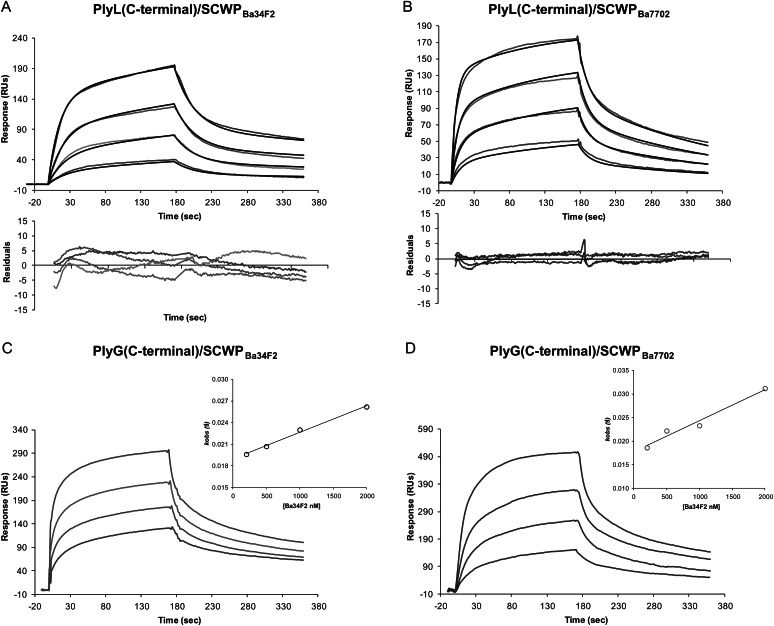
To determine whether SCWPBa is the ligand for PlyL and PlyG, we first measured the binding properties of the full-length PlyL and PlyG proteins using SPR. Figure 3 shows the BIAcore sensorgrams obtained with the immobilized proteins and increasing concentrations of the SCWPBa analyte isolated from strains B. anthracis 34F2 (SCWPBa34F2) and 7702 (SCWPBa7702). Both full-length proteins displayed strong affinities to isolated SCWPBa from the two B. anthracis strains tested and sensorgrams shifted with increasing analyte concentrations to higher response unit (RU) values. For the calculation of the binding kinetic parameters, different binding models supplied by the BIAevaluation 4.1 software were evaluated. For the PlyL proteins, a two-state interaction model was found to be the best fit (Figures 3 and 4). In contrast, for the PlyG full-length protein using global fitting, binding curves did not fit well to the different models included in the BIAevaluation software. Furthermore, because equilibrium was not reached during the association phase, the direct use of Scatchard analysis to calculate the apparent equilibrium dissociation constant was not allowed. Instead, the association rate constants (ka) were derived from the association phase using the 1:1 Langmuir association (kobs) model and a secondary plot of kobs vs analyte concentration (see inserts in Figures 3 and 4). The dissociation rate constant (kd) was calculated using the 1:1 Langmuir dissociation model, and the dissociation constant, KD, was obtained from the ratio of kd/ka as described in the method section. The dissociation constants KD calculated from these curves for PlyL and PlyG full length are in the μM range, i.e. with KD values ranging from 0.81 × 10−6 to 7.51 × 10−6 M (Table I).
Fig. 3.
Kinetics of the interaction of endolysins PlyL (full length) (A and B) and PlyG (full length) (C and D) with SCWPBa samples isolated, as indicated, from B. anthracis Sterne strains 34F2 and 7702. The SCWP concentrations used were from the top to the bottom: 2000, 1000, 500, 200 and 0 nM. The baseline response obtained with no (the 0 nM control) SCWP analyte has been subtracted from the binding curves and is not shown. For PlyL curves, a two-state interaction model gave the best fit (calculated and observed curves are superimposed with the residual curves obtained underneath); for PlyG, the association rate constants (ka) were determined from the slope of a plot of the observed association rate constant (kobs) as a function of the analyte concentration C, according to kobs = ka C + kd. (the inserts) The dissociation rate constant (kd) was calculated using the 1:1 Langmuir dissociation model from which dissociation constant, KD was obtained from the ratio of kd/ka. The SCWP samples were dissolved in HBS-EP buffer and were injected for 180 s at a flow rate of 10 μL/min. After the dissociation phase, the chip surface was regenerated with an injection of glycine (pH 2.0), followed by a pulse of 1.5 M NaCl solution. Chips were reused as long as the RU response to test samples were reproducible (for more details, see Experimental).
Fig. 4.
BIAcore analysis of the interaction of SCWPBa samples isolated from B. anthracis Sterne strains 34F2 and 7702 with the immobilized C-terminal domain of endolysin PlyL (A and B) and PlyG (C and D). Further details, the experimental conditions and the concentrations of SCWPs were the same as in Figure 3.
Table I.
Affinities, dissociation rate constants, Rmax and χ2 values obtained with SPR for the binding of the C-terminal domain and the full length protein of endolysin PlyL and PlyG to SCWP isolated from different B. anthracis and B. cereus strains (as indicated); analysis using global fitting
| SCWPs | ka (1/Ms) | kd (1/s) | KD (M) | Rmax (RU) | χ2 | ka (1/Ms) | kd (1/s) | KD (M) | Rmax (RU) | χ2 |
|---|---|---|---|---|---|---|---|---|---|---|
| PlyL (C-terminal domain) | PlyL (full-length) | |||||||||
| Ba34F2 | a1.42 × 104 | 3.23 × 10−2 | a1.14 × 10−6 | 334 | 9.52 | a2.12 × 104 | 8.88 × 10−2 | a7.51 × 10−6 | 132 | 1.67 |
| a3.96 × 10−3 | 1.51 × 10−3 | a7.14 × 10−3 | 5.67 × 10−3 | |||||||
| Ba7702 | a3.98 × 104 | 6.10 × 10−2 | a2.66 × 10−6 | 252 | 4.99 | a4.71 × 104 | 7.07 × 10−2 | a2.50 × 10−6 | 239 | 12 |
| a7.71 × 10−3 | 4.98 × 10−3 | a7.64 × 10−3 | 5.10 × 10−3 | |||||||
| BcG9241 | a9.95 × 103 | 6.49 × 10−2 | a1.68 × 10−5 | 98 | 0.65 | a1.58 × 104 | 3.74 × 10−1 | a2.63 × 10−5 | 198 | 1.76 |
| a4.27 × 10−3 | 6.76 × 10−3 | a3.35 × 10−2 | 3.92 × 10−3 | |||||||
| Bc14579 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Bc10987 | NB | NB | NB | NB | NB | NB | NB | NB | NB | NB |
| PlyG (C-terminal domain) | PlyG (full length) | |||||||||
| Ba34F2 | b3.69 × 103 | c3.01 × 10−3 | d0.81 × 10−6 | ND | ND | b5.22 × 103 | c4.25 × 10−3 | d0.81 × 10−6 | ND | ND |
| Ba7702 | b6.62 × 103 | c4.16 × 10−3 | d0.62 × 10−6 | ND | ND | b3.28 × 103 | c4.60 × 10−3 | d1.40 × 10−6 | ND | ND |
| BcG9241 | b1.1 × 104 | c3.82 × 10−3 | d0.34 × 10−6 | ND | ND | b1.17 × 103 | c2.9 × 10−3 | d2.47 × 10−6 | ND | ND |
| Bc14579 | NB | NB | NB | NB | NB | ND | ND | ND | ND | ND |
| Bc10987 | NB | NB | NB | NB | NB | ND | ND | ND | ND | ND |
NB, no binding; ND, not determined.
aAssociation and dissociation phase were fitted to a two-state reaction model.
bAssociation phases were calculated from the equation kobs= kaC+kd.
ckd was calculated using the 1:1 Langmuir dissociation model.
dKD was calculated from ka and kd values.
These KD values are in the range for what has been reported for some carbohydrate protein binding, e.g. for “bacterial” lectin to carbohydrate binding (Imberty et al. 2005). However, for lysins, stronger binding affinities have also been reported, e.g. Pseudomonas endolysin binding to PG KD = 3.4 × 10−8 M (Briers et al. 2009), or for Listeria monocytogenes bacteriophage murein hydrolase binding to bacterial cell wall carbohydrates, KD = 3.1 × 10−9 M (Loessner et al. 2002). Also, from PlyL binding affinity to whole cells of B. cereus ATCC 4342, Low et al. (2011) estimated a KD of 8 × 10−9 M, indicating a higher affinity of endolysins to whole cells as has been observed here with isolated SCWPs.
Binding of PlyL and PlyG C- and N-terminal domains to SCWPBa34F2 and SCWPBa7702
We next determined whether the proposed modular structure of PlyL and PlyG with their N-terminal catalytic domain and C-terminal CBD is reflected in the abilities of the PlyL and PlyG C- and N-terminal domains to bind the SCWPBa. Figure 4 shows the BIAcore sensorgrams obtained using the PlyL and PlyG C-terminal domains in conjunction with various concentrations of SCWPBa34F2 and SCWPBa7702. The C-terminal domains from PlyL and PlyG bound both B. anthracis SCWPBa samples in a concentration-dependent manner with increasing RU values similar to those for the respective full-length proteins (Figures 3 and 4). The binding affinities (Table I) between the PlyL C-terminal domains and both SCWPBa, as shown by the KD values, were in the micromolar range (i.e. from 1.14 × 10−6 to 2.66 × 10−6 M) and approximately in the range of the respective KD values obtained with the PlyG C-terminal domain (i.e. 0.81 × 10−6 and 0.62 × 10−6 M), thus supporting the results with the full-length proteins that showed approximately similar affinities of PlyL and PlyG toward SCWPBa.
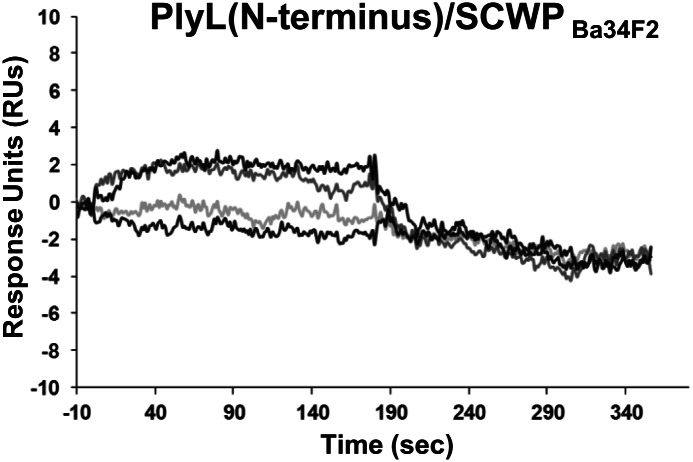
The N-terminal domains from PlyL and PlyG did not bind to the SCWPBa. Figure 5 shows an example of this result with regard to SCWPBa34F2 binding to the PlyL N-terminal domain. The same series of SCWPBa34F2 concentrations (0–2000 nM) showed no detectable binding to the PlyG N-terminal domain (data not shown). These results showed that the ability of PlyL and PlyG to bind the SCWPBa resides in their C-terminal and not their N-terminal domain. Also noteworthy, the PG-degrading activity of PlyL and PlyG that has recently been reported and that is independent of the CBD (Low et al. 2005, 2011) does not involve binding of the N-terminal domain to the SCWPBa, but seems to employ an alternative binding mechanism for CBD-independent PG binding and degrading.
Fig. 5.
BIAcore analysis of the interaction of the endolysin PlyL (N-terminal domain) with SCWPBa samples isolated from B. anthracis Sterne 34F2. The SCWP concentrations applied were 2000, 1000, 500, 200 and 0 nM (note: the different SCWP concentrations had no effect on the sensorgrams). The sensorgram obtained with the PlyG (N-terminal domain) and SCWPBa34F2 showed negative RU at all concentrations and time points (data not shown).
Comparing the PlyL and PlyG affinity for SCWPBa34F2 and SCWPBa7702
Since the SCWPBa structures of B. anthracis 34F2 and 7702 are identical with regard to the arrangement of their glycosyl residues and molecular weight (Choudhury et al. 2006; Leoff, Choudhury, et al. 2008), we expected them to show identical interactions with the Ply proteins. This is, indeed, what happened. The binding affinities of PlyL and PlyG full-length proteins to both of these SCWPs were about the same (Figure 3 and Table I). However, the overall comparison of the KD values obtained with full-length and C-terminal domains of PlyL and PlyG seems to indicate a somewhat stronger affinity of PlyG toward SCWPBa, but it remains to be seen in future binding studies if that is indeed the case. In summary, our results indicate strong binding of PlyL and PlyG to both SCWPBa samples with approximately the same binding affinity. Also, the fact that there is no significant difference in the binding affinities between PlyL and PlyG toward SCWPBa7702 and SCWPBa34F2 is consistent with our structural data which show that there is no structural difference between these two SCWPBa (Choudhury et al. 2006; Leoff, Choudhury, et al. 2008; Forsberg et al. 2012).
The specificity of PlyL and PlyG for SCWPs
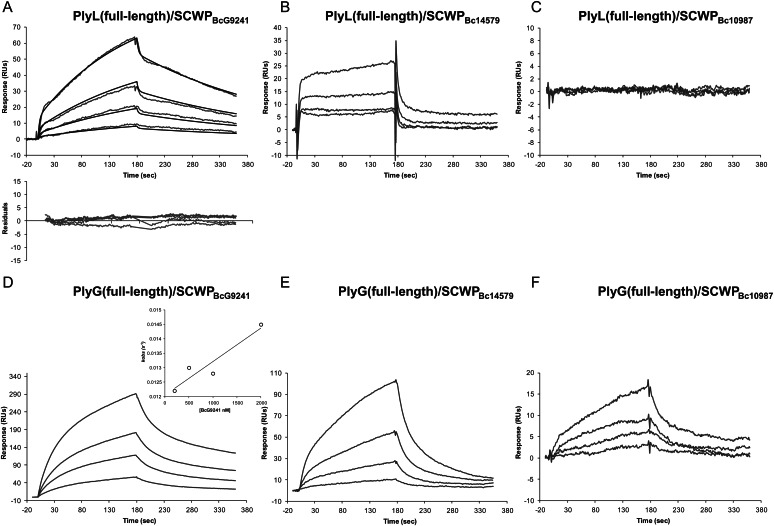
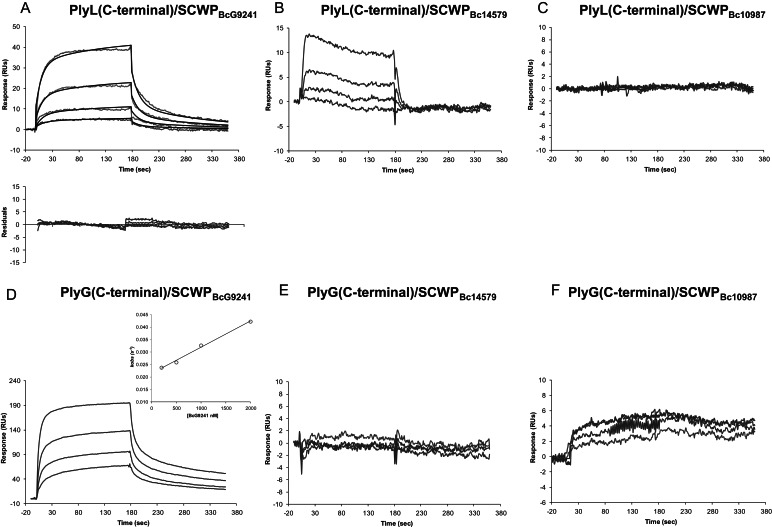
It is known that PlyG and PlyL are specific to B. anthracis strains and to a few closely related B. cereus strains (see Introduction). In order to determine if the interactions of these endolysins with the SCWPs could account for this specificity, we measured endolysin binding to a variety of recently characterized SCWPs from different B. anthracis and B. cereus strains (the SCWP structures are shown in Figure 1). The BIAcore sensorgrams in Figures 6 and 7 compare binding of the full-length proteins and their C-terminal domains to these SCWPs.
Fig. 6.
BIAcore analysis of the interaction between the full-length endolysins PlyL (top panel) and PlyG (bottom panel) and SCWPs isolated from strains B. cereus G9241 (A and D), B. cereus ATCC14579 (B and E) or B. cereus ATCC10987 (C and F). The concentrations of SCWPs used were, from the top to the bottom, 2000, 1000, 500, 200 and 0 nM. The baseline response with no SCWP analyte has been subtracted from the sensorgrams and is not shown. Further details, the experimental conditions were as described in the Figure 3 legend.
Fig. 7.
BIAcore analysis of the interaction between the C-terminal domain of endolysins PlyL (top panel) and PlyG (bottom panel) and SCWPs isolated from strains B. cereus G9241 (A and D), B. cereus ATCC14579 (B and E) or B. cereus ATCC10987 (C and F). Concentrations of SCWPs used were same as described in Figure 6 (as described there, the baseline response is also subtracted from the response curves and not shown) and experimental conditions as described in the Figure 3 legend.
With SCWPBc14579, RU values obtained for both PlyL and PlyG full-length proteins indicated some weak interaction; however, after global fitting analysis, the kinetic parameters could not be determined. In addition, the PlyG C-terminal domain showed no binding at all with SCWPBc14579, while the binding constant for the PlyL C-terminal domain could not be determined. Together, these results indicate some non-specific binding of the full-length proteins to SCWPBc14579. Moreover, neither full-length PlyL and PlyG proteins nor their C-terminal domains bound SCWPBc10987 (Figures 6 and 7 and Table I).
In contrast, SCWPBcG9241, which shares close structural similarity with B. anthracis SCWPBa, did bind to both full-length proteins. The binding of the full-length proteins to SCWPBcG9241 was also reflected in the binding of their C-terminal domains to SCWPBcG9241 (Figures 6 and 7). Both C-terminal domains showed higher binding affinities to SCWPBcG9241 when compared with their full-length counterparts. However, the measured sensorgrams indicate a difference in the binding affinities of PlyL and PlyG toward SCWPBcG9241. Although the PlyL full-length and C-terminal domain binding affinities to SCWPBcG9241 were ∼4–10-fold weaker compared with those with SCWPBa (Table I), the binding affinity of PlyG to SCWPBcG9241 was about the same as that observed with SCWPBa. Therefore, both PlyL and, in particular, PlyG bound SCWPBcG9241, SCWPBa34F2 and SCWPBa7702 with high affinity and showed only weak or no affinity to SCWPBa14579 and SCWPBc10987. This PlyG binding specificity is consistent with the reported specificity of the γ-phage endolysin for B. anthracis strains (see Introduction).
Up to now structural analysis has shown that SCWPBa34F2, SCWPBa7702 and SCWPBcG9241 all contain a →4)-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GlcNAc-(1→ trisaccharide backbone that is substituted with terminal β-Gal residues on O4 of the α-GlcNAc residue and by terminal α-Gal residues on O3 of the α- and β-GlcNAc residues. The SCWPBcG9241 structure differs from the B. anthracis structures in that 50% of the ManNAc residues are additionally substituted by a terminal α-Gal at O3 (Figure 1). The SCWPBc10987 and SCWPBc14579 structures contain GalNAc in their trisaccharide aminoglycosyl residue backbone and this backbone is substituted by a single terminal Gal and O-acetyl group (in the case of B. cereus ATCC 10987) or by various terminal hexosyl residues including Glc, ManNAc or GlcNAc (as in B. cereus ATCC 14579, Candela et al. 2011). Thus, the binding of PlyL and PlyG seems to require a -ManNAc-GlcNAc-GlcNAc- trisaccharide backbone with both GlcNAc residues being substituted with Gal as described. In addition, as the binding affinities of PlyL with SCWPBcG9241 are weaker than those observed with PlyG, the additional α-Gal residue attached to the ManNAc in the SCWPBCG9241 structure seems to lower the PlyL binding affinity but not that of PlyG.
Overall, our results with PlyL and PlyG can be summarized as follows: (i) PlyL and PlyG showed strong affinity to the SCWPBa isolates from strain B. anthracis 34F2, B. anthracis 7702 and B. cereus G9241; (ii) the C-terminal domains but not the N-terminal domains of both PlyL and PlyG bound the SCWPs; (iii) both PlyL and PlyG proteins displayed binding specificity toward SCWPBa, i.e. they did not bind SCWPBc10987 and only very weakly interacted with SCWPBc14579; (iv) PlyG and PlyL showed comparable binding affinity with both B. anthracis SCWPBa; (v) comparison of the affinity constants for the C-terminal domain–SCWP interaction with those for the full-length proteins indicates that both PlyL and PlyG have approximately the same affinity for the endolysin–SCWP ligand complex (Figures 3–7 and Table I).
Discussion
Bacteriophage endolysins are comprised of two functional domains, a catalytic domain and a domain or ligand conferring substrate recognition (Loessner 2005). To date, only a few ligands of the bacteriophage endolysin have been identified. For the pneumococcal endolysin Cp-1, it has been shown that structural repeats can bind choline, thereby anchoring the protein to choline moieties in the cell wall teichoic acid (WTA; Garcia et al. 1988; Hermoso et al. 2003, 2007). For the endolysin Lyb5 from Lactobacillus fermentum bacteriophage φPYB5, it has been shown that it contains LysM domains capable of binding PG (Hu et al. 2010). Recently, it has been shown that the Listeria bacteriophage endolysin PlyP35 contains a CBD that recognizes terminal GlcNAc residues in the WTA (Eugster et al. 2011).
The ligands for B. anthracis bacteriophage endolysins have not yet been reported. Based on the reported cell-binding properties of the PlyG and PlyL C-terminal domains (Schuch et al. 2002; Low et al. 2005, 2011; Fujinami et al. 2007; Kikkawa et al. 2007, 2008; and observations noted here), we hypothesized that the SCWPBa in the cell walls of B. anthracis comprise the ligands for endolysin binding. Using the well-characterized endolysins PlyL and PlyG, and our set of purified and structurally characterized SCWPs isolated from different B. anthracis and B. cereus strains (Figure 1), we addressed this hypothesis by using SPR analysis to determine whether PlyL and PlyG bind to the different SCWPs. In this paper, we show that the endolysins PlyL and PlyG from B. anthracis-specific bacteriophage bind to SCWPs with high affinity and specificity, thus corroborating our hypothesis that the SCWP in the B. anthracis cell wall is the ligand for PlyL and PlyG endolysin binding and in this way directs their hydrolytic activity to the PG. Moreover, our results show that the C-terminal domains of PlyL and PlyG, but not their N-terminal catalytic domains, contain the CBD that binds the B. anthracis SCWPs.
The affinity constants found for PlyL and PlyG full-length proteins and for the C-terminal domain binding (Table I) are in the range that has been reported for other protein–carbohydrate interactions (Imberty et al. 2005). They are also in the range that has been found in SPR measurements using small synthetic oligosaccharide analogs modeled after the SCWPBa structure with KD values ranging from 1.1 × 10−6 to 153.5 × 10−6 M (Mo et al. 2012). Interestingly, in that study, a synthetic hexasaccharide analog resembling the actual SCWP repeating unit was found to bind with KD values of 23 × 10−6 and 23.3 × 10−6 M for the PlyL and PlyG C-terminal domains, respectively. This 10–30-fold weaker binding of the synthetic hexasaccharide when compared with the affinities found for the isolated SCWPBa polysaccharides (each comprised in average of 10 repeating units) could indicate a polyvalent binding mode of action for the isolated SCWP ligands and may explain both (i) why a simple 1:1 binding interaction model did not work for PlyL or PlyG and (ii) why stronger affinities, compared with our SPR measurements, were found for PlyL to whole B. anthracis cells (Low et al. 2011).
With the SCWPs established as PlyL and PlyG ligands in the B. anthracis cell wall, the question was whether PlyL and PlyG specifically bound B. anthracis SCWPs. The inability or only weak binding of SCWPBc10987 and SCWPBc14579 showed that both endolysins are specific for SCWPBa. The strong Ply binding to SCWPs with a -ManNAc-GlcNAc-GlcNAc- backbone (i.e. the SCWPBa34F2, SCWPBa7702 and SCWPBcG9241) suggests that this trisaccharide, perhaps together with both GlcNAc residues being substituted with Gal, is the structural motif recognized by the Ply proteins. However, as SCWPs lacking Gal residues have not been investigated yet for binding, the importance for SCWP Gal substitution for Ply binding still needs to be established. As for SCWPBcG9241, the partial Gal substitution of the ManNAc residue evidently reduces the affinity of PlyL for this SCWP; in contrast, the binding affinity of PlyG for SCWPBcG9241 is unaffected (Table I). Despite this difference, for proteins originating from different bacteriophages, PlyL and PlyG have ligand binding specificities with a remarkable overlap in SCWP structural requirements for binding. The underlying molecular reasons for these similar binding requirements are under investigation.
Binding of both PlyL and PlyG with SCWPBa and the sensitivity of B. anthracis to the PlyG-producing γ-phage support that SCWPBa is the ligand for these endolysins. Of interest in this regard is B. cereus G9241. In this case, we have shown that PlyG (and PlyL) binds SCWPBcG9241, even though it has been shown that strain B. cereus G9241 is γ-phage resistant (Hoffmaster et al. 2006). An explanation for this could be that the phage is unable to infect B. cereus G9241 cells, e.g. if the cells lack a specific attachment site needed for the phage entry or the polysaccharide capsule of these bacteria (Hoffmaster et al. 2006; Oh et al. 2011) prevent phage binding. Although the γ-phage resistance of B. cereus G9241 still awaits an explanation, the γ-phage resistance of these bacteria despite the ability of PlyG to bind SCWPBc9241 makes it more likely that γ-phage specificity is not defined by the endolysins' ligand specificity in phage progeny release. Much remains to be learned about SCWPs and lysin action and uses (e.g. whether they have potential for detection and antibacterial uses). Further work is in progress to determine the SCWP structural features required for interaction with endolysins on PlyG lytic activity.
An observation consistent with the identical structures of SCWPBa7702 and with SCWPBa34F2 was that PlyL and PlyG bound these polysaccharides equally well. Structural analysis has shown that all the B. anthracis SCWPBa structures examined to date have identical ManNAc-GlcNAc-GlcNAc- backbones and attached Gal substitutions (Choudhury et al. 2006). Further, the distal trisaccharide repeat unit is modified by the pyruvylation of the ManNAc residue, 3-O-acetylation of the middle GlcNAc residue and de-N-acetylation of the second GlcNAc residue; the modifications are not present on the remaining SCWP repeat units (Forsberg et al. 2012). These substoichiometric non-carbohydrate modifications, in particular the pyruvyl ketal, are likely requirements of the structural epitopes that are essential for recognition and non-covalent binding to the carbohydrate-binding SLH domain of S-layer proteins and SLH domain-containing proteins (Mesnage et al. 2000; Schäffer and Messner 2005; Kern et al. 2010). However, their role in endolysin binding is unknown. We are currently examining the SCWP from putative mutants affected in these substituents, as well as the chemical modification of the SCWP to determine the role of these non-carbohydrate substituents in both endolysin binding as well and in the binding of SLH domain-containing proteins.
Experimental
Bacterial strains used in this study and culture conditions
Strains: B. anthracis Sterne 34F2 (Sterne 1937) and 7702 (Cataldi et al. 1990); B. cereus G9241 (Hoffmaster et al. 2004); B. cereus ATCC 10987 (Smith 1952) and B. cereus ATCC 14579 (Lawrence and Ford 1916). Strains B. anthracis 34F2 and the B. cereus strains were obtained from the CDC culture collection. Strain B. anthracis 7702 was obtained from T. Koehler, University of Texas/Houston Health Science Center. Strains were grown as described (Leoff, Saile, et al. 2008). Briefly, cells were cultured overnight in brain heart infusion (BHI) medium (BD BBL, Sparks, MD) containing 0.5% glycerol. Washed cells of the overnight culture were used to inoculate four 250-mL volumes of BHI medium in 1-L Erlenmeyer flasks the next morning. Cultures were grown at their optimum growth temperatures, i.e. 37°C for B. anthracis and 30°C for B. cereus strains, with shaking at 200 rpm. In a mid-log phase, cells were harvested by centrifugation (8000 × g, 4°C, 15 min), washed two times in sterile saline (0.9% w/v NaCl solution), enumerated by dilution plating on BHI agar plates and then autoclaved for 1 h at 121°C before further processing.
Preparation of bacterial cell walls
The bacterial cell walls were prepared by modification of a previously described procedure (Brown 1973; Leoff, Saile, et al. 2008). The autoclaved bacterial cells (3 × 108–9 CFU/mL) were disrupted in 40 mL sterile saline on ice by four 10-min sonication cycles; the cell disruption was checked microscopically. Unbroken cells were removed by a low-speed centrifugation run (8000 × g, 4°C, 15 min). The pellet and supernatant fractions were stored at −70°C. The cell walls were separated from the low-speed supernatants by ultracentrifugation at 100,000 × g, 4°C for 4 h. The resulting cell wall pellets were washed by suspension in cold, deionized water followed by an additional ultracentrifugation at 100,000 × g, 4°C for 4 h and lyophilized.
Release of phosphate-bound polysaccharides from the cell wall
Phosphate-bound polysaccharides were released from the cell walls by treatment with aqueous hydrogen fluoride (HF; these HF-released SCWPs are, therefore, also in the literature referred to as HF-PSs) in a modified procedure described by Ekwunife et al. (1991). Briefly, the cell walls were subjected to 47% HF under stirring at 4°C for 48 h. The reaction mixture was neutralized with NH4OH, subjected to a 10-min low-speed centrifugation (10,000 × g), and the supernatant with the released polysaccharides lyophilized, redissolved in deionized water and subjected to a chromatographic size separation on a BioGel P2 column (Bio-Rad, Hercules, CA, USA). The fractions eluting from the BioGel P2 column were monitored using a refractive index detector. Polysaccharide-containing peaks were pooled, lyophilized and analyzed by gas chromatography-mass spectrometry (GC-MS) as described in the Glycosyl composition analysis section. For size approximation, the isolated polysaccharides were chromatographed on a Superose-12 column, using α-1-6 linked dextrans as the calibration standards. Size heterogeneity was observed of the different HF-PSs with molecular masses between 12,000 and 20,000 Da. Hence, for calculations in this study, an average HF-PS/SCWP molecular mass of 16,000 Da was assumed.
Glycosyl composition analysis
The carbohydrate profiles were determined by GC-MS analysis of the trimethylsilyl methylglycosides as described previously (York et al. 1986). The SCWP fractions were subjected to methanolysis at 80°C for 18 h in methanolic HCl (1 M). The resulting methyl glycosides were N-acetylated, trimethylsilylated and then analyzed by GC-MS analysis (5890AGC-MS; Agilent Technologies, Palo Alto, CA) using a 30-m DB-1 fused silica capillary column (J&W Scientific, Folsom, CA). Inositol was used as an internal standard, and retention times were compared with the authentic standards. Composition analysis was done on each sample at least two times.
Cloning, expression and purification of the endolysins
The PlyL full-length protein and the PlyL C-terminal domain were expressed and purified as described (Low et al. 2005). The PlyL and PlyG N-terminal domains were obtained as explained in Low et al. (2005, 2011). The DNA of the PlyG C-terminal domain was codon optimized (for E. coli) and synthesized from amino acids Lys166 to Lys233, based on NCBI accession DQ221100 for the PlyG full-length protein. The resulting construct, with a methionine added in front of Lys166, was inserted into the vector pET15b (Novagen, Darmstadt, Germany) between NdeI and BamHI restriction sites. BL21DE3 (Agilent Technologies) competent cells were transformed with pET15b-PlyG-cell wall binding moiety and grown to an OD600 of 0.8, before being induced with 1 mM isopropyl-β-d-thiogalactoside for 3 h. After induction, the Escherichia coli BL21DE3 cells were harvested by centrifugation at 4°C at 6090 × g (Sorvall GS-3 rotor) for 15 min. The lysis buffer (20 mM Tris–Cl, 300 mM NaCl and 0.5% Triton X-100, at pH 7.0) was used to resuspend the cell pellet. Cells were lysed by French-press with three passes at 1000 pound per square inch and clarified by centrifugation at 38,725 × g (Sorvall SS-34) for 1 h at 4°C. The clarified lysate was loaded directly onto a 5-mL HITRAP Ni-chelating column (GE Healthcare, Piscataway, NJ, USA) equilibrated with 50 mL buffer A (20 mM Tris–Cl and 300 mM NaCl at pH 7.0). Unbound protein was eluted by applying 50 mL buffer A containing 30 mM imidazole. The His-tagged protein was eluted by 0.3 M imidazole in buffer A. A Superdex S200 16/60 (GE Healthcare) gel filtration, equilibrated with 1 × phosphate-buffered saline (PBS) (pH 7.4), was then used to further purify the target protein. The molecular weight of the final protein products (i.e. PlyL full length, 26.41 kDa; PlyL C-terminal domain, 11.02 kDa; PlyL N-terminal domain, 20.23 kDa; PlyG full length, 28.43 kDa; PlyG C-terminal domain, 10.1 kDa; PlyG N-terminal domain, 20.21 kDa; all proteins contain a His-tag with the exception of PlyL full length) was confirmed by matrix assisted laser desorption ionization-time of flight mass spectrometry.
Cell-binding assay of the PlyL C-terminal domain to B. anthracis cells
Bacillus anthracis Sterne (34F2) cells were grown in 10 mL of luria broth liquid medium to a cell density at OD600 of 1. The cells were harvested at 19,100 × g (Sorvall table-top centrifuge) for 5 min and resuspended in 10mL of PBS (pH 7.5; the cell density OD600 will be ∼1). Depending on the 6xHis-tagged PlyL C-terminal domain concentration applied (i.e. ranging from 0, 0.007 to 2.0 μM PlyL C-terminal domain), the protein was prediluted into PBS buffer before adding it to the cells. The total volume of each experiment was 1 mL. The tubes containing the cell–protein mixtures were placed on a rocking platform at room temperature for 10 min. After incubation, the cells were pelleted at 19,100 × g for 5 min using a table-top centrifuge. The pellets were resuspended and washed three times with PBS. Equal volumes of 2× sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) sample loading buffer were added to the cells and the final mixture boiled for 1 min at 100°C. Twenty microliters of the samples was loaded onto a 4–20% SDS–PAGE gel (Invitrogen, Grand Island, NY, USA). Equal loading of B. anthracis cells was confirmed making use of the intensity of the prominent band of the S-layer proteins Sap and EA1 in Coomassie-stained PAGE gels. For western blots, the penta-His antibody (Qiagen, Valencia, CA, USA) and the Goat horseradish peroxidase conjugated anti-mouse antibody (Calbiochem, Billerica, MA, USA) were used as the primary and the secondary antibodies, respectively.
SPR analysis
A BIAcore 3000 instrument under the control of BIAcore 3000 software version 3.2 was used for this analysis together with the following materials and solutions: a research grade sensor chip CM5; NHS; N-ethyl-N′-(3-diethylaminopropyl)-carbodiimide hydrochloride (EDC); 1 M ethanolamine–HCl, pH 8.5; running buffers: either HBS-EP buffer [10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 150 mM NaCl, 3.4 mM ethylenediaminetetraacetic acid (EDTA) and 0.005% (v/v) surfactant polysorbate P20 at pH 7.4, from BIAcore] or PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH 7.4, from EMD); bacteriophage endolysins PlyL and PlyG full-length, PlyL and PlyG C-terminal and PlyL and PlyG N-terminal domains (prepared as described in the Cloning, expression and purification section); SCWPs from strains B. anthracis Sterne 34F2 and 7702, B. cereus ATCC 10987, B. cereus G9241 and B. cereus ATCC 14579 (prepared as described in the section on the release of phosphate-bound polysaccharides from the cell wall); coupling buffer: 10 mM acetate buffer adjusted to appropriate pH values (see below); regeneration buffer (glycine, pH 2.0) were purchased from BIAcore.
The endolysin proteins were immobilized using the standard amine coupling procedures (Löfås and Johnsson 1990) at conditions recommended by the manufacturer: as a preliminary for protein immobilization, the pH scouting was done with coupling buffer (10 mM acetate buffer) adjusted to pH 4.0, 4.5, 5.0 or 5.5. For best protein immobilization, the pH conditions for protein coupling of the different endolysins were chosen as follows: pH 5.5 for the PlyL full length and the N terminus and pH 4.5 for the derived C-terminal protein fragment; and pH 5.0 for PlyG full length and pH 5.5 for the derived C and N terminus. The endolysin was immobilized on the surface of a CM5 sensor chip (BIAcore) using the standard amine coupling chemistry with HBS-EP [10 mM HEPES, pH 7.4, containing 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) Surfactant P20].To activate the carboxymethyl dextran surface, a freshly prepared mixture (1:1) of NHS (50 mM in water) and EDC (200 mM in water) was injected for 7 min, then the endolysin (normally 50 μg/mL in coupling buffer; some preparations were coated at a concentration of 100 μg/mL) was injected until the desired level of immobilization (∼1000 RU) was achieved (flow rate: 10 μL/min). Any remaining activated carboxylic acid groups were blocked by a 50 μL of 1 M ethanolamine, pH 8.6, followed by a 10-μL injection of 10 mM glycine at pH 2.0 to wash out any unbound endolysin. After immobilization of the ligand, buffer was passed over the surface until baseline stability was obtained. As a reference surface for the refractive index and correction of non-specific binding, a second flow cell was treated in the same manner, but without the addition of endolysin protein. For kinetics studies (adjusted from Xie et al. 2005 and Urbinati et al. 2004), the SCWP analyte samples contained 0, 200, 500, 1000 and 2000 nM of the respective SCWP dissolved in running buffer (10 mM HEPES buffer at pH7.4 or PBS). The kinetic assays were carried out at 25°C and a flow rate of 10 μL/min. For each analyte concentration, association was measured for 180 s and dissociation was measured for 300 s. After each injection, the surface was regenerated with 30 s injection of 10 mM glycine at pH 2.0, followed by a 30-s injection of 1.5 M NaCl, pH 7.4, in PBS or HEPES buffer at a flow rate of 30 μL/min. The cycle was repeated for every endolysin–SCWP pair.
Data analysis
All sensorgrams were obtained by a double-referencing procedure as described by Myszka (1999) using global fitting. The kinetic data for the different endolysin–SCWP pairs were fitted to different interaction models available in the BIAevaluation software 4.1.1. For the PlyL–SCWP interactions, a two-state binding model resulted in a good fit between calculated and observed curves The goodness of fit was determined by the χ2 values, as well as the magnitude and distribution of the residual values (defined as the difference between observed and calculated values). In contrast, the PlyG–SCWP interactions did not fit any of the binding models supplied by the BIAevaluation 4.1 software. In the latter case, the association rate constants (ka) were determined from the slope of a plot of the observed association rate constant (kobs) as a function of the analyte concentration C, according to kobs = ka C + kd (inserts in Figures 3, 4, 6 and 7; Yu et al. 2005; Chevigné et al. 2007). The dissociation rate constant (kd) was calculated using the 1:1 Langmuir dissociation model from which dissociation constant, KD, was obtained from the ratio of kd/ka.
Funding
This work was supported in part by National Institute of Health (R21 AI076753 to RWC). The Complex Carbohydrate Research Center was supported in part by Department of Energy (DE-FG02-09ER20097).
NK was funded through the Atlanta Research and Education Foundation (AREF).
Conflict of interest
None declared.
Abbreviations
ATCC, American Type Culture Collection; BHI, brain heart infusion; CBD, cell wall-binding domain; EDC, N-ethyl-N′-(3-diethylaminopropyl)-carbodiimide hydrochloride; EDTA, ethylenediaminetetraacetic acid; Gal, galactosyl; GC-MS, gas-liquid chromatography-mass spectrometry; Glc, glucosyl; HBS-EP, 10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA and 0.005% [v/v] surfactant polysorbate P20 at pH 7.4; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HF, hydrofluoric acid; HF-PS, HF-released cell wall polysaccharide; HRP, horseradish peroxidase; LB, luria broth; NHS, N-hydroxysuccinimide; PBS, phosphate-buffered saline; PG, peptidoglycan; PlyL, phage lysin L; PlyG, phage lysin G; RU, response unit; SCWP, secondary cell wall polysaccharide; SCWPBa, SCWP from B. anthracis strains; SCWPBa34F2, SCWP from strain B. anthracis 34F2; SCWPBa7702, SCWP from strain B. anthracis 7702; SCWPBc10987, SCWP from strain B. cereus ATCC 10987; SCWPBcG9241, SCWP from strain B. cereus G9241; SCWPBc14579, SCWP from strain B. cereus ATCC 14579; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SLH, S-layer homology; SPR, surface plasmon resonance; WTA, wall teichoic acid.
Acknowledgements
We thank Karen Howard and Chris Cooper for their helpful comments and suggestions to the manuscript. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers of Disease Control and Prevention. Patent Pending: University of Georgia Research Foundation, Inc., and US Centers for Disease Control and Prevention.
References
- Baillie LW. Bacillus anthracis, a story of nature subverted by man. Lett Appl Microbiol. 2005;41:227–229. doi: 10.1111/j.1472-765X.2005.01786.x. [DOI] [PubMed] [Google Scholar]
- Briers Y, Schmelcher M, Loessner MJ, Hendrix J, Engelborghs Y, Volckaert G, Lavigne R. The high-affinity peptidoglycan binding domain of Pseudomonas phage endolysin KZ144. Biochem Biophys Res Commun. 2009;383:187–191. doi: 10.1016/j.bbrc.2009.03.161. [DOI] [PubMed] [Google Scholar]
- Brown WC. Rapid methods for extracting autolysins from Bacillus subtilis. Appl Microbiol. 1973;25:295–300. doi: 10.1128/am.25.2.295-300.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ER, Cherry WB. Specific identification of Bacillus anthracis by means of a variant bacteriophage. J Inf Dis. 1955;96:34–39. doi: 10.1093/infdis/96.1.34. [DOI] [PubMed] [Google Scholar]
- Candela T, Maes E, Garenaux E, Rombouts Y, Krzewinski F, Gohar M, Guerardel Y. Environmental and biofilm-dependent changes in a Bacillus cereus secondary cell wall polysaccharide. J Biol Chem. 2011;286:31250–31262. doi: 10.1074/jbc.M111.249821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi AL, Labruyère E, Mock M. Construction and characterization of a protective antigen-deficient Bacillus anthracis strain. Mol Microbiol. 1990;4:1111–1117. doi: 10.1111/j.1365-2958.1990.tb00685.x. [DOI] [PubMed] [Google Scholar]
- Chevigné A, Barumandzadeh R, Groslambert S, Cloes B, Dehareng D, Filée P, Marx JC, Frère JM, Matagne A, Jacquet A, et al. Relationship between propeptide pH unfolding and inhibitory ability during ProDer p 1 activation mechanism. J Mol Biol. 2007;374:170–185. doi: 10.1016/j.jmb.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Choudhury B, Leoff C, Saile E, Wilkins P, Quinn CP, Kannenberg EL, Carlson RW. The structure of the major cell wall polysaccharide of Bacillus anthracis is species-specific. J Biol Chem. 2006;281:27932–27941. doi: 10.1074/jbc.M605768200. [DOI] [PubMed] [Google Scholar]
- Ekwunife FS, Singh J, Taylor KG, Doyle RJ. Isolation and purification of cell wall polysaccharide of Bacillus anthracis (delta Sterne) FEMS Microbiol Lett. 1991;66:257–262. doi: 10.1016/0378-1097(91)90270-k. [DOI] [PubMed] [Google Scholar]
- Eugster MR, Haug MC, Huwiler SG, Loessner MJ. The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol Microbiol. 2011;81:1419–1432. doi: 10.1111/j.1365-2958.2011.07774.x. [DOI] [PubMed] [Google Scholar]
- Fischetti VA. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LS, Abshire TG, Friedlander A, Quinn CP, Kannenberg EL, Carlson RW. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose-deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology. 2012;22:1103–1117. doi: 10.1093/glycob/cws080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LS, Choudhury B, Leoff C, Marston CK, Hoffmaster AR, Saile E, Quinn CP, Kannenberg EL, Carlson RW. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology. 2011;21:934–948. doi: 10.1093/glycob/cwr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A. The surface of Bacillus anthracis. Mol Aspects Med. 2009;30:374–385. doi: 10.1016/j.mam.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Fujinami Y, Hirai Y, Sakai I, Yoshino M, Yasuda J. Sensitive detection of Bacillus anthracis using a binding protein originating from gamma-phage. Microbiol Immunol. 2007;51:163–169. doi: 10.1111/j.1348-0421.2007.tb03894.x. [DOI] [PubMed] [Google Scholar]
- García E, García JL, García P, Arrarás A, Sánchez-Puelles JM, López R. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc Natl Acad Sci USA. 1988;85:914–918. doi: 10.1073/pnas.85.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermoso JA, García JL, García P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr Opin Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Hermoso JA, Monterroso B, Albert A, Galán B, Ahrazem O, García P, Martínez-Ripoll M, García JL, Menéndez M. Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure. 2003;11:1239–1249. doi: 10.1016/j.str.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hilleman MR. Overview: cause and prevention in biowarfare and bioterrorism. Vaccine. 2002;20:3055–3067. doi: 10.1016/s0264-410x(02)00300-6. [DOI] [PubMed] [Google Scholar]
- Hoffmaster AR, Hill KK, Gee JE, Marston CK, De BK, Popovic T, Sue D, Wilkins PP, Avashia SB, Drumgoole R, et al. Characterization of Bacillus cereus isolates associated with fatal pneumonias: Strains are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J Clin Microbiol. 2006;44:3352–3360. doi: 10.1128/JCM.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci USA. 2004;101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Kong J, Kong W, Guo T, Ji M. Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl Environ Microbiol. 2010;76:2410–2418. doi: 10.1128/AEM.01752-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imberty A, Mitchell EP, Wimmerová M. Structural basis of high-affinity glycan recognition by bacterial and fungal lectins. Curr Opin Struct Biol. 2005;15:525–534. doi: 10.1016/j.sbi.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Kern J, Ryan C, Faull K, Schneewind O. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J Mol Biol. 2010;401:757–775. doi: 10.1016/j.jmb.2010.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J, Schneewind O. BslA, a pXO1-encoded adhesion of Bacillus anthracis. Mol Microbiol. 2010;68:504–515. doi: 10.1111/j.1365-2958.2008.06169.x. [DOI] [PubMed] [Google Scholar]
- Kikkawa H, Fujinami Y, Suzuki S, Yasuda J. Identification of the amino acid residues critical for specific binding of the bacteriolytic enzyme of gamma-phage, PlyG, to Bacillus anthracis. Biochem Biophys Res Commun. 2007;363:531–535. doi: 10.1016/j.bbrc.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Kikkawa HS, Ueda T, Suzuki S, Yasuda J. Characterization of the catalytic activity of the gamma-phage lysin, PlyG, specific for Bacillus anthracis. FEMS Microbiol Lett. 2008;286:236–240. doi: 10.1111/j.1574-6968.2008.01280.x. [DOI] [PubMed] [Google Scholar]
- Lawrence JS, Ford WW. Studies on aerobic spore-bearing and non-pathogenic bacteria. Spore-bearing bacteria in milk. J Bacteriol. 1916;1:277–320. [PMC free article] [PubMed] [Google Scholar]
- Leoff C, Choudhury B, Saile E, Quinn CP, Carlson RW, Kannenberg EL. Structural elucidation of the nonclassical secondary cell wall polysaccharide from Bacillus cereus ATCC 10987. Comparison with the polysaccharides from Bacillus anthracis and B. cereus type strain ATCC 14579 reveals both unique and common structural features. J Biol Chem. 2008;283:29812–29821. doi: 10.1074/jbc.M803234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoff C, Saile E, Sue D, Wilkins P, Quinn CP, Carlson RW, Kannenberg EL. Cell wall carbohydrate compositions of strains from the Bacillus cereus group of species correlate with phylogenetic relatedness. J Bacteriol. 2008;190:112–121. doi: 10.1128/JB.01292-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner MJ. Bacteriophage endolysins–current state of research and applications. Curr Opin Microbiol. 2005;8:480–487. doi: 10.1016/j.mib.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Loessner MJ, Kramer K, Ebel F, Scherer S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol. 2002;44:335–349. doi: 10.1046/j.1365-2958.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- Löfås S, Johnsson B. A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J Chem Soc Chem Commun. 1990;21:1526–1528. [Google Scholar]
- Low LY, Yang C, Perego M, Osterman A, Liddington RC. The role of net charge o the catalytic domain and the influence o the cell-wall binding domain on the bactericidal activity, specificity and host-range of phage lysins. J Biol Chem. 2005;280:35433–35439. doi: 10.1074/jbc.M111.244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low LY, Yang C, Perego M, Osterman A, Liddington R. The role of electrostatic charge on the lytic activity, specificity and host-range of recombinant Gram-positive phage lysins. J Biol Chem. 2011;286:34391–34403. doi: 10.1074/jbc.M111.244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy EW. Studies on a lysogenic Bacillus strain: I. A bacteriophage specific for Bacillus anthracis. J Hyg (Lond) 1951;49:114–125. doi: 10.1017/s0022172400015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 2000;19:4473–4484. doi: 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage S, Fouet A. Plasmid-encoded autolysin in Bacillus anthracis: Modular structure and catalytic properties. J Bacteriol. 2002;184:331–334. doi: 10.1128/JB.184.1.331-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage S, Tosi-Couture E, Fouet A. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol Microbiol. 1999;31:927–936. doi: 10.1046/j.1365-2958.1999.01232.x. [DOI] [PubMed] [Google Scholar]
- Mo KF, Li X, Li H, Low LY, Quinn CP, Boons GJ. Endolysins of Bacillus anthracis bacteriophages recognize unique carbohydrate epitopes of vegetative cell wall polysaccharides with high affinity and selectivity. J Am Chem Soc. 2012;134:15556–15562. doi: 10.1021/ja3069962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Oh SY, Budzik JM, Garufi G, Schneewind O. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol Microbiol. 2011;80:455–470. doi: 10.1111/j.1365-2958.2011.07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell PK. Project BioShield: What it is, why it is needed, and its accomplishments so far. Clin Infect Dis. 2007;1(45 Suppl.):S68–S72. doi: 10.1086/518151. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Messner P. The structure of secondary cell wall polymers: How Gram-positive bacteria stick their cell walls together. Microbiology. 2005;151:643–651. doi: 10.1099/mic.0.27749-0. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci. 2012;367(1592):1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- Smith N. Aerobic spore forming bacteria. USDA Monogr. 1952;16:1–148. [Google Scholar]
- Sterne M. The effects of different carbon dioxide concentrations on the growth of virulent anthrax strains. Pathogenicity and immunity tests on guinea pigs and sheep with anthrax variants derived from virulent strains. Onderstepoort J Vet Sci An Ind. 1937;9:49–67. [Google Scholar]
- Urbinati C, Bugatti A, Oreste P, Zoppetti G, Waltenberger J, Mitola S, Ribatti D, Presta M, Rusnati M. Chemically sulfated Escherichia coli K5 polysaccharide derivatives as extracellular HIV-1 Tat protein antagonists. FEBS Lett. 2004;568:171–177. doi: 10.1016/j.febslet.2004.05.033. [DOI] [PubMed] [Google Scholar]
- Xie R, Zhuang M, Ross LS, Gomez I, Oltean DI, Bravo A, Soberon M, Gill SS. Single amino acid mutations in the cadherin receptor from Heliothis virescens affect its toxin binding ability to Cry1A toxins. J Biol Chem. 2005;280:8416–8425. doi: 10.1074/jbc.M408403200. [DOI] [PubMed] [Google Scholar]
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1986;118:3–40. [Google Scholar]
- Yu H, Muñoz EM, Edens RE, Linhardt RJ. Kinetic studies on the interactions of heparin and complement proteins using surface plasmon resonance. Biochim Biophys Acta. 2005;1726:168–176. doi: 10.1016/j.bbagen.2005.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]