Abstract
Trinucleotide repeat expansion underlies at least 17 neurologic diseases. In affected individuals, the expanded locus is characterized by dramatic changes in chromatin structure and in repeat tract length. Interestingly, recent studies show that several chromatin modifiers, including a histone acetyltransferase, a DNA methyltransferase, and the transcription factor CTCF can modulate repeat instability. Here, we propose that the unusual chromatin structure of expanded repeats directly impacts their instability. We discuss several potential models for how this might occur, including a role for DNA repair-dependent epigenetic reprogramming in increasing repeat instability, and the capacity of epigenetic marks to alter sense and antisense transcription, thereby affecting repeat instability.
Long Triplet Repeats are Unstable and Cause a Number of Degenerative Disorders
In 1991, a novel type of mutation—the expansion of trinucleotide repeats—was shown to cause two human neurological diseases: fragile X syndrome (FRAXA) and spinal and bulbar muscular atrophy (SBMA)1, 2. To date, 17 such diseases have been identified3. Normal individuals typically harbour fewer than 30 repeats, whereas patients can carry from 35 to several thousand repeats. The disease incidence can be as common as 1 in 4000 males for FRAXA and as rare as 1 in 50 000 for Friedreich ataxia (FRDA)3. Large tracts of trinucleotides can cause disease in several ways: by affecting gene expression, producing a toxic RNA species, or altering the function of the resultant protein3 (Table 1). Changes that impair gene expression or protein function are recessive (e.g. FRDA), whereas those that produce toxic RNAs or proteins are dominant (e.g. myotonic dystrophy (DM1) and Huntington disease (HD)).
Table 1.
Disorders caused by expansion of trinucleotide repeatsa
| Disease | Gene Product |
Repeat type | Pathogenic category |
Somatic instability |
|---|---|---|---|---|
| Dentatorubro-pallidoluysian atrophy (DRPLA) |
Atrophin 1 | CAG | GOFb | moderate |
| Fragile X syndrome (FXS) |
Fragile site mental retardation 1 |
CGG | LOFc | Low |
| Fragile X syndrome (FRAXE) |
Fragile site mental retardation 2 |
CGG | LOF | NDd |
| fragile-X–associated tremor/ataxia syndrome (FXTAS) |
Fragile site mental retardation 1 |
CGG | Toxic RNA | Low |
| Friedreich Ataxia (FRDA) |
Frataxin | GAA | LOF | moderate |
| Huntington disease (HD) |
Huntingtin | CAG | GOF | moderate |
| Huntington Disease Like 2 (HDL2) |
junctophilin-3 | CAG | Unknown | ND |
| Myotonic dystrophy type 1 (DM1) |
DMPK | CTG | Toxic RNA | high |
| Spinal and bulbar muscular atrophy (SBMA) | Androgen receptor | CAG | GOF | low |
| Spinocerebellar ataxia type 1 (SCA1) |
Ataxin-1 | CAG | GOF | moderate |
| Spinocerebellar ataxia type 2 (SCA2) |
Ataxin-2 | CAG | GOF | moderate |
| Spinocerebellar ataxia type 3 (SCA3) |
Ataxin-3 | CAG | GOF | moderate |
| Spinocerebellar ataxia type 6 (SCA6) |
CACNAIA | CAG | GOF | ND |
| Spinocerebellar ataxia type 7 (SCA7) |
Ataxin-7 | CAG | GOF | moderate |
| Spinocerebellar ataxia type 8 (SCA8) |
C10orf2 | CAG | Unknown | ND |
| Spinocerebellar ataxia type 12 (SCA12) |
PPP2R2B | CAG | Unknown | ND |
| Spinocerebellar ataxia type 17 (SCA17) |
TBP | CAG | GOF | limited |
Analyses of repeat tracts in patients and animal models have revealed that trinucleotide repeats often show increases in repeat number (expansions) as well as decreases (contractions) in both germline and somatic tissues. In addition, the rate of instability varies during development and between tissues within a single organism. This variability implies multiple mechanisms of instability, an idea that is supported by the identification of a number of stage specific modifiers of repeat instability in mouse models (Figure 1). The bias of instability towards expansion in the germline and the embryo accounts for the increased severity of disease symptoms in subsequent generations. This phenomenon was coined ‘anticipation’ long before its molecular basis was discovered. In affected individuals, ongoing, expansion-biased repeat instability in somatic tissues, especially those associated with the disease phenotype, might decrease the age of onset of symptoms and accelerate disease progression4.
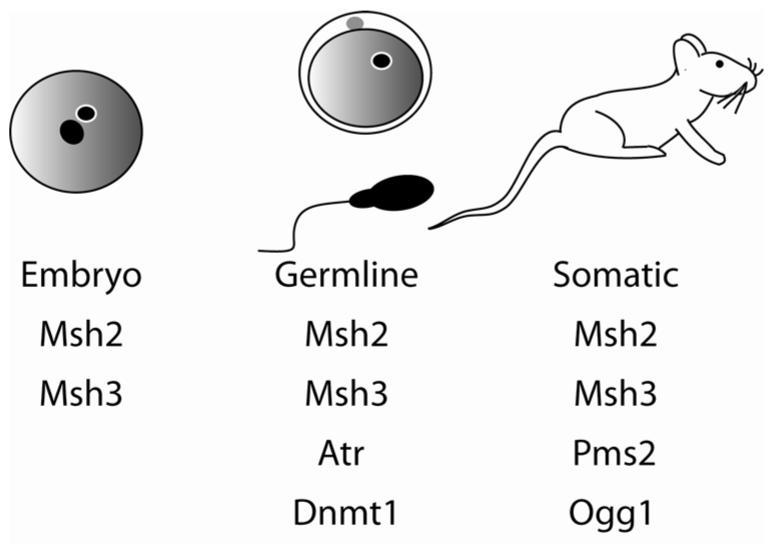
Figure 1.
Modulators of trinucleotide repeat instability. A number of gene products have been shown to modify trinucleotide repeat instability throughout mouse development42, 76-81. Msh2 and Msh3 are required for instability in all stages investigated for CAG/CTG repeats46, 76-79. Postmeiotic segregation increased 2 (Pms2)80 as well as Ogg142 are specifically involved in somatic repeat instability. Dnmt1, on the other hand, protects against expansions in an expanded CAG model only in the germlines 9. Ataxia telangiectasia and Rad3 related (Atr), a signaling kinase, prevents expansion in the female germline and age-dependent instability in somatic tissues of CGG repeats81. It should be noted that Msh6 null mice have mild triplet repeat instability phenotypes in mice, but this observation has been generally dismissed due to its effect on the stability of Msh2 and Msh3 77, 79, 82.
Instability of disease-causing trinucleotide repeats appears to be triggered by the folding of repeat sequences into abnormal secondary structures, including hairpins, slipped-strand structures, triplexes, and quadruplexes4. The mishandling of these aberrant structures by the DNA repair machinery is thought to cause changes in tract length.
Genetic studies in bacteria, yeast, flies, mammalian cells, and mice have revealed an astounding assortment of proteins whose mutation or knockdown significantly influences repeat stability4. Paradoxically, this wealth of possibilities has slowed the discovery of the major mechanisms that alter repeat stability.
To add to this complexity, the instability of repeats depends on internal influences, including the type of the repeat, its purity, and its length, as well as on the surrounding genomic context. Indeed, repeat tracts are typically much more unstable in mouse models generated through the insertion of large genomic fragments than in models that carry cDNAs, suggesting that the surrounding sequence influences repeat stability5. Examples of potential cis-acting elements include origins of replication4, transcription factor binding sites6, sense and antisense promoters7, neighboring GC-rich sequences8, and the local chromatin environment9, 10.
DNA is embedded in chromatin, which influences their metabolism (Box 1). Chromatin packing is influenced by histone tail modifications and DNA methylation at CpG dinucleotides. Recently, several studies have carefully catalogued the epigenetic marks associated with expanded GAA repeat tracts, showing that they are buried in heterochromatin6, 11, 12. In addition, several publications within the last two years have unearthed evidence that chromatin modifying enzymes and epigenetic marks contribute to repeat instability9, 10, 13. No cause-effect experiment has been done, however, to directly measure the effect of altering local chromatin structure on repeat instability. Therefore, we acknowledge at the outset that it is not yet clear whether the epigenetic status of repeats is a principal driver of repeat instability, or merely a by-product of repeat expansion.
Box 1. Chromatin structure and DNA metabolism.
Approximately 146bp of DNA is wrapped around a histone octamer to form the basic subunit of chromatin: the nucleosome. Chromosomes, however, are packaged into higher order structures, a process influenced by DNA methylation and covalent modification of histones. These epigenetic marks tend to be dynamic and vary between cells, between loci within a cell, and between different developmental stages at same locus32. Chromatin structure is broadly categorized into euchromatin and heterochromatin. Euchromatin is a looser structure that contains a high proportion of histone H3 and H4 molecules with acetylated lysine residues in their N-terminal tails. Heterochromatin is more densely packaged and is enriched in H3K9me and methylation of cytosine residues at CpG dinucleotides. Whether a given region is euchromatic or heterochromatic does not depend on any individual histone modification, but rather on collective influence of many different marks.
Chromatin regulates all DNA transactions including transcription. Transcription initiation and elongation require a complex interplay of nucleosome remodelling and chromatin modifying enzymes70. NER and BER proteins are often unable to bind nucleosomes in the absence of accessory proteins that expose damaged DNA for recognition71, 72. Origins of replication in higher eukaryotes are chiefly defined by their chromatin context, which needs to be maintained after the passage of the replication fork73. DNA damage signalling in heterochromatin has specific requirements and kinetics that differ from euchromatic regions74. Finally, the repair of double-strand breaks is accompanied by an intricate interplay of chromatin remodelling enzymes, nucleosomes eviction and modification, which allow accurate signalling, repair, and recovery from damage75. Clearly, these DNA transactions are heavily dependent on chromatin structure, and each of these processes affects triplet repeat instability.
Here, we summarize what is known about the chromatin environment of expanded repeat tracts and highlight the correlation between the epigenetic changes that occur during development and the timing of triplet repeat instability. To spark discussion, we speculate about how local chromatin structure might drive repeat instability throughout development.
The Chromatin Environment of Expanded Triplet Repeats
Several studies have identified alterations in DNA methylation, histone modification, and chromatin structure around expanded repeat tracts that are not present at the corresponding wild type alleles (Table 2). Expanded CGG repeats at the fragile X mental retardation 1 (FMR1) locus in FRAXA patients provided the first indication that such repeats are associated with heterochromatin marks. The absence of FMR1 expression, which is responsible for the mental retardation phenotype in FRAXA, is caused by extensive DNA methylation within the CGG repeat, as well as in the flanking DNA14-16. More recently, chromatin immunoprecipitation analyses of specific modified histones in the FMR1 5′ region near the expanded repeat revealed high levels of other repressive marks, including methylated histone 3 at lysine 9 (H3K9me)17, and lower levels of two marks typically encountered in euchromatic regions: acetylated (Ac) H3 and H418. These data indicate that expanded CGG repeats, unlike their normal-length counterparts, carry epigenetic marks typical of heterochromatin, and display diminished levels of euchromatic marks.
Table 2.
Epigenetic changes at expanded repeats compared to wild type alleles
| Disease | Repeat | Length | Species | Tissue | Positiond | Heterochromatic markse |
Euchromatic markse |
References |
|---|---|---|---|---|---|---|---|---|
| FRA12A | CGG | 270- 300 |
Human | Transformed lymphoblast |
U, P | mCpG ↑ | ND | 84 |
| FRAXA | CGG | 50-900 | Human | Various tissuesc | U | mCpG ↑ | ND |
14, 15, 24, 25, 50, 52,
53, 55, 56, 85 |
| 50- 200+a |
Blood | U, W | mCpG ↑ | ND | 15, 24 | |||
| NRa,b | Blood | U, W, D | mCpG ↑ | ND | 86 | |||
| NRb | Transformed lymphoblast |
P | ND | H3K4me2 ↓ H3K9Ac ↓ H4K16Ac – |
69 | |||
| NR | Transformed lymphoblast |
D | ND | H3K4me2 ↓ H3K9Ac ↓ H4K16Ac ↓ |
69 | |||
| 230- 530a |
Transformed lymphoblast |
U, W, D |
mCpG ↑ H3K9me2 ↑ |
H3K4me2 ↓ H3Ac ↓ H4Ac ↓ |
16-18, 86 | |||
| FRAXE | CGG | 100- 1500 |
Human | Blood | U,D | mCpG ↑ | ND | 87-90 |
| 130- 1500 |
Fibroblast | D | mCpG ↑ | ND | 89, 91 | |||
| 180- 733 |
Transformed lymphoblast |
D | mCpG ↑ | ND | 88, 92 | |||
| DM1 | CTG | NR | Human | Fibroblast | U | mCpG ↑ | ND | 93 |
| Fibroblast | U, W, D | H3K9me3 ↑ | H3K4me3 ↓ | 21 | ||||
| 1000- 1800 |
Blood | U, W, D | mCpG ↑ | ND | 20 | |||
| Fetal dura matter | U, W, D | mCpG ↑ | ND | 20 | ||||
| Fetal skeletal muscles |
U, W, D | mCpG ↑ | ND | 20 | ||||
| SCA1 | CAG | 143 | Mouse | Testis | P | H3 K9me2 - |
ND | 9 |
| U |
mCpG - H3K9me2 - |
H3Ac - | 9, unpublished data | |||||
| D |
mCpG ↑ H3K9me2 - |
H3Ac - | 9, unpublished data | |||||
| FRDA | GAA | 650- 750 |
Human | Brain | P |
mCpG – H3K9me2 ↑ H3K9me3 ↑ |
H3K9Ac ↓ H3K14Ac – H4K5Ac – H4K8Ac ↑ H4K12Ac – H4K16Ac – |
12 |
| U |
mCpG ↑ H3K9me2 ↑ H3K9me3 ↑ |
H3K9Ac ↓ H3K14Ac – H4K5Ac – H4K8Ac – H4K12Ac – H4K16Ac – |
12 | |||||
| D |
mCpG ↓ H3K9me2 ↑ H3K9me3 ↑ |
H3K9Ac ↓ H3K14Ac – H4K5Ac ↓ H4K8Ac ↓ H4K12Ac ↓ H4K16Ac ↓ |
12 | |||||
| Cerebellum | U | mCpG ↑ | ND | 12 | ||||
| Heart | P, U | mCpG ↑ | ND | 12 | ||||
| D | mCpG ↓ | ND | 12 | |||||
| 650- 1030a |
Transformed lymphoblasts |
P | ND | H3K9Ac – H3K14Ac – H4K5Ac – H4K8Ac ↓ H4K12Ac ↓ H4K16Ac – |
94 | |||
| U |
mCpG ↑ H3K9me1 ↓ H3K9me2 ↑ H3K9me3 ↑ |
H3K9Ac ↓ H3K14Ac – H4K5Ac ↓ H4K8Ac ↓ H4K12Ac ↓ H4K16Ac ↓ |
6, 12, 94 | |||||
| D | ND | H3K9Ac ↓ H3K14Ac – H4K5Ac ↓ H4K8Ac ↓ H4K12Ac ↓ 4K16Ac ↓ |
94 | |||||
| 580 | HEK293 | U, D | H3K9me3 ↑ | H3K9Ac ↓ H3K14Ac – H4K5Ac ↓ H4K8Ac – |
11 | |||
| 90-120 | Mouse | Brain | P |
mCpG ↑ H3K9me2 ↑ H3K9me3 ↑ |
H3K9Ac – H3K14Ac – H4K5Ac – H4K8Ac – H4K12Ac – H4K16Ac – |
14 | ||
| U |
mCpG ↑ H3K9me2 ↑ H3K9me3 ↑ |
H3K9Ac – H3K14Ac – H4K5Ac – H4K8Ac – H4K12Ac – H4K16Ac – |
12 | |||||
| 230 | U | H3 K9me3 ↑ |
H3K14Ac ↓ H4K5Ac – H4K8Ac ↓ H4K16Ac ↓ |
95 | ||||
| 90-120 | D |
mCpG – H3K9me2 ↑ H3K9me3 ↑ |
H3K9Ac ↓ H3K14Ac – H4K5Ac – H4K8Ac – H4K12Ac – H4K16Ac ↑ |
12 | ||||
| 90-120 | Cerebellum | U | mCpG ↑ | ND | 12 | |||
| Heart | P | mCpG ↓ | ND | 12 | ||||
| U | mCpG ↑ | ND | 12 | |||||
| D | mCpG – | ND | 12 |
Includes unknown repeat length alleles obtained from affected individuals or premutation
NR, repeat length not reported
A variety of somatic tissues from affected individuals was analyzed.
P, promoter; U, upstream; D, downstream; W, within the repeat tract.
↑, enriched compared to wild type levels; ↓, depleted; –, not changed; ND, not determined
Expanded CTG repeats elicit similar effects on surrounding chromatin structure in cell lines derived from patients with a severe form of myotonic dystrophy (DM1). These cells harbor over 1000 CTGs in the 3′ UTR of the dystrophia myotonica protein kinase (DMPK) gene that are largely resistant to digestion with DNaseI and methylation sensitive restriction enzymes19, 20, properties typical of heterochromatin with high levels of DNA methylation. In addition, the expanded locus contains histones enriched in H3K9me and depleted in H3Ac21. Thus, at least in the case of congenital DM1 cells, highly expanded CTG repeats, like expanded CGG repeats, appear to assume a heterochromatic conformation.
Expansion-induced changes in chromatin structure have been most extensively studied in the first intron of frataxin gene, where the expansion of a GAA repeat in the first intron gives rise to FRDA. Similar to CGG and CTG repeats, the region upstream of the expanded GAA repeat in several somatic tissues, is more highly methylated at CpG sites than it is at wild type alleles6, 12. Moreover, decreased H3Ac and H4Ac levels and increased H3K9me levels surround expanded frataxin GAA repeats 6, 11, 12.
It is not known what triggers or maintains heterochromatin at expanded triplet repeats22. In all cases, the repeat tract length must first reach a certain threshold. When the repeat nears this threshold, heterochromatin forms and spreads in a manner reminiscent of position effect variegation, the stochastic spreading of heterochromatin to adjacent euchromatic regions23. For instance, some individuals carrying FRAXA premutation alleles (harboring ~60-200 CGG repeats) display mosaic patterns of DNA methylation in different tissues24, whereas the full expansion (>200 CGGs) is associated with complete FMR1 promoter methylation25. The idea that repeats can induce heterochromatin spreading was directly tested in mice26. Whereas arrays of reporter genes were silenced only when they integrated near a heterochromatic region26, similar arrays, harboring 192 CTG or 200 GAA repeats, were silenced regardless of the transgene’s genomic location26. Thus, CGG, CTG, and GAA repeats, independent of their genomic location, seem to direct formation and spreading of heterochromatinn.
Repeat Instability during Early Embryogenesis
Several studies in FRAXA individuals indicate that instability can occur during early embryogenesis. Some individuals harbor two major FMR1 CGG repeat lengths—derivatives of the same allele—at high frequency in every examined tissue, suggesting that repeat instability arose during the first zygotic cell division27, 28. This possibility is supported by the existence of monozygotic twins who carry different allele lengths27, 29.
In support of these conclusions from patient studies, Savouret et al provided convincing evidence for instability of DM1-associated CTG repeats during early embryogenesis30. They examined the effects of knocking out the mismatch repair (MMR) gene Msh2 on intergenerational repeat instability in mice that carried >300 DMPK CTG repeats. Msh2 is central to the recognition of mismatches in the DNA31 and binds hairpins formed by CTG repeats4. Savouret et al observed ongoing instability in the germline of the transmitting parent, pointing to germline development as a period prone to repeat instability. However, if the effect of Msh2 deletion were solely confined to the parent’s germline, the genotype of the embryo should make no difference. This was not the case: whereas Msh2+/− offspring of Msh2+/− parents harbored mostly CTG expansions, Msh2−/− littermates were strongly biased toward contractions. Similarly, in a cross between Msh2−/− mice carrying the expanded allele and Msh2+/− mice, the Msh2−/− offspring had mostly contractions, but their Msh2+/− littermates displayed about equal proportions of contractions and expansions. This dependence on genotype indicates that some repeat instability must occur in the embryo. In addition, the lack of multiple repeat lengths in individual newborn mice indicates that embryonic instability must occur very early during development, probably just after fertilization30.
Vertebrate embryogenesis entails extensive epigenetic changes that are essential for resetting the developmental potential of the zygote32. This process is called reprogramming. In the mouse, the paternal chromatin is remodeled in the egg by histone exchange and acetylation and by elimination of DNA methylation via an active process that remains poorly understood32, 33. By contrast, the maternal genome is passively demethylated through replication during the first 3.5 days post fertilization. The methylation pattern is subsequently restored by de novo methyltransferases and maintained by Dnmt1 (DNA (cytosine-5-) methyltransferase 1). The temporal overlap of chromatin structure reprogramming and trinucleotide repeat instability raises the possibility of a cause-and-effect relationship between the two processes.
To probe the effects of reprogramming on repeat instability, Gorbunova et al attempted to model, in cultured cells, the demethylation that occurs during embroygenesis34. Treatments that reduced genome-wide DNA methylation levels and Dnmt1 activity stimulated CAG repeat contractions up to 1000-fold in a selection assay in mamalian cells and promoted expansions in unselected DM1 cell lines35. These data suggest that DNA demethylation provokes repeat instability, but the mechanism is unclear. Several possibilities have been eliminated, however, including homologous recombination, nonhomologous end joining, and transcription through the repeat tract36. Similar effects of CpG methylation have also been noted in bacteria: methylated CGG repeats and CAG repeats surrounded with methylated sequences are more stable than their unmethylated counterparts37. These studies provide a strong experimental link between DNA demethylation and repeat instability.
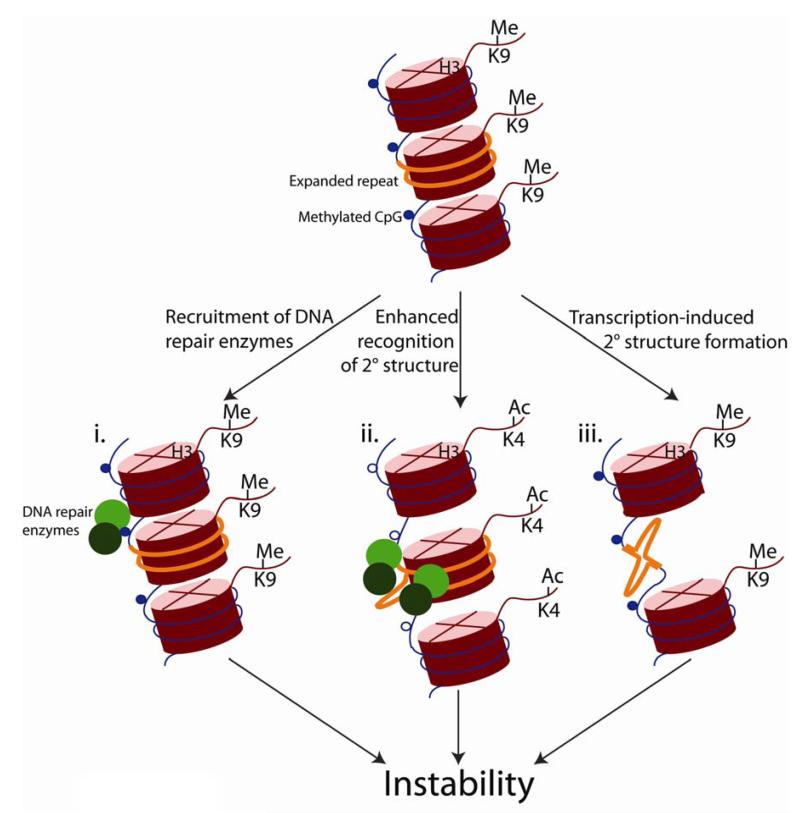
DNA demethylation during reprogramming could trigger repeat instability by engaging DNA repair pathways. Active demethylation in the paternal genome during early embryogenesis has been proposed to involve DNA repair via cytosine deaminases of the activation induced cytosine deaminase (Aid)/apolipoprotein B editing complex (Apobec1) family38, 39 in combination with the glycosylase Mbd4 (methyl-CpG binding domain protein 4) and the nucleotide excision repair (NER) component Gadd45a (growth arrest and DNA-damage-inducible alpha)39, 40. Aid and Apobec1, which are expressed in pluripotent cells, can deaminate 5-methylcytosine to thymine, creating T-G mismatches38, 39. Removal of T and its replacement by C could occur via MMR, base excision repair (BER), or even NER. Because these repair pathways introduce nicks in the DNA, and because nicks are associated with instability41, we speculate that reprogramming might cause destabilization of repeats by generating nicks at nearby methylated CpGs. Thus, methylated CpG sites might serve as signals to recruit DNA repair factors to expanded repeat tracts, leading to further instability (Figure 2i). This scenario could account for the MMR dependence of repeat instability in the early embryo30. Although no glycosylase is known to cause trinucleotide repeat instability in the embryo, there is precedence for glycosylase-induced instability in somatic tissue: removing 7,8-dihydro-8-oxo-guanine-DNA glycosylase 1 (Ogg1) greatly diminishes somatic instability in an HD mouse model42.
Figure 2.
Models for chromatin-dependent trinucleotide repeat instability. Expanded trinucleotide repeats (orange) are embedded into heterochromatin, which is enriched in H3K9me and DNA methylation at CpG sites (blue circles). Here, we present three speculative mechanisms by which chromatin environment might impact repeat instability. We imagine that these models might act in parallel at different loci, tissues, or developmental stages. i) Heterochromatin as a recruitment signal during reprogramming. During germline development and early embryogenesis, DNA is demethylated, probably through a DNA repair mechanism. DNA methylation at CpGs leads to recruitment of DNA repair factors (green circles), creating nicks that eventually lead to repeat instability. ii) Chromatin structure as a cis-acting factor in triplet repeat instability. Loose euchromatin could facilitate the access of DNA repair factors to aberrantly structured repeat tract. Promoting access would enhance gratuitous repair of the repeat tract and therefore instability (open circles are unmethylated CpG sites). iii) Transcription in triplet repeat instability. Sense, antisense, or convergent transcription through an expanded repeat could promote secondary DNA structure formation and therefore instability.
Repeat Instability during Germline Development
Instability in the germline and during gametogenesis is well documented in humans and mice. FRAXA patients, for example, show a striking parent-of-origin effect. Expanded alleles are often observed in children whose asymptomatic mothers harbor premutation alleles. Male fetuses from these mothers carrythe full mutation in all tested tissues except fetal testes, in which contractions were observed43. Similar analyses of female fetuses support the conclusion that CGG expansions occur in the female germline43.
Several studies have addressed the timing of germline instability. In HD patients, a substantial fraction of all instability of the CAG tract—mostly expansions—is generated pre-meiotically44. Similarly, in a DM1 mouse model, repeat instability increased with age, and all instability was already present in spermatogonia, a cell type found at the earliest stage of spermatogenesis, and no further increase was apparent at later stages of gametogenesis45. In contrast, using a mouse model for HD, Kovtun and McMurray46 detected instability in males only after the development of spermatozoa. The basis for the difference is unclear.
Analogous studies in the female germline of spinocerebellar ataxia type 1(SCA1) mouse models found that moderate to large contractions dominated transmissions from the maternal parent and the size of the contractions increased with age9, 47, 48. Thus, CAG repeat instability arises during both spermatogenesis and oogenesis..
As is the case during embryogenesis, instability of repeat tracts in the male and female germlines coincides with epigenetic changes to the genome. In the mouse, primordial germ cells are demethylated between days 8.5 and 13.5 of embryonic development, with the bulk of DNA demethylation occurring between E11.5 and E12.5, concurrent with substantial changes in histone modification and content32. DNA remethylation in the male germline starts at E1532. In the female germline, the genome is remethylated only after birth, during oocyte growth 32. Thus, epigenetic changes occur throughout germline development, at all stages where repeat instability has been identified.
The rapid loss of methylation in the developing germline suggests that demethylation is active and possibly dependent on DNA repair, as proposed for embryogenesis49. But germline reprogramming has not been studied as extensively as early embryogenesis, where evidence for a role of DNA repair in reprogramming is rapidly accumulating33. Nonetheless, we suggest that a spike in DNA repair activity near the repeat tract during reprogramming might account for the increased instability (Figure 2i). Alternatively, as reprogramming decreases the density of heterochromatic regions, DNA repair enzymes might gain freer access to the repeat tract, leading to enhanced recognition of secondary structures, gratuitous repair, and subsequent instability (Figure 2ii).
To probe the effects of DNA methylation on repeat stability, we removed one copy of Dnmt1 in a SCA1 mouse model carrying 143 CAG repeats at the endogenous locus9. Both male and female Dnmt1+/− SCA1 mice passed an expansion to their progeny 3 to 4 times more frequently than did Dnmt1+/+ SCA1 mice. In addition, the effects in Dnmt1+/− mice were confined to the germline; repeat instability was not observed in the early embryo or in somatic tissues9.
Analyses of testes and ovaries from Dnmt1+/− SCA1 mice did not reveal any global differences in DNA methylation of repetitive elements relative to Dnmt1+/+ SCA1 mice9, suggesting that Dnmt1 deficiency had little effect on genome-wide methylation. At CpG sites 17 and 20bp upstream of the repeat tract, however, ovaries displayed significantly reduced methylation levels, whereas testes showed significantly elevated levels. In the same region, testes of Dnmt1+/− mice also displayed variegated H3K9me2 levels, suggesting that the local chromatin environment underwent alterations. Collectively, these observations suggest that Dnmt1 deficiency enhances an expansion-biased process early on in the development of both the male and female germlines, at a time overlapping, and perhaps perturbing, the period of hypomethylation associated with reprogramming.
Studies of intergenerational instability in a fly model of CAG repeat instability suggest that histone acetylation is also involved13. Haploinsufficiency of CREB-binding protein (CBP), which encodes a histone acetyltransferase, increases triplet repeat instability13. By contrast, treatment with the histone deacetylase inhibitor, trichostatin A (TSA), decrease triplet repeat instability13. Although genome-wide levels of acetylated histones differed between the CBP-mutant flies or TSA-treated flies and their wild type counterparts, changes in histone acetylation were not observed near the repeat tract, nor was transcription through the repeat affected13. These results implicate histone acetylation as a modifier of triplet repeat instability but the effect, in this case, appears to be indirect.
Repeat Instability in Somatic Tissues
Somatic instability of CGG repeats, which, unlike CAGs and GAAs, can be methylated, is low in patients harboring the full mutation (>200 CGGs)50, 51. By contrast, FRAXA males, who express FMR1 although they harbor a large expansion, have unmethylated repeats with a high degree of instability in somatic tissues52, 53. One such individual displayed instability in some tissues and not in others, a pattern that correlated perfectly with methylation status of the repeat52. Cultured cells from FRAXA patients show the same absolute correlation: methylated CGG repeats are stable, whereas unmethylated repeats are unstable54-56. Importantly, in cultured primate cells, DNA methylation of plasmids containing CGG repeats stabilized the repeat tract, specifically implicating DNA methylation within or near repeat tracts as a key modifier of triplet repeat instability37.
In addition to epigenetic modifiers, transcription has emerged as an important player in triplet repeat instability. Transcription through a repeat tract can stimulate repeat instability in human cells and in fly models of CAG repeat diseases57. Lin et al 7, 58 showed that transcription through a repeat of 95 CAGs caused substantial contractions of the repeat tract7. Moreover, transcription-induced destabilization required MMR and transcription-coupled NER (TC-NER)57. Thus, heterochromatin might reduce repeat instability by lowering the rate of transcription through the repeat tract, thereby decreasing the potential for TC-NER (Figure 2iii). The absence of transcription through the highly expanded CGG repeats at the FMR1 locus could account for their surprising stability in somatic tissues.
Many repeat loci, however, are more unstable than their shorter counterparts even though they are transcribed at reduced levels57. For these loci, another mechanism must be at work. We suggest that this mechanism might be antisense transcription. Indeed, a surprisingly high fraction of human genes are associated with antisense transcripts, including most wild type alleles of triplet repeat-associated genes59 (Table 3). At the FMR1 locus, heterochromatin appears to shut-down both sense and antisense transcription60. By contrast, at the SCA8 and DM1 loci, the levels of antisense transcription are higher on the expanded alleles21, 61. At least at the DMPK locus, high levels of antisense transcription are associated with heterochromatin marks21.
Table 3.
Sense and antisense transcripts at disease loci with normal length repeatsa
| Disease | Repeat | Gene | Cell Lineb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBMC | HCT116 | Jurkat | MaPaC | MRC5 | ||||||||
| Sc | ASd | S | AS | S | AS | S | AS | S | AS | |||
| DM1 | CTG | DMPK | 34 | 0 | 35 | 2 | 72 | 13 | 35 | 1 | 307 | 11 |
| DRPLA | CAG | ATN1 | 31 | 5 | 67 | 2 | 113 | 19 | 75 | 1 | 455 | 3 |
| FRAXA | CGG | FMR1 | 69 | 0 | 16 | 1 | 98 | 0 | 9 | 0 | 35 | 0 |
| FRAXE | CCG | AFF2 | 10 | 8 | 2 | 4 | 1242 | 9 | 2 | 4 | 9 | 7 |
| FRDA | GAA | FXN | 16 | 0 | 16 | 1 | 25 | 0 | 13 | 0 | 32 | 2 |
| HD | CAG | HTT | 569 | 6 | 223 | 10 | 879 | 15 | 211 | 2 | 672 | 10 |
| HDL2 | CTG | JPH3 | 5 | 41 | 2 | 15 | 16 | 92 | 0 | 9 | 8 | 31 |
| SBMA | CAG | AR | 7 | 1 | 3 | 0 | 1 | 0 | 1 | 0 | 91 | 2 |
| SCA1 | CAG | ATXN1 | 758 | 15 | 107 | 8 | 212 | 12 | 6 | 2 | 1458 | 22 |
| SCA2 | CAG | ATXN2 | 97 | 5 | 60 | 1 | 235 | 1 | 42 | 1 | 325 | 5 |
| SCA3 | CAG | ATXN3 | 36 | 2 | 24 | 4 | 77 | 6 | 3 | 2 | 37 | 6 |
| SCA6 | CAG | CACNA1A | 9 | 11 | 4 | 8 | 11 | 14 | 11 | 2 | 207 | 13 |
| SCA7 | CAG | ATXN7 | 380 | 9 | 107 | 2 | 473 | 11 | 26 | 3 | 327 | 9 |
| SCA8 | CTG | C10orf2 | 11 | 0 | 41 | 2 | 19 | 1 | 16 | 0 | 8 | 1 |
| SCA12 | CAG | PPP2R2B | 232 | 13 | 8 | 4 | 11 | 9 | 2 | 2 | 11 | 7 |
| SCA17 | CAG | TBP | 14 | 1 | 5 | 0 | 30 | 1 | 4 | 0 | 13 | 0 |
Data compiled from59. In these experiments, RNA was isolated from 6 different cell lines and subjected to deamination to differentiate between sense and antisense strands. cDNAs generated from the deaminated transcripts were deep sequenced59. The numbers reported in the table refer to the number of different sequencing reads that mapped to a given transcript. We highlighted the cases where the number of antisense sequences found for a gene amounts to at least 10% of the number of sense sequencing reads for the same gene. Data for the Jurkat and MRC5 cell lines have been summed from duplicate experiments. Note that in the case of HDL2 antisense transcripts were found to be more abundant than the sense transcript in all cell lines tested.
PBMCs are primary peripheral blood mononuclear cells; the HCT116 is a colorectal cancer cell line; Jurkat 1 is T cell leukemia cell line; MaPac is a pancreatic cancer line; MRC51 is a lung fibroblast cell line.
S is the number of sense sequences that map to a single gene.
AS is the number of antisense sequences that map to a single gene.
At the DM1 locus, the expanded CTG repeat tract is associated with methylation of a CCCTC-binding factor (CTCF) binding site, which appears to insulate the repeat from antisense transcription19. When the CTCF site is methylated, however, CTCF cannot bind, allowing enhanced antisense transcription through the expanded repeat19. Thus, paradoxically, heterochromatinization allows for higher levels of antisense transcription through the repeat, which could promote instability via TC-NER (Figure 2iii).
Libby et al investigated the role of the CTCF binding site in triplet repeat instability in a mouse model for SCA710. In these mice, CTCF binding sites flank the 92 CAG repeats in the transgene. Mutation of the downstream CTCF site dramatically increased instability in the germline. Furthermore, the kidney, cortex, brainstem, and liver of mice mutant for the CTCF binding site also displayed a higher level of CAG instability compared to mice with a wild type binding site10. Intriguingly, one mouse that carried a wild type CTCF binding site showed elevated levels of instability only in its kidney. Further investigation showed an aberrantly methylated CTCF binding site in this tissue. DNA methyaltion prevented CTCF binding its target sequence in vitro. The authors concluded that CTCF binding, which is influenced by DNA methylation, contributes directly to preventing repeat instability. These results are akin to those for the expanded CTG repeat in DM1 cells, where methylation of the CTCF binding sites correlate with loss of CTCF binding and increased antisense transcription19, 21. In the SCA7 mice, however, the role of antisense transcription has not been analyzed. Taken together, these data provide a tantalizing link between CAG repeat instability, CG-rich sequences, DNA methylation, and perhaps antisense transcription through the locus Antisense transcription could promote repeat instability in other ways. Simultaneous sense and antisense transcription might lead to head-on collisions between converging RNA polymerases, which could amplify the problems that arise when a repeat is transcribed in a single direction (e.g. enhance formation of secondary structure, persistence of RNA/DNA hybrids, and increased gratuitous TC-NER) thereby elevating instability.
Antisense transcription could also generate double-stranded RNA, which might trigger the RNA interference (RNAi). RNAi is a specialized pathway that allows post transcriptional repression of specific mRNAs62. Double stranded RNAs, such as microRNAs, are processed by the ribonuclease Dicer into 21nucleotide fragments. Short RNAs are subsequently loaded in the RNA-induced silencing complex (RISC), which targets mRNAs for degradation or inhibits their translation62. In fission yeast, the RNAi pathway prevents the appearance of centromeric transcripts by facilitating formation and maintenance of centromeric heterochromatin62. Whether an analogous process operates in mammalian cells is not yet clear. Nonetheless, repeat-containing transcripts can be processed by Dicer, and enter the RNAi pathway22, 63, 64. Thus, the RNAi pathway could promote heterochromatin formation at disease loci22, which could impact instability.
RNAs perform a variety of other functions that might be relevant to repeat instability: they can serve as a template for DNA synthesis during DNA repair65, guide programmed chromosome rearrangements66, and help remove T-G mismatches generated during reprogramming67. One of these functions might ultimately be found to play a role in repeat instability.
Concluding Remarks
The instability of trinucleotide repeats presents a surprisingly complex puzzle. One key point of agreement is that instability follows from the capacity of unstable repeats to form secondary structures, which in turn engage a variety of DNA repair activities in an attempt to regenerate a normal Watson-Crick duplex. In the past decade, most of the effort geared towards elucidating the mechanisms of triplet repeat instability has focused on knocking out or knocking down individual candidate genes and observing the resultant instability phenotype. A high fraction of tested genes can modulate triplet repeat instability, including elements of the replication machinery, DNA damage checkpoint components, and proteins involved in MMR, NER, BER, recombination, and transcription4, 57.
The chromatin environment in which repeat tracts are embedded adds yet another layer of complexity. The distinctive heterochromatic structure at expanded repeats might prevent or promote instability depending on the developmental stage and nearby cis elements. The well characterized links between chromatin structure and DNA metabolism require an expansion of the discussion of the mechanisms of triplet repeat instability to include the possible roles of epigenetics and chromatin.
In the future, it will be important to catalogue the changes in epigenetic marks that occur at expanded repeat tracts during development. To move beyond correlation to causation will undoubtedly prove challenging. In addition, a better understanding of the relationship between chromatin structure at repeat tracts and repeat instability might guide the design of new therapeutic approaches. For example, histone deacetylase inhibitors can reactivate gene expression at the FMR1 and FRDA loci68, 69. It will be interesting to test whether these molecules also alter the stability of repeat tracts. Drugs that can preferentially induce repeat contractions could lead to permanent improvements to patient health.
Acknowledgments
We thank the members of the Wilson laboratory for helpful discussion, Susan Gasser for support and F. Hamaratoğlu, H. Ferreira, S. Kueng, B. Pike, B. Towbin, M. Bühler, R. Waterland, and T. Punga for critical reading of the manuscript. V.D. is currently supported by a post-doctoral fellowship from the Terry Fox Foundation through The Canadian Cancer Society Research Institute. Work on triplet repeats in the Wilson laboratory is supported by NIH grant GM38219.
References
- 1.Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 2.La Spada AR, et al. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 3.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 4.Pearson CE, et al. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 5.Libby RT, et al. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum Mol Genet. 2003;12:41–50. doi: 10.1093/hmg/ddg006. [DOI] [PubMed] [Google Scholar]
- 6.Greene E, et al. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, et al. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 8.Brock GJ, et al. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum Mol Genet. 1999;8:1061–1067. doi: 10.1093/hmg/8.6.1061. [DOI] [PubMed] [Google Scholar]
- 9.Dion V, et al. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum Mol Genet. 2008;17:1306–1317. doi: 10.1093/hmg/ddn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Libby RT, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soragni E, et al. Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;36:6056–6065. doi: 10.1093/nar/gkn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Mahdawi S, et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 13.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 14.Sutcliffe JS, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 15.Oberle I, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 16.Hornstra IK, et al. High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Hum Mol Genet. 1993;2:1659–1665. doi: 10.1093/hmg/2.10.1659. [DOI] [PubMed] [Google Scholar]
- 17.Coffee B, et al. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am J Hum Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffee B, et al. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 19.Filippova GN, et al. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 20.Steinbach P, et al. The DMPK gene of severely affected myotonic dystrophy patients is hypermethylated proximal to the largely expanded CTG repeat. Am J Hum Genet. 1998;62:278–285. doi: 10.1086/301711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho DH, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Kumari D, Usdin K. Chromatin remodeling in the noncoding repeat expansion diseases. J Biol Chem. 2009;284:7413–7417. doi: 10.1074/jbc.R800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 24.Genc B, et al. Methylation mosaicism of 5′-(CGG)(n)-3′ repeats in fragile X, premutation and normal individuals. Nucleic Acids Res. 2000;28:2141–2152. doi: 10.1093/nar/28.10.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell MV, et al. Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell. 1991;64:861–866. doi: 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- 26.Saveliev A, et al. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 27.Devys D, et al. Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. Am J Med Genet. 1992;43:208–216. doi: 10.1002/ajmg.1320430134. [DOI] [PubMed] [Google Scholar]
- 28.Zeesman S, et al. Paternal transmission of fragile X syndrome. Am J Med Genet A. 2004;129A:184–189. doi: 10.1002/ajmg.a.30191. [DOI] [PubMed] [Google Scholar]
- 29.Helderman-van den Enden AT, et al. Monozygotic twin brothers with the fragile X syndrome: different CGG repeats and different mental capacities. J Med Genet. 1999;36:253–257. [PMC free article] [PubMed] [Google Scholar]
- 30.Savouret C, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. Embo J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 32.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 33.Gehring M, et al. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Gorbunova V, et al. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol Cell Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorbunova V, et al. Genome-wide demethylation destabilizes CTG.CAG trinucleotide repeats in mammalian cells. Hum Mol Genet. 2004;13:2979–2989. doi: 10.1093/hmg/ddh317. [DOI] [PubMed] [Google Scholar]
- 36.Dion V, et al. Genome-wide demethylation promotes triplet repeat instability independently of homologous recombination. DNA Repair (Amst) 2008;7:313–320. doi: 10.1016/j.dnarep.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichol K, Pearson CE. CpG methylation modifies the genetic stability of cloned repeat sequences. Genome Res. 2002;12:1246–1256. doi: 10.1101/gr.74502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan HD, et al. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 39.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 41.Panigrahi GB, et al. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 42.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malter HE, et al. Characterization of the full fragile X syndrome mutation in fetal gametes. Nat Genet. 1997;15:165–169. doi: 10.1038/ng0297-165. [DOI] [PubMed] [Google Scholar]
- 44.Yoon SR, et al. Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc Natl Acad Sci U S A. 2003;100:8834–8838. doi: 10.1073/pnas.1331390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savouret C, et al. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol Cell Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 47.Kaytor MD, et al. Increased trinucleotide repeat instability with advanced maternal age. Hum Mol Genet. 1997;6:2135–2139. doi: 10.1093/hmg/6.12.2135. [DOI] [PubMed] [Google Scholar]
- 48.Watase K, et al. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Hum Mol Genet. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- 49.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reyniers E, et al. Postmortem examination of two fragile X brothers with an FMR1 full mutation. Am J Med Genet. 1999;84:245–249. doi: 10.1002/(sici)1096-8628(19990528)84:3<245::aid-ajmg16>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 51.Tassone F, et al. Strong similarities of the FMR1 mutation in multiple tissues: postmortem studies of a male with a full mutation and a male carrier of a premutation. Am J Med Genet. 1999;84:240–244. [PubMed] [Google Scholar]
- 52.Taylor AK, et al. Tissue heterogeneity of the FMR1 mutation in a high-functioning male with fragile X syndrome. Am J Med Genet. 1999;84:233–239. [PubMed] [Google Scholar]
- 53.Wohrle D, et al. Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet. 1998;35:103–111. doi: 10.1136/jmg.35.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glaser D, et al. Mitotic behavior of expanded CGG repeats studied on cultured cells: further evidence for methylation-mediated triplet repeat stability in fragile X syndrome. Am J Med Genet. 1999;84:226–228. [PubMed] [Google Scholar]
- 55.Salat U, et al. Increase of FMRP expression, raised levels of FMR1 mRNA, and clonal selection in proliferating cells with unmethylated fragile X repeat expansions: a clue to the sex bias in the transmission of full mutations? J Med Genet. 2000;37:842–850. doi: 10.1136/jmg.37.11.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burman RW, et al. Fully expanded FMR1 CGG repeats exhibit a length- and differentiation-dependent instability in cell hybrids that is independent of DNA methylation. Hum Mol Genet. 1999;8:2293–2302. doi: 10.1093/hmg/8.12.2293. [DOI] [PubMed] [Google Scholar]
- 57.Lin Y, et al. Transcription destabilizes triplet repeats. Mol Carcinog. 2009;48:350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y, et al. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ladd PD, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 61.Moseley ML, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 62.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krol J, et al. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 64.Handa V, et al. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Storici F, et al. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nowacki M, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boland MJ, Christman JK. Characterization of Dnmt3b:thymine-DNA glycosylase interaction and stimulation of thymine glycosylase-mediated repair by DNA methyltransferase(s) and RNA. J. Mol. Biol. 2008;379:492–504. doi: 10.1016/j.jmb.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gottesfeld JM. Small molecules affecting transcription in Friedreich ataxia. Pharmacol Ther. 2007;116:236–248. doi: 10.1016/j.pharmthera.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biacsi R, et al. SIRT1 inhibition alleviates gene silencing in Fragile X mental retardation syndrome. PLoS Genet. 2008;4:e1000017. doi: 10.1371/journal.pgen.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reid G, et al. Marking time: the dynamic role of chromatin and covalent modification in transcription. Int J Biochem Cell Biol. 2009;41:155–163. doi: 10.1016/j.biocel.2008.08.028. [DOI] [PubMed] [Google Scholar]
- 71.Reed SH. Nucleotide excision repair in chromatin: the shape of things to come. DNA Repair (Amst) 2005;4:909–918. doi: 10.1016/j.dnarep.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 72.Jagannathan I, et al. Base excision repair in nucleosome substrates. Chromosome Res. 2006;14:27–37. doi: 10.1007/s10577-005-1020-7. [DOI] [PubMed] [Google Scholar]
- 73.Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 74.Goodarzi AA, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 75.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37:1363–1377. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manley K, et al. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 77.van den Broek WJ, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 78.Wheeler VC, et al. Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 79.Foiry L, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 80.Gomes-Pereira M, et al. Pms2 is a genetic enhancer of trinucleotide CAG{middle dot}CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 81.Entezam A, Usdin K. ATR protects the genome against CGG. Fragile X premutation mice. Nucleic Acids Res. 2008;36:1050–1056. doi: 10.1093/nar/gkm1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dragileva E, et al. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao R, et al. Instability of expanded CAG/CAA repeats in spinocerebellar ataxia type 17. Eur J Hum Genet. 2008;16:215–222. doi: 10.1038/sj.ejhg.5201954. [DOI] [PubMed] [Google Scholar]
- 84.Winnepenninckx B, et al. CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am J Hum Genet. 2007;80:221–231. doi: 10.1086/510800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stoger R, et al. Epigenetic variation illustrated by DNA methylation patterns of the fragile-X gene FMR1. Hum Mol Genet. 1997;6:1791–1801. doi: 10.1093/hmg/6.11.1791. [DOI] [PubMed] [Google Scholar]
- 86.Hansen RS, et al. Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum Mol Genet. 1992;1:571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- 87.Biancalana V, et al. Expansion and methylation status at FRAXE can be detected on EcoRI blots used for FRAXA diagnosis: analysis of four FRAXE families with mild mental retardation in males. Am J Hum Genet. 1996;59:847–854. [PMC free article] [PubMed] [Google Scholar]
- 88.Knight SJ, et al. Triplet repeat expansion at the FRAXE locus and X-linked mild mental handicap. Am J Hum Genet. 1994;55:81–86. [PMC free article] [PubMed] [Google Scholar]
- 89.Lo Nigro C, et al. FRAXE mutation in a mentally retarded subject and in his phenotypically normal twin brother. Eur J Hum Genet. 2000;8:157–162. doi: 10.1038/sj.ejhg.5200425. [DOI] [PubMed] [Google Scholar]
- 90.Mila M, et al. Screening for FMR1 and FMR2 mutations in 222 individuals from Spanish special schools: identification of a case of FRAXE-associated mental retardation. Hum Genet. 1997;100:503–507. doi: 10.1007/s004390050542. [DOI] [PubMed] [Google Scholar]
- 91.Gecz J, et al. FMR2 expression in families with FRAXE mental retardation. Hum Mol Genet. 1997;6:435–441. doi: 10.1093/hmg/6.3.435. [DOI] [PubMed] [Google Scholar]
- 92.Knight SJ, et al. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell. 1993;74:127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- 93.Otten AD, Tapscott SJ. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proc Natl Acad Sci U S A. 1995;92:5465–5469. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herman D, et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 95.Rai M, et al. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]