Abstract
Cationic polymers are one of the major nonviral gene delivery vectors investigated in the past decade. In this study, we synthesized several cationic copolymers using atom transfer radical polymerization (ATRP) for gene delivery vectors: pluronic F127-poly(dimethylaminoethyl methacrylate) (PF127-pDMAEMA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate) (PF127-p(DMAEMA-tBA)), and pluronic F127-poly(dimethylaminoethyl methacrylate-acrylic acid) (PF127-p(DMAEMA-AA)). The copolymers showed high buffering capacity and efficiently complexed with plasmid deoxyribonucleic acid (pDNA) to form nanoparticles 80–180 nm in diameter and with positive zeta potentials. In the absence of 10% fetal bovine serum, PF127-p(DMAEMA-AA) showed the highest gene expression and the lowest cytotoxicity in 293T cells. After acrylic acid groups had been linked with a fluorescent dye, the confocal laser scanning microscopic image showed that PF127-p(DMAEMA-AA)/pDNA could efficiently enter the cells. Both clathrin-mediated and caveolae-mediated endocytosis mechanisms were involved. Our results showed that PF127-p(DMAEMA-AA) has great potential to be a gene delivery vector.
Keywords: nonviral vector, pluronic F127, dimethylaminoethyl methacrylate, copolymer, atom transfer radical polymerization
Introduction
Gene therapy has for some time been considered a promising treatment for various diseases. Designing a gene delivery vector with low cytotoxicity and high loading ability of a gene drug, such as plasmid deoxyribonucleic acid (pDNA), signal interfering ribonucleic acid, and antisense oligodeoxynucleotide, is currently a challenging task.1 Over the past decade, gene carriers have been developed in two systems: nonviral and viral gene delivery systems. Nonviral gene delivery systems using cationic polymers are highly attractive for treating genetic diseases and cancer because they improve drawbacks encountered with viral carriers, such as cytotoxicity and immunogenicity. Positively charged groups of cationic polymers complexed with negatively charged gene drugs via electrostatic interactions not only protect the gene drugs from the nuclease degradation, but also facilitate efficient delivery of the gene drugs into cells.2
Polyethylenimine (PEI),3 poly(2-dimethyl amino)ethyl methacrylate (PDMAEMA),4 poly(lysine) (PLL),5 and polyamidoamine (PAAM) are commonly-used cationic polymers for gene delivery carriers.6 Among them, PDMAEMA is the easiest one to manipulate its macromolecular architectures (star, liner, graft, and block). In addition, its monomer can be polymerized in differently controlled manners like atom transfer radical polymerization (ATRP),7–9 and reversible addition-fragmentation chain transfer polymerization.10,11 However, the transfection efficiency and cytotoxicity of PDMAEMA go hand-in-hand with increasing the molecular weight. This situation limits PDMAEMA application in gene delivery systems.12 Several methods have been adopted to reduce cytotoxicity without sacrificing the transfection efficiency of PDMAEMA, including PEGylation of PDMAEMA by preparing copolymers with polyethylene glycol (PEG) or with hydrophobic and hydrophilic monomers.13–15 In addition, the incorporation of multiple functionalities into the PDMAEMA copolymers have been extensively developed for gene delivery systems.16
Pluronic copolymers consist of hydrophobic poly(propylene) (PPO) segments and hydrophilic poly(ethylene oxide) (PEO) segments. They can self-assemble into micelles with a hydrophobic core by PPO and a hydrophilic shell by PEO.17 The pluronic copolymers have been modified with cationic polymers, such as PEI,18 PLL,19 and PDMAEMA,20–22 to improve gene transfection as well as to reduce cytotoxicity. The hydrophobic PPO segments of pluronic polymers enhance the stability of polyplexes between polymers and deoxyribonucleic acid (DNA),23,24 and promote the cellular uptake of polyplexes because of the increased interaction between the cell membrane and polyplexes.20
A successful example was reported in the improvement of transfection efficiency using a pluronic P85-based polymer instead of using a PEG-based polymer comprising poly(N-[N-(2-aminoethyl)-2-aminoethyl]) aspartamide cationic blocks as a gene vector. This fact is attributable to the idea that pluronic P85 interacts with the plasma membrane and promotes the cellular uptake of biomacromolecules.20 In this study, we tried to copolymerize pluronic F127 (PF127) with PDMAEMA to reduce the cytotoxicity of PDMAEMA as a gene vector. However, when we prepared pentablock copolymers of PF127-pDMAEMA with different PDMAEMA block lengths via ATRP (unpublished data), we found that the pDNA-condensed ability and its transfection efficiency were mainly dependent on a PDMAEMA chain length. The higher block length of PDMAEMA indeed showed the higher transfection efficiency but resulted in a higher cytotoxicity as well. To overcome the cytotoxicity caused by the high chain length of PDMAEMA >100 repeating units, we tried to modify the internal structure of PF127-pDMAEMA by copolymerizing a negatively charged acrylic acid (AA) to produce PF127-p(DMAEMA-AA). We hope that the carboxyl groups of PF127-p(DMAEMA-AA) can neutralize the positively charged PDMAEMA and reduce its inherited cytotoxicity without sacrificing its gene transfection ability. In addition, the AA groups can be used to react with a fluorescent dye for cellular trafficking. Thus, the merits of PF127-p(DMAEMA-AA) as a gene vector include (1) cationic DMAEMA segments for condensing pDNA; (2) PF127 segments for reducing cytotoxicity and self-assembly; and (3) AA functional groups linked with a fluorescent dye for internalization trafficking.
Three artificial copolymers were synthesized and characterized using Fourier-transform infrared (FTIR) spectroscopy and proton nuclear magnetic resonance (1H-NMR) spectroscopy. The cytotoxicity was determined by the viability of transfected cells using a 3-(4,5-Dimethyl-thiazol-2yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. The hydrodynamic diameters and zeta potentials were measured using dynamic light scattering (DLS). The morphologies were obtained using a transmission electron microscope. To test the potency of copolymers as a gene carrier, the gene transfection efficiency of polyplexes were studied in 293T cells. The mechanism of cellular uptake was studied using a flow cytometer, and the in vitro cell internalization of pDNA into 293T cells was directly visualized using a confocal laser scanning microscope (CLSM).
Materials and methods
Materials
PF127, MTT, and N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDAC) were purchased from Sigma-Aldrich (St Louis, MO, USA). Then, 2,2′-bipyridine (Bpy), 2-bromo-2-methylpropionyl bromide, Amberlite® IR120, DMAEMA, and Copper (I) bromide (CuBr) were purchased from Acros Organics (Morris Plains, NJ, USA). Aluminum oxide neutral (Al2O3) was from Seedchem Company Pty Ltd (Melbourne, VIC, Australia). Ethidium bromide (EtBr) was purchased from MP Biomedicals, LLC (Verona, Italy). Tert-butyl acrylate (tBA) was purchased from Alfa Aesar (Ward Hill, MA, USA). The PGL3-control and luciferase assay kit were from Promega (Promega Corporation, Fitchburg, WI, USA). Fetal bovine serum (FBS) was purchased from Biological Industries (Beit Haemek, Israel). Agarose, Dulbecco’s Modified Eagle’s medium (DMEM) was purchased from Invitrogen (Life Technologies, Carlsbad, CA, USA). All other unstated chemicals were from Sigma-Aldrich and used without further purification.
Preparing macroinitiator (PF127-Br)
In a two-neck round-bottom flask, PF127 (12.6 g, 1 mmol) was dissolved in 20 mL dichloromethane at room temperature. The solution was cooled to 0°C and triethylamine (0.7 mL, 5 mmol) was added with stirring. After 20 minutes, 2-bromo-2-methylpropionyl bromide (0.6 mL, 5 mmol) was slowly injected into the reaction flask under argon. After 48 hours reaction at room temperature, the bromide-modified product, di-2-bromoisobutyryl-pluronic F127 (PF127-Br), was obtained by precipitation in excess n-hexane and dried under vacuum.
Synthesizing PF127-p(DMAEMA-tBA) and PF127-pDMAEMA copolymers
PF127-Br (250 mg, 0.02 mmol), DMAEMA (0.65 mL, 4 mmol), with or without tBA (0.14 mL, 1 mmol), 2-propanol (1.6 mL), and double-deionized (DD) water (0.4 mL) were added to a two-neck round-bottom flask. The solution was degassed by five consecutive standard freeze–pump–thaw cycles. Next, CuBr (5.6 mg, 0.04 mmol) and Bpy (6.1 mg, 0.04 mmol) were quickly added to the mixture under argon. After 4 hours at room temperature, the reaction was stopped by diluting with DD water. The product was purified by dialysis against DD water using an weight-averaged molecular weight (Mw) cut-off 3500 membrane (Spectrum® Laboratories, Inc, Rancho Dominguez, CA, USA) for 2 days. The aqueous solution was removed and freeze-dried. The freeze-dried product was dissolved in toluene and passed through aluminum oxide column and Amberlite® IR120 (Sigma-Aldrich) to remove catalyst complexes. The obtained copolymer was precipitated in excess n-hexane and dried under vacuum.
Preparing PF127-p(DMAEMA-AA) copolymer
One hundred milligrams of PF127-p(DMAEMA-tBA) were dissolved in 10 mL DD water and hydrolyzed by adding 0.2 mL of hydrogen chloride (12 N HCl) in the solution at 40°C. After 24 hours of hydrolysis, the solution was dialyzed against DD water using the Mw cut-off 3500 membrane for 2 days. The aqueous solution was collected and freeze-dried. The PF127-p(DMAEMA-AA) product was stored at room temperature until used.
Synthesizing PF127-p(DMAEMA-AA)-rhodamine 123
PF127-p(DMAEMA-AA) (100 mg, 0.043 mmol) and EDAC (8.2 mg, 0.043 mmol) were dissolved in 10 mL DD water. The solution was adjusted to pH 7.4 with 0.1 N sodium hydroxide (NaOH) and stirred for 1 hour at room temperature. Rhodamine 123 (R123) (16.4 mg, 0.043 mmol) in 10 mL DD water was added to the above solution. The reaction solution was protected from light and kept reacting at room temperature for 12 hours. The product was purified by dialysis against DD water using the M cut-off 3500 membrane for 2 days. The aqueous solution was removed and freeze-dried. The final product was stored at −20°C until used.
Characterization
The chemical structure of copolymers was determined using 1H-NMR and FTIR spectroscopy. 1H-NMR spectra were obtained from a Varian Mercury plus-200 spectrometer (Varian, Inc, Palo Alto, CA, USA), using heavy water as a solvent. FTIR spectra were acquired using Perkin-Elmer System 2000 (PerkinElmer, Waltham, MA, USA). The molecular weights of copolymers were measured by gel permeation chromatography using an Agilent 1100 series (Agilent Technologies, Santa Clara, CA, USA) equipped with a Shodex-KF804 column. The sample was dissolved in tetrahydrofuran at a concentration of 5 mg/mL and filtered through a 0.45 μm filter prior to injection. The tetrahydrofuran was used as an eluent at a flow rate of 1 mL/minute. Ten polystyrene standards were used to generate a calibration curve.
Acid–base titration was carried out using a computer-controlled system assembled with a 702 SM Titroprocessor, a 728 stirrer, and a PT-100 combination pH electrode (Metrohm AG, Herisau, Switzerland). Approximately 20 mg of each copolymer was dissolved in 20 mL of 150 mM sodium chloride solution. The pH value of the solution was adjusted to 2 using 0.1023 N hydrochloric acid followed by back-titration to pH 11 using 0.0998 N NaOH.
Preparing pDNA
pGL3-control plasmid with Hind III/Xba I firefly luciferase cDNA fragment were introduced into the E. coli strain DH5α (Gibco®-BRL, Life Technologies, Carlsbad CA, USA) and purified using a kit (Maxi-V500 Plasmid Kit; Viogene-Biotek Corporation, Sunnyvale, CA, USA). The purity of the plasmid DNA was certified by the absorbance ratio at OD260/OD280 and by distinctive bands of DNA fragments at corresponding base pairs in gel electrophoresis after restriction enzyme treatment of DNA. pDNA was stored at −20°C until used.
Preparing copolymer/pDNA polyplexes
Each copolymer was dissolved in DD water to a final concentration of 2 mg/mL at pH 5 to pursue the protonated amino groups of PDMAEMA. The pDNA concentration was fixed at 3 μg/100 μL in DD water to measure pDNA binding assay and 1 μg/100 μL for other studies. Equal volumes of pDNA and copolymer solution with different N/P ratios ranging from 1–20 were immediately vortexed at high speed for 60 seconds. The polyplexes were kept at room temperature for 10 minutes for complete complexation before analysis.
Characterizing copolymer/pDNA polyplexes
The DNA binding ability of polyplexes was evaluated using an agarose gel electrophoresis. The stability of the copolymer/pDNA polyplexes with and without 10% FBS was evaluated using a gel electrophoresis with 0.8% agarose in Tris acetate–ethylenediaminetetraacetic acid with EtBr (1 μg/mL). A current of 100 V was applied to the gels for 35 minutes, and DNA retention was visualized under ultraviolet illumination at 365 nm.
The hydrodynamic diameters and zeta potentials of the polyplexes were measured using laser Doppler anemometry with a Zetasizer Nano ZS instrument (Malvern Instruments, Malvern, UK). Light scattering measurements were done with a laser at 633 nm and a 90° scattering angle. The particle sizes and zeta potentials were measured three times. The transmission electron microscopy (JEM-2000 EXII; JEOL, Tokyo, Japan) was used to observe the morphology. The polyplexes (N/P = 9) in DD water were placed on the copper grid and allowed to dry for 5 days at room temperature. The nanoparticles were analyzed by Image J software (National Institutes of Health, Bethesda, MD, USA).
Cell experiments
HEK 293T cells (human embryonic kidney 293T cell line) were cultivated at 37°C under humidified 5% carbon dioxide (CO2) in DMEM, supplemented with 10% FBS and 100 μg/mL penicillin–streptomycin. The medium was replenished every 3 days, and the cells were subcultured after they had reached confluence.
Cytotoxicity was tested in 293T cells using an MTT assay. The cells were seeded in 96-well culture plates at a density of 5 × 103 cells per well in DMEM containing 10% FBS for 24 hours. The number of viable cells was obtained by the estimation of their mitochondrial reductase activity using the tetrazolium-based colorimetric method.
The transfection assay was evaluated using a pGL-3 plasmid in 293T cells. The transfection efficiencies of polyplexes were compared with those of naked DNA as a negative control, and Lipofectamine® 2000 (LIPO; Life Technologies, Carlsbad, CA, USA) and branched PEI (25 KDa, N/P = 10) as positive controls. The 293T cells were seeded at a density of 1 × 105 cells/well in twelve well tissue culture plates and incubated in DMEM medium containing 10% FBS for 24 hours before transfection. When the cells were at 50%–70% confluence, the culture medium was replaced with 1 mL of DMEM with or without 10% FBS. Polyplexes with N/P ratios ranging from 1–20 were prepared using different amounts of copolymers and a fixed pDNA amount of 1 μg, to a final volume of 100 μL. After being left to stand for 10 minutes, the polyplexes were added to each well containing the cells and incubated for 4 hours. The medium was replaced with 1 mL of fresh DMEM and the cells were incubated for 44 hours posttransfection. The transfected cells were rinsed gently with 1 mL of 0.1 M phosphate buffered saline (PBS) (twice) and added to a 200 μL/well of lysis buffer (0.1M Tris-HCl, 2 mM ethylenediaminetetraacetic acid and 0.1% Triton X-100, pH 7.8). The luciferase activity was monitored using a microplate scintillation and luminescence counter after mixing the contents of a 50 μL well of supernatant with the contents of 50 μL well of luciferase assay reagent (Promega Corporation, Fitchburg, WI, USA). The total protein content of the cell lysate was examined using a bicinchoninic acid protein assay kit (Pierce Protein Biology Products, Rockford IL, USA).
Endocytosis inhibition using a flow cytometer
In addition, 293T cells were seeded at a density of 2 × 105 cells/well in 6-well plates in DMEM with 10% FBS and incubated for 24 hours. The cells were pretreated with inhibitors at concentrations: 50 nM wortamannin,25 10 μg/mL chlorpromazine,26 and 200 μM genistein for 30 minutes,27 respectively. After 30 minutes of incubation, the medium containing the inhibitors was changed to fresh DMEM, and the cells were treated with R123-linked copolymer/pDNA (N/P = 9) and incubated for another 2 hours. Next, the cells were trypsinized centrifuged and resuspended in 1 mL of PBS and then analyzed using a flow cytometer (EPICS XL, Beckman Coulter, Inc, Brea, CA, USA).
Intracellular trafficking using a CLSM
The intracellular trafficking of a polyplex at an N/P ratio of 9 was studied in 293T cells using a CLSM. The cells were seeded at a density of 1 × 105 cells/well in 12-well plates containing one glass cover slip/well, in DMEM supplemented with 10% FBS, and incubated for 24 hours. Subsequently, the polyplex was added and incubated for 4 hours. The cover slips were removed and washed three times with PBS. The cell nuclei and endolysosome were stained with 5 μg/mL Hoechst 33342 and 100 nM LysoTracker® Green (Invitrogen, Life Technologies, Carlsbad, CA, USA) for 30 minutes. Next, the cells were fixed with 3.7% paraformaldehyde for 30 minutes and the cells on the cover slips were washed three times with PBS and mounted with fluorescent mounting medium on glass slides. A CLSM (Fv 1000; Olympus Corporation, Tokyo, Japan) was used for cell imaging.
Statistical analysis
Means, standard deviations, and standard errors of the data were calculated. Comparison between groups was tested using Student–Newman–Keuls’ test and P < 0.05 was considered significant.
Results and discussion
Preparing and characterizing copolymers
The chemical structure of PF127-Br was confirmed from the 1H-NMR spectrum (Supplementary materials, Figure S1) using the relative peak intensity at 1.92 ppm (a, C(Br)–CH3 of the 2-bromo-2-methylpropionyl groups) and 1.18 ppm (b, methyl protons of PPO block). The degree of halogenation was determined to be ~95%. The copolymers composed of DMAEMA, DMAEMA-co-tBA, and DMAEMA-co-AA, were prepared via ATRP using PF127-Br as a center block (Figure S2). PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA), were controlled with a similar DMAEMA block length.
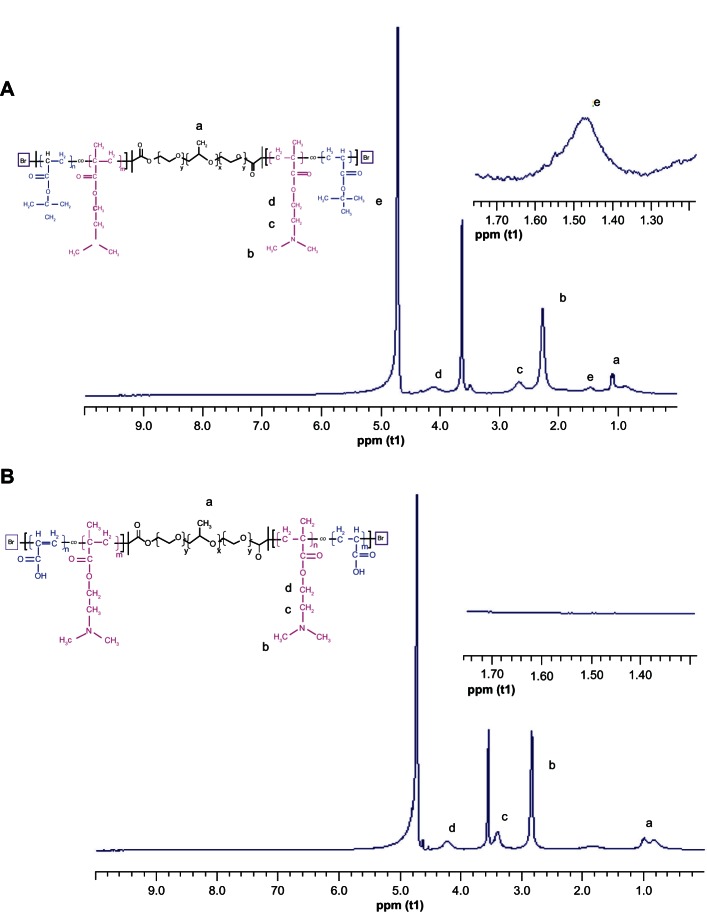
The chemical structure of PF127-pDMAEMA was measured from the 1H-NMR spectrum (Figure S3). Figure 1A shows the 1H-NMR spectrum of PF127-p(DMAEMA-tBA). The chemical shift at 1.48 ppm is mainly attributable to the methyl protons (e, –C(CH3)) of the tBA segments. The chemical shifts in the region of 2.25–2.63 ppm are mainly associated with the methyl (b, N–CH3) and methylene (c, N–CH2) protons of the DMAEMA segments. The chemical shift at 4.12 ppm is associated with the methylene protons adjacent to the oxygen moieties of the ester linkages (d, H2C–O–C=O). The numbers of tBA in the PF127-p(DMAEMA-tBA) copolymer were determined using the relative peak intensity at 1.48 ppm (e, –C(CH3)) of the tBA segments, and 1.18 ppm methyl protons of the PF127 segments, which are ~15 tBA repeat units. PF127-p(DMAEMA-AA) was obtained by hydrolyzing PF127-p(DMAEMA-tBA) in an acidic condition to remove tert-butyl groups.28,29 Figure 1B shows the chemical shift of the tert-butyl groups at 1.48 ppm, which completely disappeared after PF127-p(DMAEMA-tBA) had been hydrolyzed. In addition, the peaks associated with the methyl protons (b, N–CH3) and methylene protons (c, N–CH2) of DMAEMA units shifted to 2.85 and 3.42 ppm, respectively, because of protonation of the amino groups on PDMAEMA.30
Figure 1.
1H-NMR spectra of PF127-p(DMAEMA-tBA) and PF127-p(DMAEMA-AA). (A) PF127-p(DMAEMA-tBA) and (B) PF127-p(DMAEMA-AA).
Notes: a–CH3 is the methyl protons of PPO units; bN-CH3 is the methyl protons of DMAEMA units; cN–CH2 is the methylene protons of DMAEMA units; dH2C–O–C=O is the methylene protons of DMAEMA units; e–C(CH3) is the methyl protons of tBA units.
Abbreviations: 1H-NMR, proton nuclear magnetic resonance; PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); ppm (t1), parts per million (t1-longitudinal [spin-lattice] relaxation time).
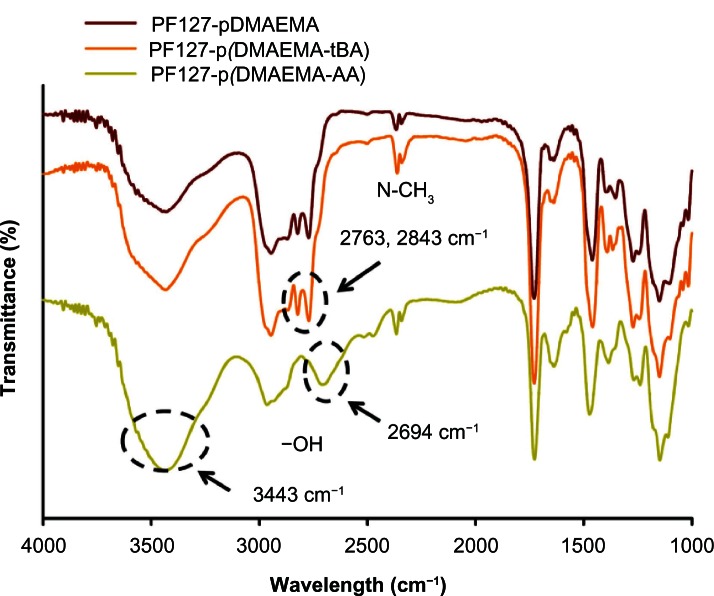
No distinguishable FTIR spectra between PF127-pDMAEMA and PF127-p(DMAEMA-tBA) were obtained (Figure 2). After PF127-p(DMAEMA-tBA) had been hydrolyzed, the characteristic absorption peaks of the –COOH groups in PF127-p(DMAEMA-AA) were observed at 2694 and 3443 cm−1 (O–H stretching).31 From 1H-NMR and FTIR results, we ensured the successful hydrolysis of tBA to AA. The molecular weights and their distributions measured by gel permeation chromatography and the chain lengths measured by 1H-NMR of PF127-pDMAEMA and PF127-p(DMAEMA-tBA) were summarized in Table 1. To evaluate the optimized ratio between DMAEMA and AA, several PF127-p(DMAEMA-tBA) copolymers were prepared by fixing the molar ratio of DMAEMA/PF127 at 200 and varying tBA/PF127 molar ratios of 10, 50, and 100. The DMAEMA length of PF127-p(DMAEMA-tBA) decreased remarkably as the feeding monomer ratio of tBA/PF127 was 100. In contrast, the intrinsic cytotoxicity of PDMAEMA remained using a tBA/PF127 ratio of 10 (the number of AA units in the copolymer was 5). Thus, a tBA/PF127 molar ratio of 50 was chosen to prepare the PF127-p(DMAEMA-tBA) copolymer, whereas the chain lengths of DMAEMA and tBA were ~130 and 15, respectively.
Figure 2.
FTIR spectra of PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA).
Abbreviations: FTIR, Fourier transform-infrared; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid).
Table 1.
Molecular weights and chain lengths of copolymers
| Sample | Mn (g/mol)a | DP (DMAEMA)b | DP (tBA)b | PDIc |
|---|---|---|---|---|
| PF127-Br | 10600 | 1.45 | ||
| PF127-pDMAEMA | 33823 | 135 | 1.34 | |
| PF127-p(DMAEMA-tBA) | 34957 | 130 | 15 | 1.40 |
Notes:
The number-averaged molecular weight (Mn) of copolymers was done by gel permeation chromatography;
the number of DMAEMA (or tBA) per copolymer chain was estimated by proton nuclear magnetic resonance;
PDI = Mw/Mn measured by GPC, where Mw is the weight-averaged molecular weight.
Abbreviations: DP, degree of polymerization; DMAEMA, (2-dimethyl amino)ethyl methacrylate; tBA, tert-butyl acrylate; PDI, polydispersity index; PF127-Br, di-2-bromoisobutyryl-pluronic F127; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); GPC, gel permeation chromatography.
Figure 3 shows the titration profiles of PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA). PF127-pDMAEMA and PF127-p(DMAEMA-tBA) show a similar buffer capacity and are obviously better than PF127-p(DMAEMA-AA) because a higher amount of NaOH is needed to change the pH from 5.5 to 7.4, which has been defined as a measurement of buffer capacity.32 The apparent pKa values of amino groups are 7.3, 7.4, and 7.2, for PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA), respectively. The introduction of anionic AA to PDMAEMA slightly reduces the apparent pKa value. All values are close to the reported value for PDMAEMA, 7.0–7.5.33 The pKa value of a tertiary amine function is within 10–11, which shifts to a lower value because of the neighboring group effect upon polymerization.34 Increasing a buffering effect to assist the endosomal rupture of complexes is one of the key points to design a good gene delivery vector.35 Since the buffer capacity of PF127-p(DMAEMA-AA) decreases with an increased amount of AA groups,36 the use of 15 AA groups is an optimized condition to retain a good transfection efficiency in a later study.
Figure 3.
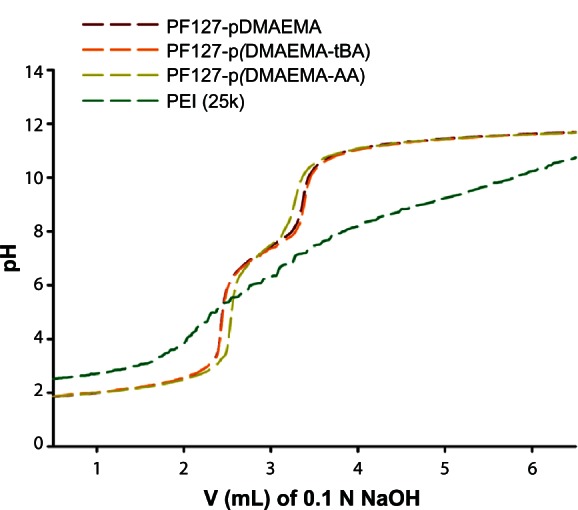
Titration of an acidified solution of PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA).
Abbreviations: PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); PEI, polyethylenimine; V, volume; N, normality.
Characterizing copolymer/pDNA polyplexes
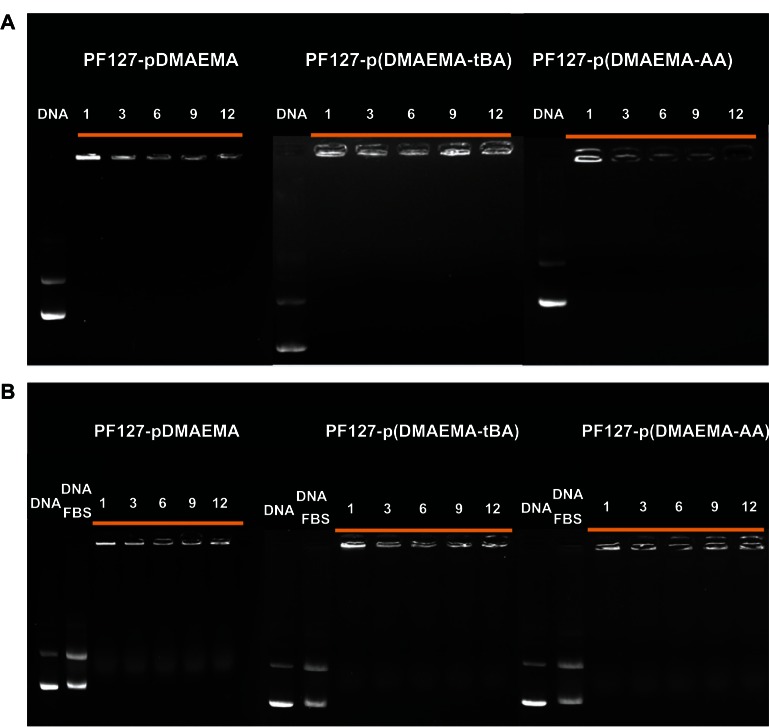
The gel retardation assay was performed to study the binding ability of copolymers and pDNA at various N/P ratios. Because no exposed pDNA was stained by EtBr at every N/P ratio ranging from 1–12, PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA) had excellent binding ability with pDNA, both with and without 10% FBS (Figure 4). The introduced 15 AA units in PF127-p(DMAEMA-AA) did not reduce the binding ability with pDNA.
Figure 4.
Gel electrophoresis to test pDNA retention in copolymer/pDNA polyplexes at various N/P ratios. (A) Without 10% FBS and (B) with 10% FBS.
Note: Naked pDNA was used as a reference, and numerals of each graph indicate an N/P ratio.
Abbreviations: pDNA, plasmid deoxyribonucleic acid; N/P, nitrogen/phosphate; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); DNA, deoxyribonucleic acid; FBS, fetal bovine serum.
The hydrodynamic diameters of copolymer/pDNA are within 75–180 nm and decrease with an increase in the N/P ratio (Figure 5A). At the same N/P ratio, the hydrodynamic diameters of PF127-pDMAEMA/pDNA and PF127-p(DMAEMA-tBA)/pDNA are similar, and larger than that of PF127-p(DMAEMA-AA)/pDNA. The zeta potentials of copolymer/pDNA are positive and increase with an increase in the N/P ratio (Figure 5B). At an N/P ratio > 9, the zeta potentials of PF127-p(DMAEMA-AA)/pDNA are clearly lower than those of PF127-pDMAEMA/pDNA and PF127-p(DMAEMA-tBA)/pDNA. As can be seen in Figure 5C, the transmission electron microscope micrographs also show that PF127-p(DMAEMA-AA)/pDNA has the smallest particle size. The particle sizes of PF127-pDMAEMA/pDNA, PF127-p(DMAEMA-tBA)/pDNA, and PF127-p(DMAEMA-AA)/pDNA (averaged from ten particles) are 125 nm, 122 nm, and 108 nm, respectively. This result is consistent with the DLS finding. The decrease in zeta potential and particle size in PF127-p(DMAEMA-AA) is because of electrostatic interactions between DMAEMA and AA segments, which not only neutralize the counter charges but also physically crosslink the shell compartment.
Figure 5.
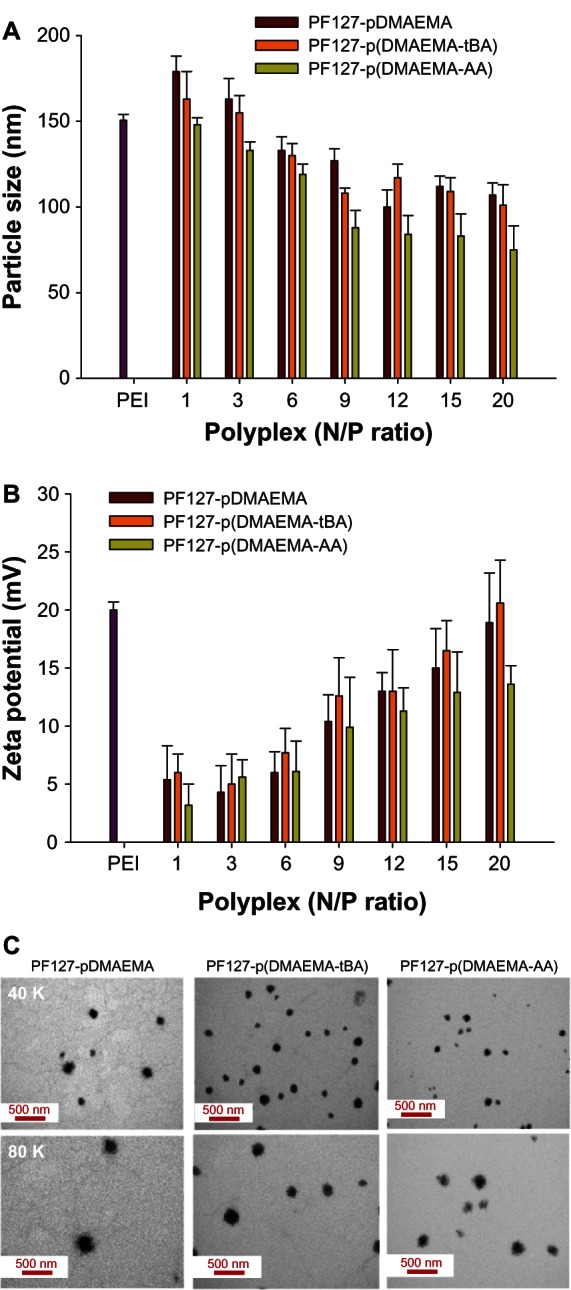
Copolymer/pDNA polyplexes at various MZN/P ratios. (A) Hydrodynamic diameters and (B) zeta potentials of copolymer/pDNA polyplexes at various N/P ratios (n = 3); (C) TEM images of copolymer/pDNA polyplexes at N/P = 9.
Note: The N/P ratio of PEI (25K)/pDNA was 10.
Abbreviations: pDNA, plasmid deoxyribonucleic acid; N/P, nitrogen/phosphate; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); PEI, polyethylenimine; TEM, transmission electron microscope.
To test the stability of polyplexes with time, the particle sizes of the polyplexes at N/P = 9 were measured by DLS after they had been left to stand in DD water with 10% FBS for 4 hours, or without 10% FBS for 10 minutes and 4 hours (Table 2). Without 10% FBS, all polyplexes are stable with a narrow size distribution in DD water even after 4 hours standing. There is no obvious change in particle size between 10 minutes and 4 hours standing. In the presence of 10% FBS, both size and its distribution remarkably increase after 4 hours standing. This may be attributable to positively charged polyplexes and the negatively charged FBS resulting in aggregation.37 With 10% FBS, the particle diameter of PF127-p(DMAEMA-tBA)/pDNA is much smaller than that of PF127-pDMAEMA and PF127-p(DMAEMA-AA)/pDNA. This may be due to the hydrophobic segments of tBA preventing the formation of large aggregates with FBS.38
Table 2.
Hydrodynamic diameters of copolymer/pDNA polyplexes in DD water with and without 10% FBS after left to stand for 10 minutes or 4 hours
| Polyplex (N/P = 9) | Dh (nm) at 10 minutes | PDI | Dh (nm) at 4 hours | PDI | Dh (nm) at 4 hours with FBS | PDI |
|---|---|---|---|---|---|---|
| PF127-pDMAEMA/pDNA | 102 ± 11 | 0.28 ± 0.02 | 103 ± 10 | 0.27 ± 0.02 | 225 ± 30 | 0.35 ± 0.11 |
| PF127-p(DMAEMA-tBA)/pDNA | 97 ± 8 | 0.30 ± 0.03 | 108 ± 9 | 0.26 ± 0.03 | 142 ± 15 | 0.33 ± 0.09 |
| PF127-p(DMAEMA-AA)/pDNA | 82 ± 7 | 0.26 ± 0.02 | 92 ± 5 | 0.25 ± 0.03 | 284 ± 47 | 0.41 ± 0.12 |
Note: n = 3.
Abbreviations: pDNA, plasmid deoxyribonucleic acid; DD water, double-deionized water; FBS, fetal bovine serum; N/P, nitrogen/phosphate; Dh, hydrodynamic diameters; PDI, polydispersity index; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid).
Cytotoxicity
The cytotoxicities of PF127-pDMAEMA and PF127-p(DMAEMA-tBA) increase remarkably at a concentration of 12.5 μg/mL. Their cell viabilities are ~50% and 40%, respectively. In contrast, PF127-p(DMAEMA-AA) remains 90% viable, indicating a significantly improved cell viability after AA modification (Figure 6A). The dosages required to inhibit cell proliferation by 50% (IC50) of PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA) are 7.9 μg/mL, 7.7 μg/mL, and 35.5 μg/mL, respectively. After formed polyplexes with pDNA, at an N/P of copolymers/pDNA <9, the cell viabilities of three polyplexes are higher than those of LIPO/pDNA and PEI(25K)/pDNA at an N/P of 10. At N/P = 9, the cell viabilities are 72%, 63%, and 84%, respectively, and at N/P = 20, they are 21%, 20%, and 45%, for PF127-pDMAEMA/pDNA, PF127-p(DMAEMA-tBA)/pDNA, and PF127-p(DMAEMA-AA)/pDNA, respectively. PF127-p(DMAEMA-AA)/pDNA shows significantly lower cytotoxicity at an N/P ratio ≥9 as compared with PF127-pDMAEMA/pDNA and PF127-p(DMAEMA-tBA)/pDNA (Figure 6B). The incorporation of hydrophilic AA groups indeed reduces the inherited cytotoxicity of PDMAEMA. This result is consistent with the finding in our previous study in incorporating a polysaccharide (chondroitin sulfate) into PDMAEMA.39 The neutralization between the negatively-charged chondroitin sulfate and pDMAEMA significantly reduced the cytotoxicity of chondroitin sulfate-modified PDMAEMA copolymers.
Figure 6.
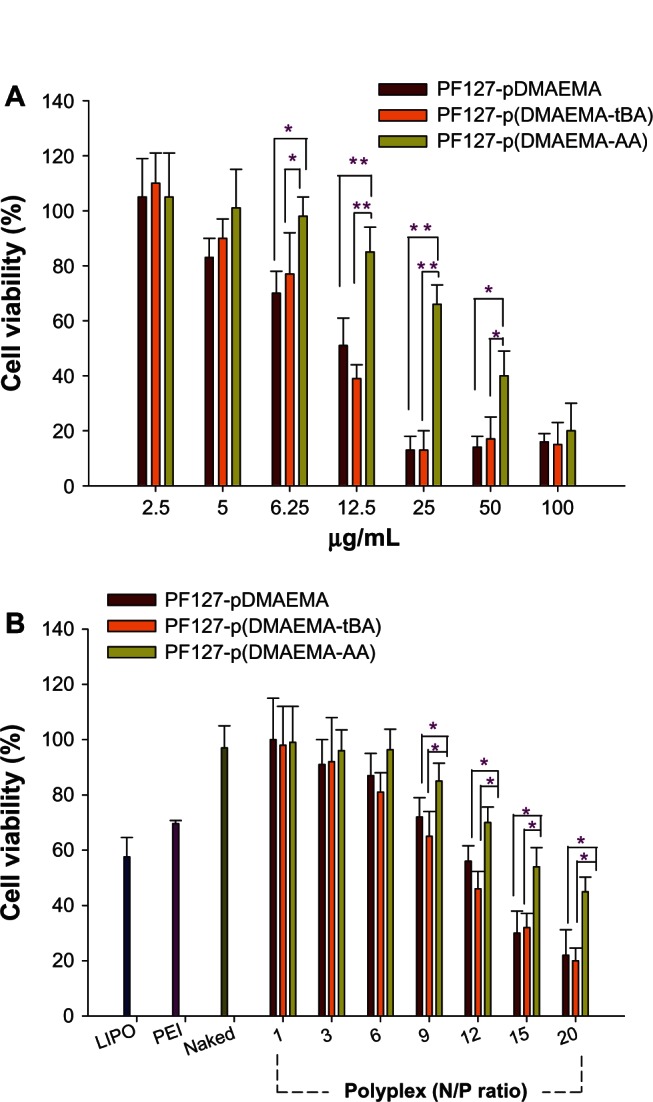
Cell viabilities of copolymers and their copolymer/pDNA polyplexes in 293T cells as a function of copolymer concentrations and polyplexes at various N/P ratios. (A) Copolymer concentrations (n = 8, *P < 0.05, **P < 0.001), and (B) polyplexes at various N/P ratios (n = 3, *P < 0.05).
Note: The N/P ratio of PEI (25K)/pDNA was 10.
Abbreviations: pDNA, plasmid deoxyribonucleic acid; N/P, nitrogen/phosphate; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); LIPO, Lipofectamine® 2000; PEI, polyethylenimine; N/P, nitrogen/phosphate.
In vitro gene transfection
The transfection ability of copolymers was assayed by pGL3-control plasmid for luminescence measurement. The gene transfection efficiencies of PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA) complexed with pDNA at various N/P ratios were compared with those of LIPO/pDNA, PEI(25K)/pDNA at N/P = 10, and naked pDNA. Without 10% FBS, the transfection efficiencies of three polyplexes at any N/P ratios are comparable with that of PEI(25K)/pDNA, but slightly poorer than that of LIPO/pDNA (Figure 7A). As an N/P ratio increases to ≥15, the transfection efficiencies of PF127-p(DMAEMA-AA)/pDNA are higher than those of PF127-pDMAEMA/pDNA and PF127-p(DMAEMA-tBA)/pDNA (P < 0.05). The high charge density of cationic polymer/DNA polyplex is suggested to be a major restriction for intracellular release of DNA and cytotoxicity.4 We speculate that the PF127-p(DMAEMA-AA) copolymer forms intramolecular electrostatic interactions between the amino groups and the carboxyl groups, which facilitates the DNA dissociation from the endolysosomal compartment; as well, it reduces the high cationic charge density of PDMAEMA to increase cell viability.
Figure 7.
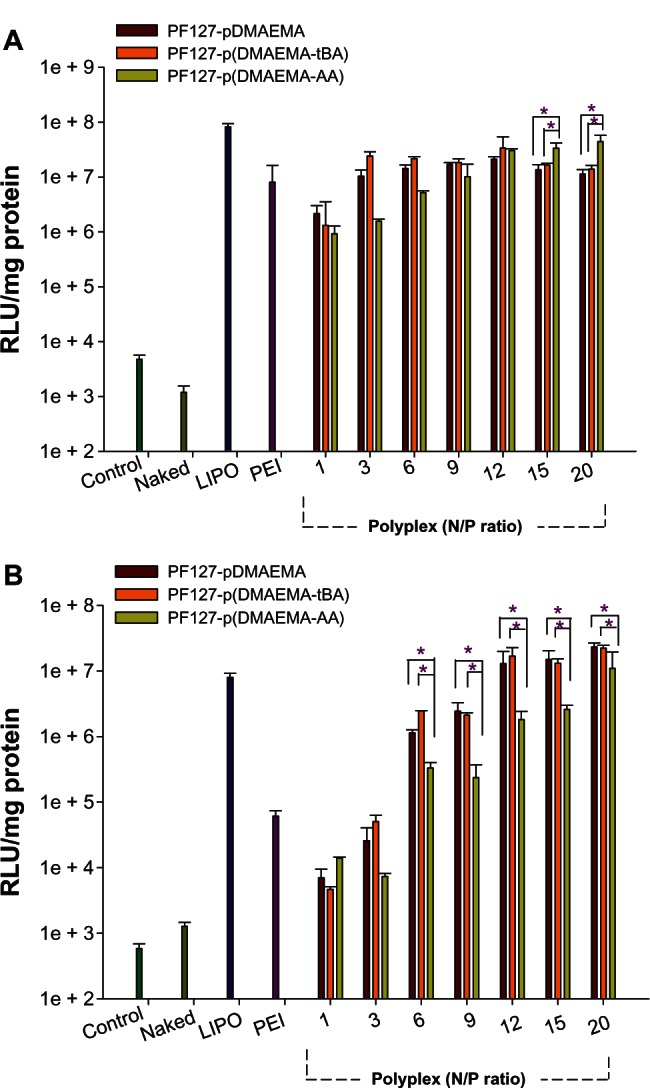
In vitro gene expression of copolymer/pDNA polyplexes in 293T cells using various N/P ratios. (A) Without 10% FBS and (B) with 10% FBS (n = 3, *P < 0.05).
Note: The N/P ratio of PEI (25K)/pDNA was 10.
Abbreviations: pDNA, plasmid deoxyribonucleic acid; N/P, nitrogen/phosphate; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); RLU, relative luminescence units; LIPO, Lipofectamine® 2000; PEI, polyethylenimine; FBS, fetal bovine serum.
The transfection efficiency was also done with 10% FBS. Because of the competition between FBS and pDNA with cationic polymers, which may reduce the pDNA concentration in the polyplexes or the FBS adsorption on the polyplex surface inhibiting transgene expression,36,40,41 the lower transfection efficiencies were obtained. The transfection efficiencies of both PF127-pDMAEMA/pDNA and PF127-p(DMAEMA-tBA)/pDNA are similar at every N/P ratio, but better than that of PEI/pDNA when an N/P was adjusted to ≥3, and comparable to that of LIPO/pDNA when an N/P was adjusted to N/P ≥12. The transfection efficiencies of PF127-p(DMAEMA-AA)/pDNA are significantly lower than those of PF127-pDMAEMA/pDNA and PF127-p(DMAEMA-tBA)/pDNA, when compared at the same N/P ratio of >1.
The good transfection ability of polyplexes is correlated to high surface charges,42 small particle sizes,43 and enhanced association with cellular uptake via the hydrophobic interactions.44 From Table 2, the particle diameters of PF127-p(DMAEMA-AA)/pDNA are 284 nm and 92 nm after 4 hours standing with and without 10% FBS, respectively. In the presence of FBS, the remarkable increase in the particle size may limit the cellular uptake of PF127-p(DMAEMA-AA)/pDNA, which results in low transfection efficiency (Figure S4). However, although the particle diameters of PF127-pDMAEMA/pDNA also increase from 103 nm without FBS to 225 nm with FBS, the high surface charges may allow it to retain high transfection efficiency.
Cellular internalization
The PF127-p(DMAEMA-AA) copolymer has multiple functions including (1) cationic DMAEMA segments for condensing pDNA; (2) PF127 segments for reducing cytotoxicity and self-assembly; and (3) AA functional groups linked with a fluorescent dye for internalization trafficking. R123 was used as a model fluorescent probe and attached to PF127-p(DMAEMA-AA) through a chemical reaction between the amino groups of R123 and the carboxylic groups of AA to produce PF127-p(DMAEMA-AA)-R123. The PF127-p(DMAEMA-AA)-R123/pDNA polyplex was used to study the cellular internalization mechanism in 293T cells.
A variety of formations of endocytosis have been displayed to be involved in the cellular uptake of polyplexes. The interactions between polyplexes and cell membranes were examined by treating the cells with different chemical inhibitors: wortmannin for macropinocytosis;25 chlorpromazine for clathrin-mediated endocytosis;26 and genistein for caveolae-mediated endocytosis.27 The cytotoxicity of the chemical inhibitors was tested and the inhibitors were found to be nontoxic to 293T cells at the concentrations used (Figure S5). As compared with the polyplex-treated cells (control), the flow cytometric diagrams clearly shifted left when the cells were pretreated with genistein and chlorpromazine, and remained intact when the cells were pretreated with wortmannin (Figure 8A). The reduced fluorescence intensity was calculated based on the control group. The percentages of inhibition are 32% for the cells pretreated with chlorpromazine and 27% for the cells pretreated with genistein, respectively (Figure 8B). In addition, the luciferase gene expression of the polyplex decreased significantly in the presence of chlorpromazine, genistein, and both together (Figure S6). The flow cytometric and gene expression data suggest that both clathrin-mediated and caveolae-mediated endocytosis are two possible pathways in the cellular uptake of copolymer/pDNA.
Figure 8.
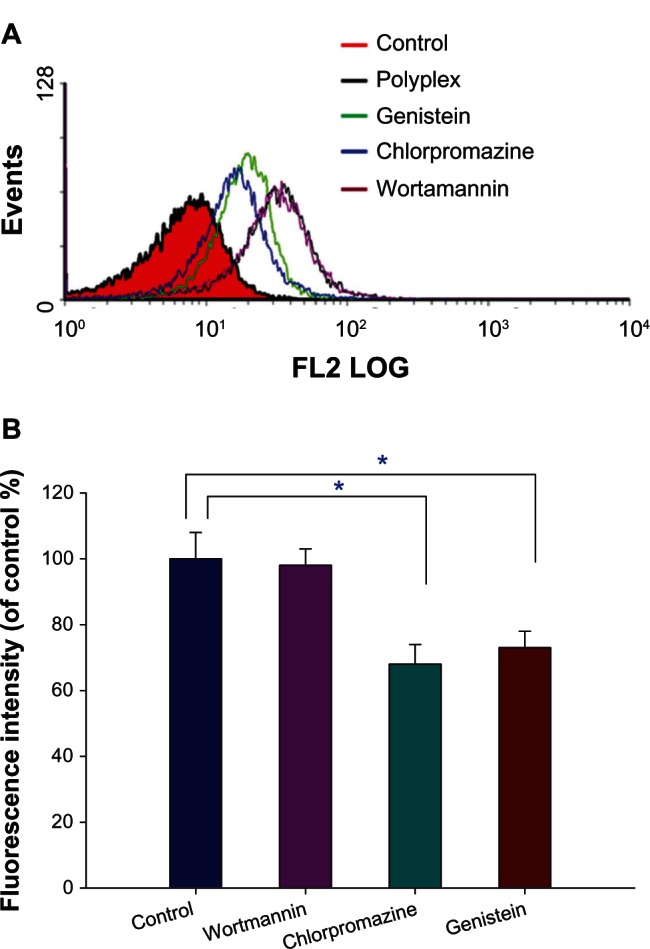
The effect of inhibitors on internalization of PF127-p(DMAEMA-AA)-R123/pDNA at N/P = 9 in 293T cells. (A) Flow cytometric diagrams and (B) mean fluorescence intensities (n = 3, *P < 0.05).
Note: The polyplex-treated cells were used as a control group.
Abbreviations: FL2 LOG, flow cytometric channel that is 575nm band pass filter and the fluorescence intensity measured in Log scale; PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); R123, rhodamine 123; pDNA, plasmid deoxyribonucleic acid; N/P, nitrogen/phosphate.
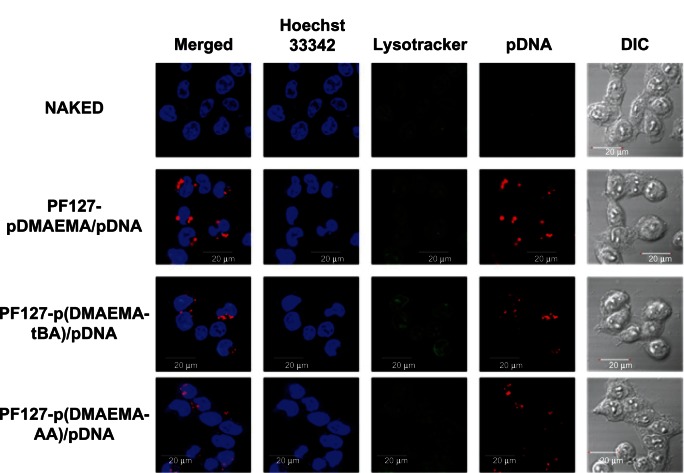
To directly visualize pDNA internalization into 293T cells, we labeled pGL3-control plasmid with fluorescent Cy5 dye and traced the Cy5-labeled pGL3 using a CLSM. The nuclei were stained with Hoechst 33342 in blue and the endolysosomes with LysoTracker® in green. Intracellular uptake of pDNA was traced in red (Figure 9). After 4 hours of incubation, all three polyplexes show more red regions than does the naked pDNA, implying that pDNA can be efficiently released from the polyplexes into the cytoplasm and most of the pDNA molecules reach the perinuclear region in the 293T cells.
Figure 9.
CLSM images of 293T cells exposed to copolymer/Cy5-labeled pGL3 polyplexes at N/P = 9 for 4 hours of incubation.
Note: Blue: nuclei (Hoechst 33342), green: endolysosome (Lysotracker), and red: Cy5-labeled pGL3 plasmid DNA.
Abbreviations: CLSM, confocal laser scanning microscope; N/P, nitrogen/phosphate; pDNA, plasmid deoxyribonucleic acid; DIC, differential interference contrast; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate); PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid).
Conclusion
We successfully synthesized PF127-pDMAEMA, PF127-p(DMAEMA-tBA), and PF127-p(DMAEMA-AA) copolymers with similar DMAEMA chain lengths via ATRP. Three copolymers showed the similar buffering capacity and complexed well with pDNA. The particle diameters decreased and the zeta potentials increased with increasing N/P ratios. PF127-p(DMAEMA-AA)/pDNA formed the smallest particle sizes and the highest cell viabilities among three polyplexes. At an N/P ratio of 9, all three polyplexes were spherical in shape. Without FBS, three polyplexes had the similar transfection efficiencies, which were comparable to PEI(25K)/pDNA, but with FBS, they were better than PEI(25K)/pDNA and comparable to LIPO/pDNA when N/P ratios were adjusted to ≥6.
We optimized the 15 AA units in PF127-p(DMAEMA-AA) to retain good transfection efficiency with low cytotoxicity. PF127-p(DMAEMA-AA) was a multifunctional copolymer because the DMAEMA segment could form electrostatic interactions with pDNA for a gene delivery vector; the carboxylic groups of AA could be utilized to react with functionalized molecules such as fluorescent dyes, and targeting ligands. R123 was used as a probe for cellular internalization study. Both clathrin-mediated and caveolae-mediated endocytosis pathways were involved in cellular uptake of polyplexes. The pDNA could be released from polyplexes to perform its transfection action.
Supplementary figures
1H-NMR spectrum of the PF127-Br.
Abbreviations: 1H-NMR, proton nuclear magnetic resonance; PF127-Br, di-2-bromoisobutyryl-pluronic F127; ppm (t1), parts per million (t1-longitudinal [spin-lattice] relaxation time).
Chemical reaction steps to produce PF127-p(DMAEMA-AA) and PF127-p(DMAEMA-AA)-R123.
Abbreviations: PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); R123, rhodamine 123; PF127-Br, di-2-bromoisobutyryl-pluronic F127; tBA, tert-butyl acrylate; CuBr/bpy, copper(I) bromide/2,2’- Bipyridine; HCl, hydrochloric acid; Dna, deoxyribonucleic acid; EDAC, N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride; PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate).
1H-NMR spectrum of the PF127-pDMAEMA.
Abbreviations: 1H-NMR, proton nuclear magnetic resonance; PF127-pDMAEMA, pluronic F127-poly(dimethylaminoethyl methacrylate); ppm (t1), parts per million (t1-longitudinal [spin-lattice] relaxation time).
CLSM images of 293T cells exposed to PF127-p(DMAEMA-AA)/Cy5-labeled pGL3 polyplexes at N/P = 9 for 4 hours of incubation.
Notes: Notes: Blue: nuclei (Hoechst 33342), green: endolysosome (Lysotracker), and red: Cy5-labeled pGL3 pDNA.
Abbreviations: CLSM, confocal laser scanning microscope; PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); N/P, nitrogen/phosphate; DNA, deoxyribonucleic acid; FBS, fetal bovine serum; pDNA, plasmid deoxyribonucleic acid.
The cytotoxicity of the chemical inhibitors in 293T cells (n = 3).
Abbreviations: n, number.
The gene expression of PF127-p(DMAEMA-AA)/pDNA in the presence of the chemical inhibitors in 293T cells (n = 3).
Abbreviations: PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); n, number; pDNA, plasmid deoxyribonucleic acid; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate).
Acknowledgments
We are grateful for the financial support from the National Science Foundation of Taiwan (grant numbers NSC-98-2221-E037-001-MY3, NSC-100-2320-B037-003-MY3, and NSC-101-2325-B037-006).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Behr JP. Synthetic gene transfer vectors II: back to the future. Acc Chem Res. 2012;45(7):980–984. doi: 10.1021/ar200213g. [DOI] [PubMed] [Google Scholar]
- 2.Samal SK, Dash M, Van Vlierberghe S, et al. Cationic polymers and their therapeutic potential. Chem Soc Rev. 2012;41(21):7147–7194. doi: 10.1039/c2cs35094g. [DOI] [PubMed] [Google Scholar]
- 3.Jäger M, Schubert S, Ochrimenko S, Fischer D, Schubert US. Branched and linear poly(ethylene imine)-based conjugates: synthetic modification, characterization, and application. Chem Soc Rev. 2012;41(13):4755–4767. doi: 10.1039/c2cs35146c. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal S, Zhang Y, Maji S, Greiner A. PDMAEMA based gene delivery materials. Materials Today. 2012;15(9):388–393. [Google Scholar]
- 5.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58(14):1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Wen Y, Guo Z, Du Z, et al. Serum tolerance and endosomal escape capacity of histidine-modified pDNA-loaded complexes based on polyamidoamine dendrimer derivatives. Biomaterials. 2012;33(32):8111–8121. doi: 10.1016/j.biomaterials.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 7.Majewski AP, Schallon A, Jérôme V, Freitag R, Müller AH, Schmalz H. Dual-responsive magnetic core-shell nanoparticles for non-viral gene delivery and cell separation. Biomacromolecules. 2012;13(3):857–866. doi: 10.1021/bm2017756. [DOI] [PubMed] [Google Scholar]
- 8.Dai F, Sun P, Liu Y, Liu W. Redox-cleavable star cationic PDMAEMA by arm-first approach of ATRP as a nonviral vector for gene delivery. Biomaterials. 2010;31(3):559–569. doi: 10.1016/j.biomaterials.2009.09.055. [DOI] [PubMed] [Google Scholar]
- 9.Xu FJ, Li H, Li J, Zhang Z, Kang ET, Neoh KG. Pentablock copolymers of poly(ethylene glycol), poly((2-dimethyl amino)ethyl methacrylate) and poly(2-hydroxyethyl methacrylate) from consecutive atom transfer radical polymerizations for non-viral gene delivery. Biomaterials. 2008;29(20):3023–3033. doi: 10.1016/j.biomaterials.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 10.Guo S, Huang Y, Zhang W, et al. Ternary complexes of amphiphilic polycaprolactone-graft-poly (N, N-dimethylaminoethyl methacrylate), DNA and polyglutamic acid-graft-poly(ethylene glycol) for gene delivery. Biomaterials. 2011;32(18):4283–4292. doi: 10.1016/j.biomaterials.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 11.Zhu C, Jung S, Luo S, et al. Co-delivery of siRNA and paclitaxel into cancer cells by biodegradable cationic micelles based on PDMAEMA-PCL-PDMAEMA triblock copolymers. Biomaterials. 2010;31(8):2408–2416. doi: 10.1016/j.biomaterials.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 12.Synatschke CV, Schallon A, Jérôme V, Freitag R, Müller AH. Influence of polymer architecture and molecular weight of poly(2-(dimethylamino)ethyl methacrylate) polycations on transfection efficiency and cell viability in gene delivery. Biomacromolecules. 2011;12(12):4247–4255. doi: 10.1021/bm201111d. [DOI] [PubMed] [Google Scholar]
- 13.Sharma R, Lee JS, Bettencourt RC, Xiao C, Konieczny SF, Won YY. Effects of the incorporation of a hydrophobic middle block into a PEG-polycation diblock copolymer on the physicochemical and cell interaction properties of the polymer-DNA complexes. Biomacromolecules. 2008;9(11):3294–3307. doi: 10.1021/bm800876v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funhoff AM, Monge S, Teeuwen R, et al. PEG shielded polymeric double-layered micelles for gene delivery. J Control Release. 2005;102(3):711–724. doi: 10.1016/j.jconrel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Lam JK, Ma Y, Armes SP, Lewis AL, Baldwin T, Stolnik S. Phosphorylcholine-polycation diblock copolymers as synthetic vectors for gene delivery. J Control Release. 2004;100(2):293–312. doi: 10.1016/j.jconrel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 16.Husseini GA, Pitt WG. Micelles and nanoparticles for ultrasonic drug and gene delivery. Adv Drug Deliv Rev. 2008;60(10):1137–1152. doi: 10.1016/j.addr.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabanov AV, Batrakova EV, Alakhov VY. Pluronic block copolymers as novel polymer therapeutics for drug and gene delivery. J Control Release. 2002;82(2–3):189–212. doi: 10.1016/s0168-3659(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Wu B, Lu P, Cloer C, Tucker JD, Lu Q. Polyethylenimine-modified pluronics (PCMs) improve morpholino oligomer delivery in cell culture and dystrophic mdx mice. Mol Ther. 2013;21(1):210–216. doi: 10.1038/mt.2012.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon E, Kim HD, Kim JS. Pluronic-grafted poly-(L)-lysine as a new synthetic gene carrier. J Biomed Mater Res A. 2003;66(4):854–859. doi: 10.1002/jbm.a.10012. [DOI] [PubMed] [Google Scholar]
- 20.Lai TC, Kataoka K, Kwon GS. Pluronic-based cationic block copolymer for forming pDNA polyplexes with enhanced cellular uptake and improved transfection efficiency. Biomaterials. 2011;32(20):4594–4603. doi: 10.1016/j.biomaterials.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal A, Unfer RC, Mallapragada SK. Dual-role self-assembling nanoplexes for efficient gene transfection and sustained gene delivery. Biomaterials. 2008;29(5):607–617. doi: 10.1016/j.biomaterials.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Valeeva YK, Dorodnykh TY, Alexandrova NA, et al. Block copolymers of Pluronics and poly-(2-dimethylaminoethyl methacrylate) for delivery of oligonucleotides into tumor cells. J Drug Deliv Sci Technol. 2006;16(4):245–251. [Google Scholar]
- 23.Mishra S, Peddada LY, Devore DI, Roth CM. Poly(alkylene oxide) copolymers for nucleic acid delivery. Acc Chem Res. 2012;45(7):1057–1066. doi: 10.1021/ar200232n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan MM, Zhang X, Qin J, Li BJ, Sun X, Zhang S. Self-assembly pluronic and β-cyclodextrin to hollow nanospheres for enhanced gene delivery. Macromol Rapid Commun. 2011 Jul 22; doi: 10.1002/marc.201100272. Epub. [DOI] [PubMed] [Google Scholar]
- 25.Araki N, Johnson MT, Swanson JA. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J Cell Biol. 1996;135(5):1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E, Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol Ther. 2006;14(5):745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Perumal OP, Inapagolla R, Kannan S, Kannan RM. The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials. 2008;29(24–25):3469–3476. doi: 10.1016/j.biomaterials.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 28.Yu K, Wang H, Xue L, Han Y. Stimuli-responsive polyelectrolyte block copolymer brushes synthesized from the Si wafer via atom-transfer radical polymerization. Langmuir. 2007;23(3):1443–1452. doi: 10.1021/la062159g. [DOI] [PubMed] [Google Scholar]
- 29.Feng CL, Vancso GJ, Schönherr H. Interfacial reactions in confinement: kinetics and temperature dependence of the surface hydrolysis of polystyrene-block-poly(tert-butyl acrylate) thin films. Langmuir. 2005;21(6):2356–2363. doi: 10.1021/la047490j. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Li GL, Zhang ZG, Li J, Neoh KG, Kang ET. Self-assembly of pH-responsive and fluorescent comb-like amphiphilic copolymers in aqueous media. Polymer. 2010;51(15):3377–3386. [Google Scholar]
- 31.Baek KY, Lee SH, Hwang SS. Synthesis of amphiphilic star block copolymer with photosensitive core by ATRP. Macromolecular Research. 2011;19(5):461–467. [Google Scholar]
- 32.Tseng WC, Tang CH, Fang TY. The role of dextran conjugation in transfection mediated by dextran-grafted polyethylenimine. J Gene Med. 2004;6(8):895–905. doi: 10.1002/jgm.572. [DOI] [PubMed] [Google Scholar]
- 33.De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm Res. 2000;17(2):113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 34.van Duijvenbode RC, Borkovec M, Koper GJM. Acid-base properties of poly(propylene imine)dendrimers. Polymer. 1998;39(12):2657–2664. [Google Scholar]
- 35.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151(3):220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Oskuee RK, Philipp A, Dehshahri A, Wagner E, Ramezani M. The impact of carboxyalkylation of branched polyethylenimine on effectiveness in small interfering RNA delivery. J Gene Med. 2010;12(9):729–738. doi: 10.1002/jgm.1490. [DOI] [PubMed] [Google Scholar]
- 37.Alshamsan A, Hamdy S, Samuel J, El-Kadi AO, Lavasanifar A, Uludağ H. The induction of tumor apoptosis in B16 melanoma following STAT3 siRNA delivery with a lipid-substituted polyethylenimine. Biomaterials. 2010;31(6):1420–1428. doi: 10.1016/j.biomaterials.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Jones SP, Gabrielson NP, Wong CH, et al. Hydrophobically modified dendrons: developing structure-activity relationships for DNA binding and gene transfection. Mol Pharm. 2011;8(2):416–429. doi: 10.1021/mp100260c. [DOI] [PubMed] [Google Scholar]
- 39.Lo YL, Wang YS, Wang LF. The Copolymer of Poly(2-dimethylaminoethyl methacrylate) and Methacrylated Chondroitin Sulfate with Low Cytotoxicity for Gene Delivery. Adv Healthc Mater. 2013 doi: 10.1002/adhm.201200373. In press. [DOI] [PubMed] [Google Scholar]
- 40.Réthoré G, Mathew A, Naik H, Pandit A. Preparation of chitosan/polyglutamic acid spheres based on the use of polystyrene template as a non-viral gene carrier. Tissue Eng Part C Methods. 2009;15(4):605–613. doi: 10.1089/ten.TEC.2008.0581. [DOI] [PubMed] [Google Scholar]
- 41.Morris VB, Sharma CP. Folate mediated histidine derivative of quaternised chitosan as a gene delivery vector. Int J Pharm. 2010;389(1–2):176–185. doi: 10.1016/j.ijpharm.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 42.Hanzlíková M, Ruponen M, Galli E, et al. Mechanisms of polyethylenimine-mediated DNA delivery: free carrier helps to overcome the barrier of cell-surface glycosaminoglycans. J Gene Med. 2011;13(7–8):402–409. doi: 10.1002/jgm.1587. [DOI] [PubMed] [Google Scholar]
- 43.Sunshine JC, Peng DY, Green JJ. Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol Pharm. 2012;9(11):3375–3383. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang B, He C, Tang C, Yin C. Effects of hydrophobic and hydrophilic modifications on gene delivery of amphiphilic chitosan based nanocarriers. Biomaterials. 2011;32(20):4630–4638. doi: 10.1016/j.biomaterials.2011.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H-NMR spectrum of the PF127-Br.
Abbreviations: 1H-NMR, proton nuclear magnetic resonance; PF127-Br, di-2-bromoisobutyryl-pluronic F127; ppm (t1), parts per million (t1-longitudinal [spin-lattice] relaxation time).
Chemical reaction steps to produce PF127-p(DMAEMA-AA) and PF127-p(DMAEMA-AA)-R123.
Abbreviations: PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); R123, rhodamine 123; PF127-Br, di-2-bromoisobutyryl-pluronic F127; tBA, tert-butyl acrylate; CuBr/bpy, copper(I) bromide/2,2’- Bipyridine; HCl, hydrochloric acid; Dna, deoxyribonucleic acid; EDAC, N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride; PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate).
1H-NMR spectrum of the PF127-pDMAEMA.
Abbreviations: 1H-NMR, proton nuclear magnetic resonance; PF127-pDMAEMA, pluronic F127-poly(dimethylaminoethyl methacrylate); ppm (t1), parts per million (t1-longitudinal [spin-lattice] relaxation time).
CLSM images of 293T cells exposed to PF127-p(DMAEMA-AA)/Cy5-labeled pGL3 polyplexes at N/P = 9 for 4 hours of incubation.
Notes: Notes: Blue: nuclei (Hoechst 33342), green: endolysosome (Lysotracker), and red: Cy5-labeled pGL3 pDNA.
Abbreviations: CLSM, confocal laser scanning microscope; PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); N/P, nitrogen/phosphate; DNA, deoxyribonucleic acid; FBS, fetal bovine serum; pDNA, plasmid deoxyribonucleic acid.
The cytotoxicity of the chemical inhibitors in 293T cells (n = 3).
Abbreviations: n, number.
The gene expression of PF127-p(DMAEMA-AA)/pDNA in the presence of the chemical inhibitors in 293T cells (n = 3).
Abbreviations: PF127-p(DMAEMA-AA), pluronic F127-poly (dimethylaminoethyl methacrylate-acrylic acid); n, number; pDNA, plasmid deoxyribonucleic acid; PF127-pDMAEMA, pluronic F127-poly (dimethylaminoethyl methacrylate); PF127-p(DMAEMA-tBA), pluronic F127-poly (dimethylaminoethyl methacrylate-tert-butyl acrylate).