Abstract
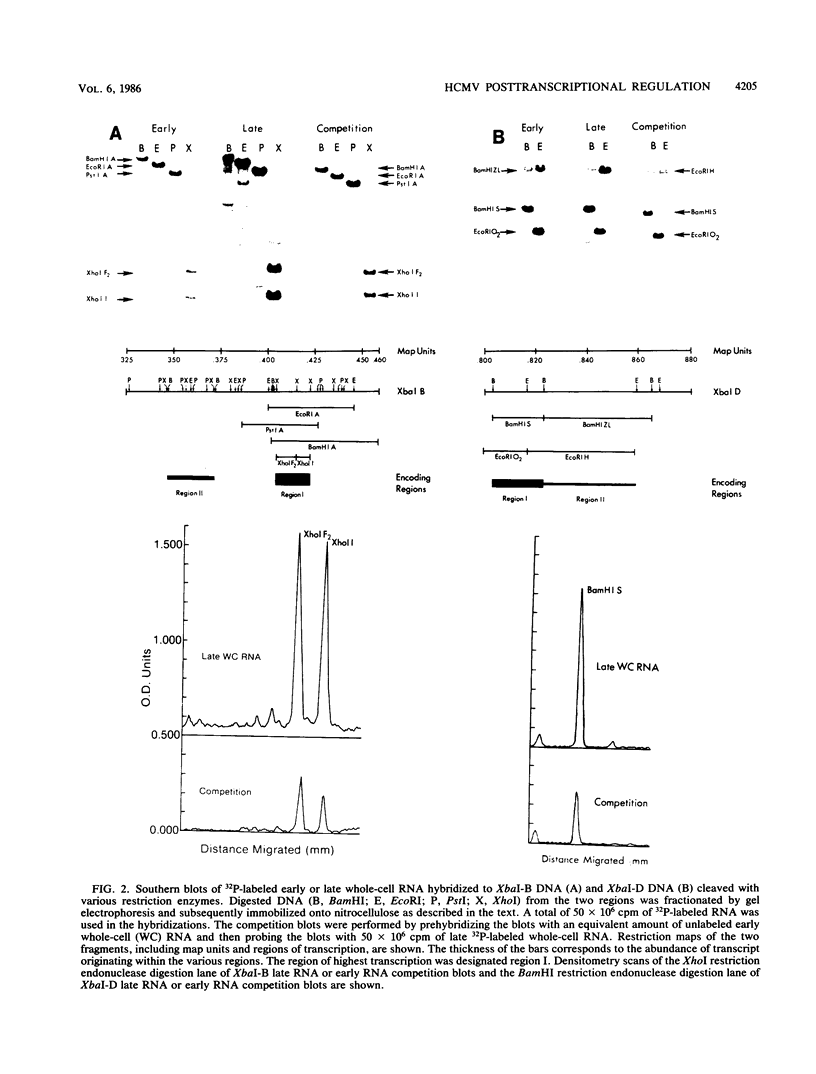
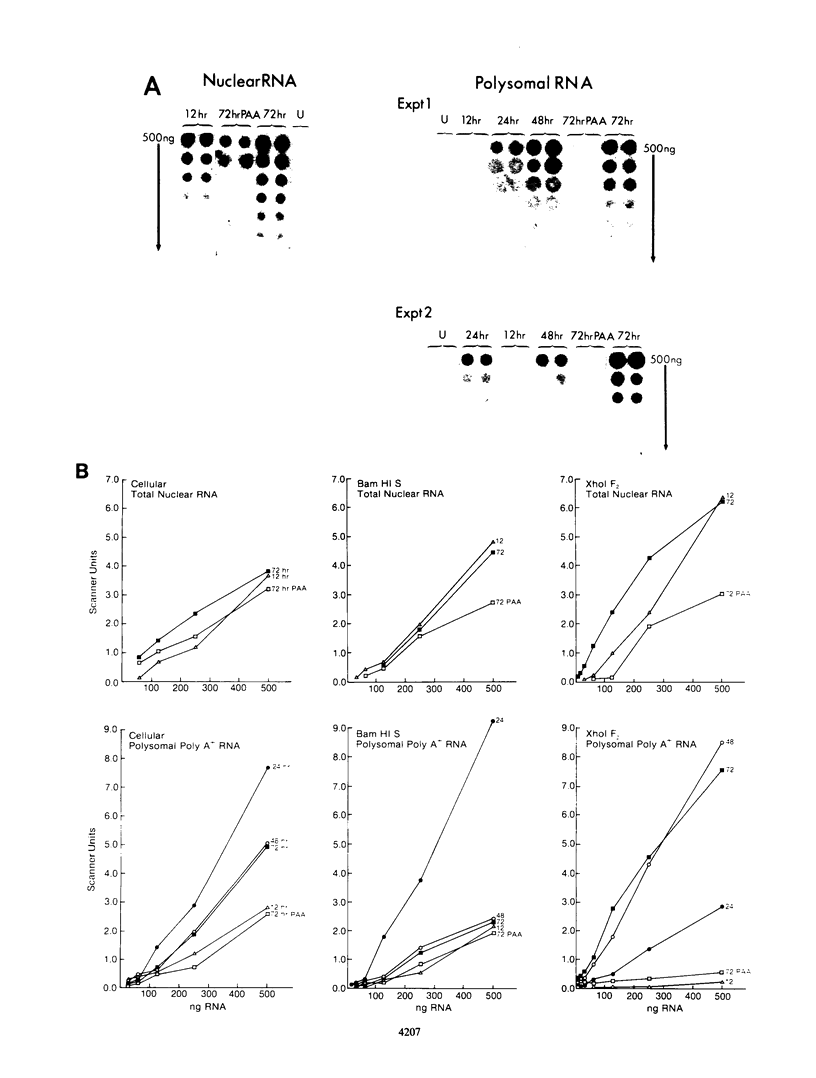
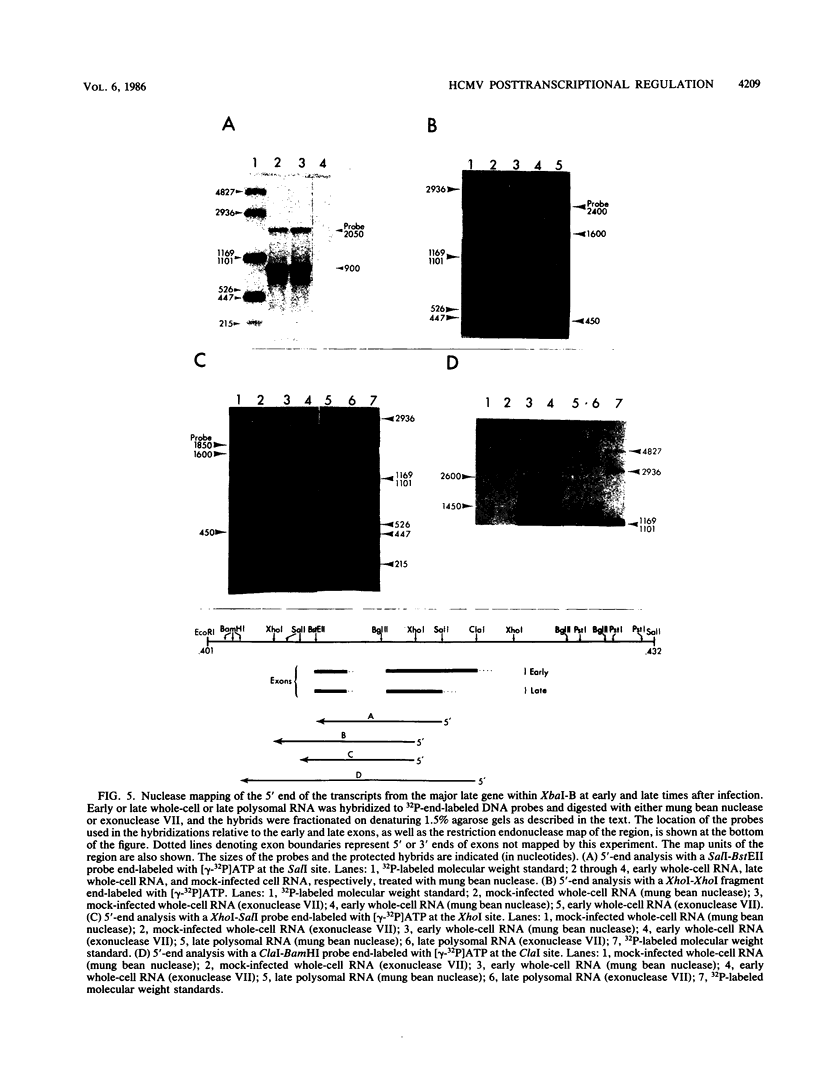
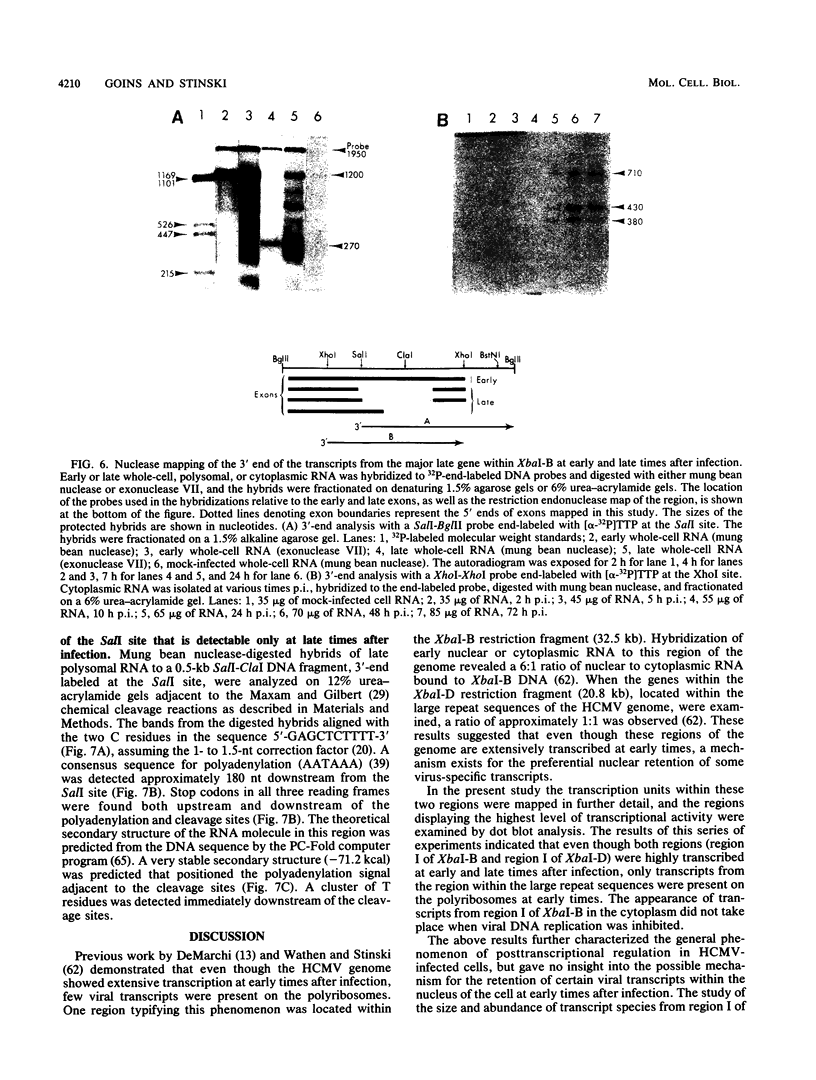
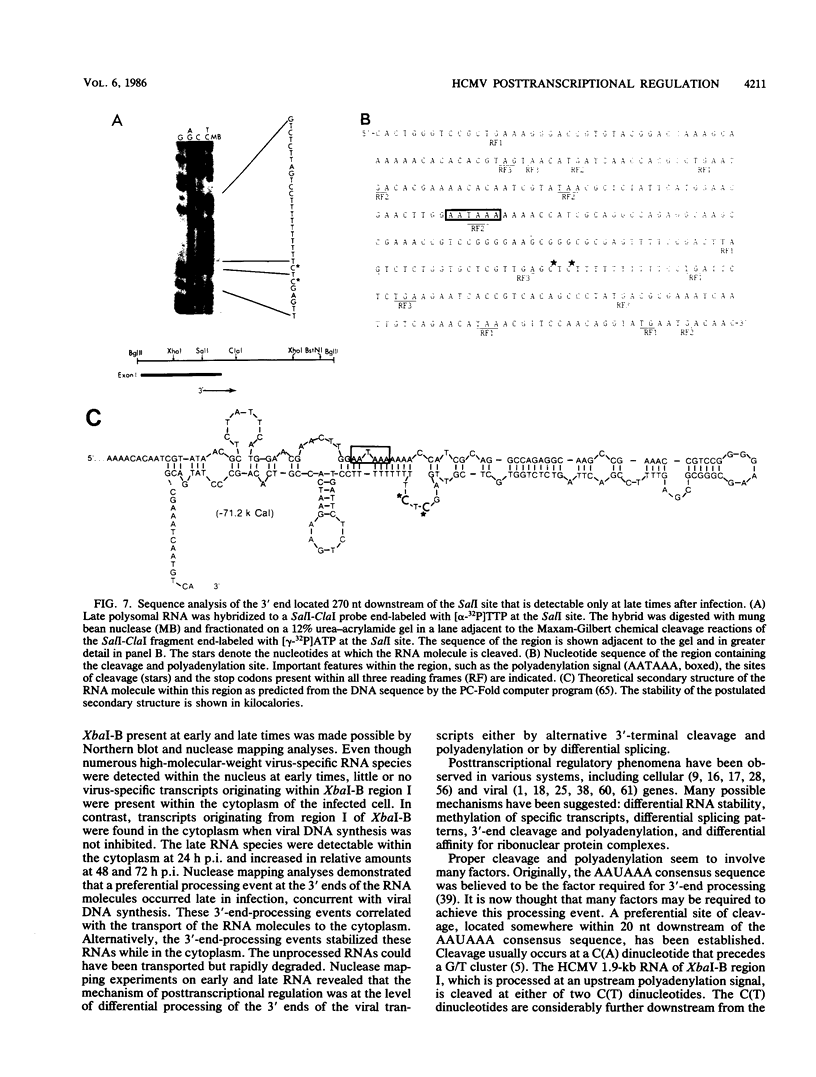
A phenomenon of posttranscriptional regulation has been previously identified in cytomegalovirus-infected human fibroblast cells (Wathen and Stinski, J. Virol. 41:462, 1982). A region typifying this phenomenon has been located within the large unique component of the viral genome (map units 0.408 to 0.423). Even though this transcriptional unit was highly transcribed at early times after infection, mRNAs from this region were only detectable on the polyribosomes after viral DNA replication. Thus, this region is believed to code for a late gene. Single-strand-specific nuclease mapping experiments of viral transcripts established that the transcriptional initiation sites and the 5' ends of a downstream exon were identical at early and late times. However, the late transcripts differed from the early transcripts by the processing of the 3' end of the viral RNAs. This involved either the removal of a distinct region of the transcript by the selection of an upstream cleavage and polyadenylation site or the differential splicing of the RNA molecule. The upstream cleavage and polyadenylation site was identified by nuclease mapping analyses and DNA sequencing. The 3'-end processing of these transcripts is necessary for the detection of these viral RNAs within the cytoplasm of the infected cell. We propose that human cytomegalovirus either codes for a factor(s) that is involved in the 3'-end-processing event at late times after infection or stimulates the synthesis of a host cell factor(s) involved in this complex regulatory event. This level of regulation may have an influence on the types of cells that permit productive cytomegalovirus replication.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babiss L. E., Ginsberg H. S., Darnell J. E., Jr Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985 Oct;5(10):2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Boldogh I., Beth E., Huang E. S., Kyalwazi S. K., Giraldo G. Kaposi's sarcoma. IV. Detection of CMV DNA, CMV RNA and CMNA in tumor biopsies. Int J Cancer. 1981 Oct 15;28(4):469–474. doi: 10.1002/ijc.2910280412. [DOI] [PubMed] [Google Scholar]
- Bächi B., Arber W. Physical mapping of BglII, BamHI, EcoRI, HindIII and PstI restriction fragments of bacteriophage P1 DNA. Mol Gen Genet. 1977 Jun 24;153(3):311–324. doi: 10.1007/BF00431596. [DOI] [PubMed] [Google Scholar]
- Chua C. C., Carter T. H., St Jeor S. Transcription of the human cytomegalovirus genome in productively infected cells. J Gen Virol. 1981 Sep;56(Pt 1):1–11. doi: 10.1099/0022-1317-56-1-1. [DOI] [PubMed] [Google Scholar]
- Clanton D. J., Jariwalla R. J., Kress C., Rosenthal L. J. Neoplastic transformation by a cloned human cytomegalovirus DNA fragment uniquely homologous to one of the transforming regions of herpes simplex virus type 2. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3826–3830. doi: 10.1073/pnas.80.12.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. F., Weiss M., Darnell J. E., Jr Liver-specific RNA metabolism in hepatoma cells: variations in transcription rates and mRNA levels. Mol Cell Biol. 1985 Oct;5(10):2633–2641. doi: 10.1128/mcb.5.10.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Raskas H. J. Effect of cycloheximide on RNA metabolism early in productive infection with adenovirus 2. J Virol. 1974 Jul;14(1):26–32. doi: 10.1128/jvi.14.1.26-32.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. G., Huang E. S. Nucleotide sequence of a human cytomegalovirus DNA fragment encoding a 67-kilodalton phosphorylated viral protein. J Virol. 1985 Oct;56(1):7–11. doi: 10.1128/jvi.56.1.7-11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarchi J. M., Blankenship M. L., Brown G. D., Kaplan A. S. Size and complexity of human cytomegalovirus DNA. Virology. 1978 Sep;89(2):643–646. doi: 10.1016/0042-6822(78)90209-x. [DOI] [PubMed] [Google Scholar]
- DeMarchi J. M. Post-transcriptional control of human cytomegalovirus gene expression. Virology. 1983 Jan 30;124(2):390–402. doi: 10.1016/0042-6822(83)90355-0. [DOI] [PubMed] [Google Scholar]
- DeMarchi J. M. Post-transcriptional control of human cytomegalovirus gene expression. Virology. 1983 Jan 30;124(2):390–402. doi: 10.1016/0042-6822(83)90355-0. [DOI] [PubMed] [Google Scholar]
- DeMarchi J. M., Schmidt C. A., Kaplan A. S. Patterns of transcription of human cytomegalovirus in permissively infected cells. J Virol. 1980 Aug;35(2):277–286. doi: 10.1128/jvi.35.2.277-286.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi H., Takahashi Y., Sekiya T., Sato S., Nagase S., Sugimura T. Presence of albumin mRNA precursors in nuclei of analbuminemic rat liver lacking cytoplasmic albumin mRNA. Proc Natl Acad Sci U S A. 1982 Feb;79(3):734–738. doi: 10.1073/pnas.79.3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag S. O., Beaudet A. L., Bock H. G., O'Brien W. E. Molecular structure of the human argininosuccinate synthetase gene: occurrence of alternative mRNA splicing. Mol Cell Biol. 1984 Oct;4(10):1978–1984. doi: 10.1128/mcb.4.10.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe A. P., Leach F. S., Mocarski E. S. Regulation of cytomegalovirus late gene expression: gamma genes are controlled by posttranscriptional events. J Virol. 1986 Mar;57(3):864–874. doi: 10.1128/jvi.57.3.864-874.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Green M. R., Roeder R. G. Definition of a novel promoter for the major adenovirus-associated virus mRNA. Cell. 1980 Nov;22(1 Pt 1):231–242. doi: 10.1016/0092-8674(80)90171-3. [DOI] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Nevins J. R. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985 Dec;43(3 Pt 2):677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974 Jul;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E. S., Chen S. T., Pagano J. S. Human cytomegalovirus. I. Purification and characterization of viral DNA. J Virol. 1973 Dec;12(6):1473–1481. doi: 10.1128/jvi.12.6.1473-1481.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn G., Knust E., Schmolla H., Sarre T., Nelson J. A., McDougall J. K., Fleckenstein B. Predominant immediate-early transcripts of human cytomegalovirus AD 169. J Virol. 1984 Feb;49(2):363–370. doi: 10.1128/jvi.49.2.363-370.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeor S. C., Albrecht T. B., Funk F. D., Rapp F. Stimulation of cellular DNA synthesis by human cytomegalovirus. J Virol. 1974 Feb;13(2):353–362. doi: 10.1128/jvi.13.2.353-362.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M., Roizman B. Regulation of herpesvirus macromolecular synthesis: nuclear retention of nontranslated viral RNA sequences. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4322–4326. doi: 10.1073/pnas.71.11.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Leys E. J., Kellems R. E. Control of dihydrofolate reductase messenger ribonucleic acid production. Mol Cell Biol. 1981 Nov;1(11):961–971. doi: 10.1128/mcb.1.11.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McDonough S. H., Spector D. H. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology. 1983 Feb;125(1):31–46. doi: 10.1016/0042-6822(83)90061-2. [DOI] [PubMed] [Google Scholar]
- McDonough S. H., Staprans S. I., Spector D. H. Analysis of the major transcripts encoded by the long repeat of human cytomegalovirus strain AD169. J Virol. 1985 Mar;53(3):711–718. doi: 10.1128/jvi.53.3.711-718.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Clements J. B. A 3' co-terminus of two early herpes simplex virus type 1 mRNAs. Nucleic Acids Res. 1982 Jan 22;10(2):501–512. doi: 10.1093/nar/10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. A., Fleckenstein B., Galloway D. A., McDougall J. K. Transformation of NIH 3T3 cells with cloned fragments of human cytomegalovirus strain AD169. J Virol. 1982 Jul;43(1):83–91. doi: 10.1128/jvi.43.1.83-91.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. A., Fleckenstein B., Jahn G., Galloway D. A., McDougall J. K. Structure of the transforming region of human cytomegalovirus AD169. J Virol. 1984 Jan;49(1):109–115. doi: 10.1128/jvi.49.1.109-115.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilder S., Moore M., Logan J., Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986 Feb;6(2):470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Connelly S., Manley J. L., Alwine J. C. Identification of a sequence element on the 3' side of AAUAAA which is necessary for simian virus 40 late mRNA 3'-end processing. Mol Cell Biol. 1985 Oct;5(10):2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiki M., Hattori S., Hirayama Y., Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Faye G. Steps in processing of the mitochondrial cytochrome oxidase subunit I pre-mRNA affected by a nuclear mutation in yeast. Proc Natl Acad Sci U S A. 1984 Jan;81(1):8–12. doi: 10.1073/pnas.81.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. D., De Harven E. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. I. Sequence of viral replication. J Virol. 1973 Oct;12(4):919–930. doi: 10.1128/jvi.12.4.919-930.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stenberg R. M., Thomsen D. R., Stinski M. F. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol. 1984 Jan;49(1):190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg R. M., Witte P. R., Stinski M. F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985 Dec;56(3):665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M. F., Roehr T. J. Activation of the major immediate early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J Virol. 1985 Aug;55(2):431–441. doi: 10.1128/jvi.55.2.431-441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M. F. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J Virol. 1978 Jun;26(3):686–701. doi: 10.1128/jvi.26.3.686-701.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M. F. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol. 1977 Sep;23(3):751–767. doi: 10.1128/jvi.23.3.751-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M. F., Thomsen D. R., Rodriguez J. E. Synthesis of human cytomegalovirus-specified RNA and protein in interferon-treated cells at early times after infection. J Gen Virol. 1982 Jun;60(Pt 2):261–270. doi: 10.1099/0022-1317-60-2-261. [DOI] [PubMed] [Google Scholar]
- Stinski M. F., Thomsen D. R., Stenberg R. M., Goldstein L. C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983 Apr;46(1):1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Furukawa T., Plotkin S. A. Human cytomegalovirus stimulates host cell RNA synthesis. J Virol. 1975 Feb;15(2):297–304. doi: 10.1128/jvi.15.2.297-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therwath A., Scherrer K. Post-transcriptional suppression of globin gene expression in cells transformed by avian erythroblastosis virus. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3776–3780. doi: 10.1073/pnas.75.8.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen D. R., Stinski M. F. Cloning of the human cytomegalovirus genome as endonuclease XbaI fragments. Gene. 1981 Dec;16(1-3):207–216. doi: 10.1016/0378-1119(81)90077-9. [DOI] [PubMed] [Google Scholar]
- Vannice J. L., Taylor J. M., Ringold G. M. Glucocorticoid-mediated induction of alpha 1-acid glycoprotein: evidence for hormone-regulated RNA processing. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4241–4245. doi: 10.1073/pnas.81.14.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal L. P., Carr S. Genetic test for involvement of intervening sequences in transport of nuclear RNA. Mol Cell Biol. 1982 Dec;2(12):1550–1557. doi: 10.1128/mcb.2.12.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen A., Pettersson U. Organization of early region 1B of human adenovirus type 2: identification of four differentially spliced mRNAs. J Virol. 1985 May;54(2):383–391. doi: 10.1128/jvi.54.2.383-391.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathen M. W., Stinski M. F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982 Feb;41(2):462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathen M. W., Thomsen D. R., Stinski M. F. Temporal regulation of human cytomegalovirus transcription at immediate early and early times after infection. J Virol. 1981 May;38(2):446–459. doi: 10.1128/jvi.38.2.446-459.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. A., Bancroft F. C. Cytoplasmic dot hybridization. Simple analysis of relative mRNA levels in multiple small cell or tissue samples. J Biol Chem. 1982 Aug 10;257(15):8569–8572. [PubMed] [Google Scholar]