Abstract
Background
Radiotherapy is used routinely to treat testicular cancer. Testicular cells vary in radio-sensitivity and the aim of this study was to investigate cellular and molecular changes caused by low dose irradiation of mice testis and to identify transcripts from different cell types in the adult testis.
Methods
Transcriptome profiling was performed on total RNA from testes sampled at various time points (n = 17) after 1 Gy of irradiation. Transcripts displaying large overall expression changes during the time series, but small expression changes between neighbouring time points were selected for further analysis. These transcripts were separated into clusters and their cellular origin was determined. Immunohistochemistry and in silico quantification was further used to study cellular changes post-irradiation (pi).
Results
We identified a subset of transcripts (n = 988) where changes in expression pi can be explained by changes in cellularity. We separated the transcripts into five unique clusters that we associated with spermatogonia, spermatocytes, early spermatids, late spermatids and somatic cells, respectively. Transcripts in the somatic cell cluster showed large changes in expression pi, mainly caused by changes in cellularity. Further investigations revealed that the low dose irradiation seemed to cause Leydig cell hyperplasia, which contributed to the detected expression changes in the somatic cell cluster.
Conclusions
The five clusters represent gene expression in distinct cell types of the adult testis. We observed large expression changes in the somatic cell profile, which mainly could be attributed to changes in cellularity, but hyperplasia of Leydig cells may also play a role. We speculate that the possible hyperplasia may be caused by lower testosterone production and inadequate inhibin signalling due to missing germ cells.
Keywords: Mice, Testis, Irradiation, Gene expression, Transcriptomics, Microarray, Clustering, Leydig cell, Hyperplasia
Background
Radiotherapy is used routinely to treat testicular cancer. Cells in the testis display different radio-sensitivity and the degree of radiation-induced damage depends on dose and fractionation of the radiation [1]. Some cells are very sensitive and undergo apoptosis after low dose irradiation, others are only partially affected and presumably recover after a period of time, while yet others seem completely unaffected. Spermatogonial stem cells (SSCs) are the most radio-resistant germ cells, probably due to their slow turnover. Contrarily, A spermatogonia have the highest turnover and are the germ cells most susceptible to irradiation-induced damage [2,3] followed by Intermediate and B spermatogonia [4,5]. The more differentiated germ cells such as spermatocytes and spermatids seem unaffected by low dose irradiation and continue their maturation toward spermatozoa [1].
The somatic cells in the testis are also to some extent affected by irradiation. Human Leydig cells are damaged by the irradiation doses used to treat testicular cancer patients (16–20 Gy) [1,6,7], although in many cases the testosterone production is maintained. Rodent Leydig cells have shown dysfunction after high dose irradiation (5 Gy) leading to low testosterone and increased serum luteinizing hormone (LH) levels [8]. Similarly, it has also been reported that rodent Sertoli cells are affected by high dose irradiation (5 Gy) [9].
Low dose irradiation of adult rodent testes kills A through B spermatogonia, which creates a “gap” in the germinal epithelium [10]. The gap moves through the differentiation stages of the germ cells as spermatogenesis continues and SSCs re-populate the testis. The cellular and molecular changes caused by the movement of the gap can be used to study gene expression in adult spermatogenesis. However, by studying the whole testis we investigate changes in the relative gene expression, i.e. the contribution from each cell type to the total RNA pool, which is determined by the concentration of the RNA in the specific cell type and the percentage of the total volume that the cell type occupies - the cellularity [11].
In this study, we identified five clusters of cell type-specific transcripts by transcriptome profiling. The transcripts in these clusters all had gene expression profiles that could be explained by changes in the cellularity during recovery from irradiation. We observed large expression changes in the somatic cell cluster and further investigations by immunohistochemistry (IHC) revealed that low dose irradiation seemed to cause hyperplasia of the Leydig cells, whereas peritubular myoid (PTM) and Sertoli cells seemed unaffected.
Methods
Mice testis preparation
Treatment of mice and preparation of testicular RNA and sections were performed as in Shah et al., [10]. In brief, male C3H/He mice from Japan SLC (Shizuoka, Japan) were maintained under controlled conditions (22 ± 2°C, 55 ± 5% humidity, 12 h light/dark cycle, lights on 0600 h) with laboratory chow (CE-2, Japan Crea, Tokyo, Japan) and water ad libitum. Eleven-week old mice were anesthetised with pentobarbital and covered with lead sheeting except for the scrotum. The testes were locally exposed to X-ray irradiation with 1 Gy. Testes from one to four mice were sampled and weighted regularly during recovery on days 0, 3, 7, 10, 14, 17, 21, 24, 28, 31, 35, 38, 42, 45, 48, 52, 56, and 59. One testis was fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, overnight at 4°C and subsequently dehydrated in graded series of ethanol and embedded in paraffin for IHC. The contralateral testis was snap-frozen in liquid nitrogen for later preparation of total RNA.
The Japanese Pharmacological Society approved the animal study. The animals were treated according to generally accepted guidelines for animal experimentation at St. Marianna University Graduate School of Medicine and guiding principles for the care and use of laboratory animals.
Gene expression microarrays
Total RNA was purified from whole testis samples collected pi day 3 to day 59 (n = 17) with NucleoSpin RNA II (Macherey-Nagel, Dueren, Germany) according to the manufacturer’s protocol. RNA quality was determined using Bioanalyzer nano kit (Agilent Technologies, Santa Clara, California, US). The samples were amplified (one round) using the MessageAmp II aRNA Amplification Kit (Applied Biosystems, Carlsbad, California, US) and the aRNA was applied to Agilent whole mouse genome 4 × 44K oligo microarrays. Hybridisation and scanning of the one-colour arrays were done as described by the manufacturer (Agilent Technologies, Santa Clara, California, US). The microarray data analysis was performed in the R software [12] where the gProcessedSignals were loaded into the limma R/Bioconductor package [13,14] and normalised between arrays using the quantile normalisation method [15]. The probes were collapsed for each systematic transcript ID by the median expression value.
Enrichment score
Transcripts with expression profiles potentially explained by changes in cellularity due to the recovery from irradiation were identified and selected for further analysis based on the enrichment score (ES) calculated as follows:
where GEi and GE(i + 1) is the expression value for a transcript at time point i and the neighbouring time point i + 1, respectively, and is the mean expression value for a transcript through the time series.
The false discovery rate (FDR) of the ES was calculated for each transcript based on ten calculations on shuffled time points. A subset of transcripts was chosen for further analysis all with a cumulative minimum of the FDR ≤ 30% and a standard deviation (SD) of the most extreme expression value in the time series ≤ 15%.
Cluster analysis
Cluster analyses were performed on the subset of transcripts selected as described above. A distance matrix was generated with the pair-wise correlation variances of the gene expression values during the time series centred around 1. The transcripts were clustered according to the distance matrix using partitioning around medoids (PAM) clustering, which was performed multiple times with different number of clusters. The number of clusters that separated most unique transcript clusters was chosen as the best separation.
Association of transcript clusters to specific testis cells
To determine the cellular origin of the transcript clusters, we compared their gene expression patterns to the expression profiles of cell markers determined in our previous study of the same testis samples where we used differential display and in situ hybridisation [10]. We also mapped testicular cell-specific markers to the clusters to confirm their cellular origin [16-18].
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed on the transcripts in each cluster separately using DAVID [19,20]. All genes represented on the array were used as background. The cut off for statistical significance was set to the Bonferroni corrected p-value ≤ 0.01.
Immunohistochemistry (IHC)
The following primary antibodies were used: Vimentin/HRP (Dako, Glostrup, Denmark; U7034), Transforming growth factor β receptor III (Tgfbr3) 1:75 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; sc-6199), 3β-hydroxysteroid-dehydrogenase (Hsd3b) 1:6000 (R1484 a gift from Prof. J. Ian Mason, Edinburgh), and Smooth muscle actin (SMA) 1:600 Abcam ab5694 (330 Cambridge Science Park, Cambridge CB4 OFL, UK).
Vimentin was used according to the manufacturer’s protocol. In short, the sections were deparaffinised, rehydrated and blocked for endogen peroxidase with H2O2, washed in tap water, placed 5 min in TBS (0.5 M Tris/HCl, 0.15 M NaCl, pH 7.6) at 37°C, incubated with 1:10 Trypsin (Gibson 15400) in TBS 15 min at 37°C, and exposed to the antibody for 1 h at room temperature. Development was performed with 3-amino-9-ethylcarbazole (AEC). Thorough washing with TBS was performed after each individual step and finally the sections were washed in water before a short staining with Meyers haematoxylin.
The three remaining antibodies, against Tgfbr3, Hsd3b and SMA, were used as in a protocol based on a Zymed histostain kit (Invitrogen, Carlsbad, CA, USA). In short, the sections were deparaffinised, rehydrated and blocked for endogen peroxidase as described above, followed by microwave treatment for 15 min in TEG buffer (10 mM Tris, 0.5 mM EGTA, pH 9.0) for Tgfbr3 and Hsd3b, whereas Citrate buffer (10 mM, pH 6) was used for SMA. Cross reactivity of the antibodies was minimized by treatment with 0.5% milk powder diluted in TBS. Sections were exposed to the primary antibodies over night at 5°C and 1 h at room temperature, then incubated with biotinylated goat anti-rabbit IgG or with biotinylated donkey anti-goat IgG 1:400 in TBS (The binding site Ltd., Birmingham, UK; AB360) for Tgfbr3 before exposure to a peroxidase-conjugated streptavidin complex. Finally, the sections were developed with AEC and counter stained with Meyers haematoxylin. Thorough washing with TBS was performed after each individual step. Slides without addition of primary antibodies were used as controls.
In silico quantification of Leydig cells
In order to obtain a measure of the amount of Leydig cells relative to other cells in the testis, we quantified the area covered by Leydig cells relative to the area covered by other testicular cells. Three different areas, each representing more than ten seminiferous tubules, from sections of the testis of representative days pi were stained with the Leydig cell marker Hsd3b, scanned and evaluated in silico with the VisioMorph add-in in the VIS program (Visiopharm A/S, Hørsholm, Denmark). A red colour was assigned to the red Hsd3b staining, blue to other cells (Meyers haematoxylin) and white to the background (see Additional file 1: Figure S1). A Baysian classification of the colour space from the stained sections was applied and subsequently red areas smaller than 20 μm2 assigned as blue. The areas of red, blue and white were measured and the ratio of red to blue used as a measure of the amount of Leydig cells relative to the amount of other testis cells.
Results
Clusters of transcripts affected by low dose irradiation
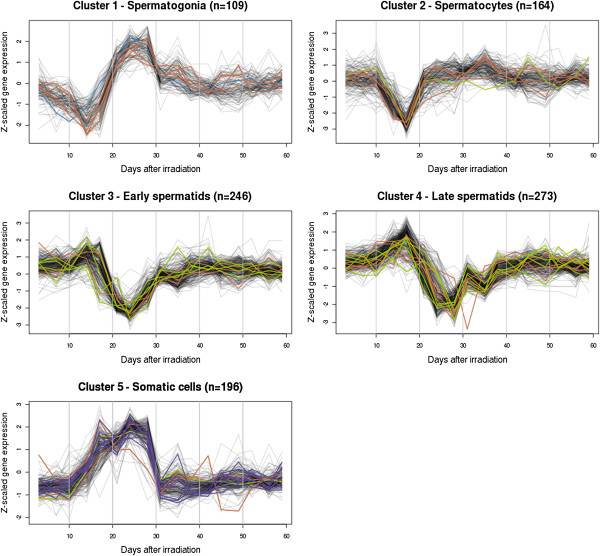
Gene expression profiling was performed on total RNA from mice testis (n = 17) from a time series following low dose irradiation (1 Gy). In total, 988 transcripts were selected for further analysis. These transcripts all had expression profiles during the time series that potentially were caused by the recovery from irradiation, i.e. transcripts with large overall expression changes during the time series, but small expression changes between neighbouring time points. We performed PAM clustering of the distance matrix of the pair-wise correlation variances and identified five clusters with unique expression patterns. This was the maximum number of clusters separable in this data set. The names of the transcripts in each cluster are available in additional material (see Additional file 2: Table S1, Additional file 3: Table S2, Additional file 4: Table S3, Additional file 5: Table S4, Additional file 6: Table S5).
Assignment of transcript clusters to specific testicular cells
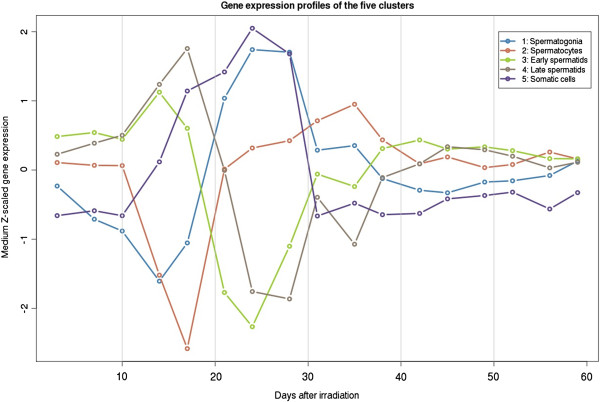
To identify the cellular origin of the clusters, we compared the expression patterns of the five clusters with the expression patterns of cell markers investigated in our previous study of the same testicular material [10]. We found that four of the clusters identified in this study independently had similar expression profiles as four cell-specific markers identified in our previous study. The four markers were in our previous study associated with spermatogonia (cluster 1 in the current study; n = 109), spermatocytes (cluster 2; n = 164), spermatids (cluster 3; n = 246) and Sertoli cells (cluster 5; n = 196). Since transcripts in the early spermatid cluster showed a decrease in expression with a trough already at pi day 24, we assigned the fifth novel cluster, with transcripts that showed a decrease with a trough in expression at pi day 27, to late spermatids (cluster 4; n = 273). The expression level of most transcripts returned to pre-irradiation levels around pi day 40 where a complete round of spermatogenesis had taken place [21].
To add further evidence of the association of cell types to the clusters, we identified previously published cell-specific markers in the five clusters [16-18]. In total, 39 markers were identified in the five clusters representing spermatogonia (n = 1), spermatocytes (n = 19), spermatids (n = 11), Leydig cells (n = 3), PTM cells (n = 1), and Sertoli cells (n = 4) (Table 1, Figure 1 and Figure 2). The single marker associated with spermatogonia was found in the corresponding cluster (cluster 1). The spermatocyte markers were evenly distributed in all five clusters, but comparing the expression patterns to the profiles identified earlier [10,16] strongly indicated that this cluster (cluster 2) with a reduced expression at pi day 17, mainly contained transcripts from spermatocytes. Most spermatid markers were present in the two clusters that we associated with early and late spermatids (cluster 3 and 4). The transcripts in the somatic cell cluster contained all somatic cell markers (cluster 5), which confirmed that the cluster represented all somatic cells and not only transcripts from Sertoli cells as suggested in our earlier study [10].
Table 1.
Testis cell-specific markers mapped to the transcript clusters
| Gene symbol | Gene name | Associated cell | Identified cluster # |
|---|---|---|---|
| Dazl |
Deleted in azoospermia-like |
Spermatogonia |
1 |
| 1500011H22Rik |
RIKEN cDNA 1500011H22 gene |
Spermatocytes |
2 |
| 1700029G01Rik |
RIKEN cDNA 1700029G01 gene |
Spermatocytes |
3 |
| 2410022L05Rik |
RIKEN cDNA 2410022 L05 gene |
Spermatocytes |
2 |
| Akap12 |
A kinase (PRKA) anchor protein (gravin) 12 |
Spermatocytes |
5 |
| Aldoa-ps1 |
Aldolase 1, A isoform, retrogene 2 |
Spermatocytes |
4 |
| Atl3 |
Atlastin GTPase 3 |
Spermatocytes |
4 |
| Cypt3 |
Cysteine-rich perinuclear theca 3 |
Spermatocytes |
3 |
| D030056L22Rik |
RIKEN cDNA D030056L22 gene |
Spermatocytes |
1 |
| Gsg2 |
Germ cell-specific gene 2 |
Spermatocytes |
3 |
| H3f3b |
H3 histone, family 3B |
Spermatocytes |
1 |
| Hnrpa2b1 |
Heterogeneous nuclear ribonucleoprotein A2/B1 |
Spermatocytes |
1 |
| Lyar |
Ly1 antibody reactive clone |
Spermatocytes |
2 |
| Nt5c1b |
5′-nucleotidase, cytosolic IB |
Spermatocytes |
4 |
| Pgk2 |
Phosphoglycerate kinase 2 |
Spermatocytes |
4 |
| Slc2a3 |
Solute carrier family 2 (facilitated glucose transporter), member 3 |
Spermatocytes |
2 |
| Spert |
Spermatid associated |
Spermatocytes |
4 |
| Stard10 |
START domain containing 10 |
Spermatocytes |
4 |
| Stmn1 |
Stathmin 1 |
Spermatocytes |
1 |
| Vkorc1 |
Vitamin K epoxide reductase complex, subunit 1 |
Spermatocytes |
5 |
| Acrv1 |
Acrosomal vesicle protein 1 |
Spermatids |
3 |
| Actl7a |
Actin-like 7a |
Spermatids |
4 |
| Akap4 |
A kinase (PRKA) anchor protein 4 |
Spermatids |
4 |
| Marcksl1 |
MARCKS-like 1 |
Spermatids |
5 |
| Pdpk1 |
3-phosphoinositide dependent protein kinase-1 |
Spermatids |
4 |
| Pdzk1 |
PDZ domain containing 1 |
Spermatids |
4 |
| Prm1 |
Protamine 1 |
Spermatids |
2 |
| Spag4l |
Sperm associated antigen 4-like |
Spermatids |
3 |
| Tctex1d1 |
Tctex1 domain containing 1 |
Spermatids |
4 |
| Tnp2 |
Transition protein 2 |
Spermatids |
3 |
| Zpbp |
Zona pellucida binding protein |
Spermatids |
3 |
| Hsd17b3 |
Hydroxysteroid (17-beta) dehydrogenase 3 |
Leydig cells |
5 |
| Hsd3b1 |
Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
Leydig cells |
5 |
| Cyp17a1 |
Cytochrome P450, family 17, subfamily a, polypeptide 1 |
Leydig cells |
5 |
| Acta2 |
Actin, alpha 2, smooth muscle, aorta |
PTM cells |
5 |
| Cldn11 |
Claudin 11 |
Sertoli cells |
5 |
| Clu |
Clusterin |
Sertoli cells |
5 |
| Ctsl |
Cathepsin L |
Sertoli cells |
5 |
| Vim | Vimentin | Sertoli cells | 5 |
Figure 1.
Gene expression profiles of the five transcript clusters during recovery from irradiation. The clusters contain the subset of 988 transcripts changing in expression during recovery from irradiation, which was separated into clusters by PAM. The number of transcripts in each cluster is stated in the title of each plot. The cell specific markers are coloured as following: Blue: Spermatogonia; Red: Spermatocytes; Green: Spermatids; Purple: Somatic cells.
Figure 2.
The median z-scaled gene expression of the five transcript clusters during recovery from irradiation. The clusters are coloured as following: Blue: Spermatogonia (cluster 1); Red: Spermatocytes (cluster 2); Green: Early spermatids (cluster 3); Brown: Late spermatids (cluster 4); Purple: Somatic cells (cluster 5).
Gene set enrichment analysis
GSEA was performed on the genes from each cluster separately to gain knowledge of overrepresented functional categories in each cluster. Statistical significant categories are listed in Table 2. Of particular interest, this analysis revealed overrepresentation of genes involved in mRNA processing, spliceosome and nucleus for the transcripts in the spermatogonia cluster. The transcripts in the spermatocyte cluster were enriched in male gamete generation, spermatogenesis and cell cycle process. The early spermatid cluster differed from the late spermatid cluster in enrichment for acrosomal vesicle. The somatic cell cluster was enriched in processes such as lipid metabolic process, response to oxidative stress and lipid biosynthetic process. All enriched Gene Ontology categories are available in the additional material (see Additional file 7: Table S6, Additional file 8: Table S7, Additional file 9: Table S8, Additional file 10: Table S9 and Additional file 11: Table S10).
Table 2.
| Cluster | Category | Term | Count | % | p-value | Fold enrichment | Bonferroni p-value |
|---|---|---|---|---|---|---|---|
| 1: Spermatogonia |
BP |
mRNA processing |
11 |
11.83 |
6.12E-07 |
8.22 |
3.11E-04 |
| mRNA metabolic process |
11 |
11.83 |
2.46E-06 |
7.05 |
0.001 |
||
| RNA splicing |
9 |
9.68 |
6.05E-06 |
8,87 |
0.003 |
||
| CC |
Ribonucleoprotein complex |
19 |
20.43 |
4.15E-12 |
8.28 |
6.56E-10 |
|
| Macromolecular complex |
34 |
36.56 |
5.83E-09 |
2.75 |
9.21E-07 |
||
| Intracellular organelle |
60 |
64.52 |
2.24E-08 |
1.54 |
3.54E-06 |
||
| Organelle |
60 |
64.52 |
2.32E-08 |
1.54 |
3.67E-06 |
||
| Intracellular part |
64 |
68.82 |
4.10E-08 |
1.41 |
6.48E-06 |
||
| Intracellular |
64 |
68.82 |
2.70E-07 |
1.36 |
4.26E-05 |
||
| Intracellular organelle part |
35 |
37.63 |
5.02E-07 |
2.25 |
7.93E-05 |
||
| Organelle part |
35 |
37.63 |
5.95E-07 |
2.24 |
9.40E-05 |
||
| Spliceosome |
8 |
8.60 |
2.23E-06 |
12.97 |
3.53E-04 |
||
| Nucleus |
38 |
40.86 |
1.60E-05 |
1.85 |
0.003 |
||
| Intracellular membrane-bounded organelle |
51 |
54.84 |
4.71E-05 |
1.47 |
0.007 |
||
| Membrane-bounded organelle |
51 |
54.84 |
4.90E-05 |
1.47 |
0.008 |
||
| MF |
RNA binding |
21 |
22.58 |
2.81E-12 |
7.11 |
4.35E-10 |
|
| Nucleic acid binding |
31 |
33.33 |
1.36E-06 |
2.36 |
2.11E-04 |
||
| 2: Spermatocytes |
BP |
Sexual reproduction |
18 |
12.41 |
1.02E-09 |
6.70 |
7.80E-07 |
| Gamete generation |
16 |
11.03 |
7.42E-09 |
6.94 |
5.67E-06 |
||
| Male gamete generation |
14 |
9.66 |
2.03E-08 |
7.89 |
1.55E-05 |
||
| Spermatogenesis |
14 |
9.66 |
2.03E-08 |
7.89 |
1.55E-05 |
||
| Multicellular organism reproduction |
16 |
11.03 |
1.22E-07 |
5.62 |
9.34E-05 |
||
| Reproductive process in a multicellular organism |
16 |
11.03 |
1.22E-07 |
5.62 |
9.34E-05 |
||
| Reproductive process |
18 |
12.41 |
1.01E-06 |
4.17 |
7.72E-04 |
||
| Reproduction |
18 |
12.41 |
1.11E-06 |
4.14 |
8.44E-04 |
||
| Cell cycle process |
14 |
9.66 |
2.88E-06 |
5.12 |
0.002 |
||
| CC |
Intracellular organelle part |
45 |
31.03 |
1.01E-06 |
2.03 |
2.04E-04 |
|
| Macromolecular complex |
39 |
26.90 |
1.16E-06 |
2.21 |
2.36E-04 |
||
| Organelle part |
45 |
31.03 |
1.22E-06 |
2.02 |
2.48E-04 |
||
| Intracellular part |
90 |
62.09 |
1.67E-06 |
1.35 |
3.39E-04 |
||
| Intracellular membrane-bounded organelle |
74 |
51.03 |
6.04E-06 |
1.47 |
0.001 |
||
| Membrane-bounded organelle |
74 |
51.03 |
6.04E-06 |
1.47 |
0.001 |
||
| Intracellular organelle |
80 |
55.17 |
6.71E-06 |
1.41 |
0.001 |
||
| Organelle |
80 |
55.17 |
6.87E-06 |
1.41 |
0.001 |
||
| 3: Early spermatids |
BP |
Sexual reproduction |
19 |
9.60 |
2.17E-10 |
6.69 |
1.19E-07 |
| Male gamete generation |
13 |
6.57 |
2.33E-07 |
7.03 |
1.28E-04 |
||
| Spermatogenesis |
13 |
6.57 |
2.33E-07 |
7.03 |
1.28E-04 |
||
| Reproductive process |
19 |
9.60 |
3.10E-07 |
4.21 |
1.70E-04 |
||
| Reproduction |
19 |
9.60 |
3.51E-07 |
4.17 |
1.93E-04 |
||
| Gamete generation |
14 |
7.07 |
5.99E-07 |
5.81 |
3.29E-04 |
||
| Multicellular organism reproduction |
14 |
7.07 |
6.43E-06 |
4.69 |
0.004 |
||
| Reproductive process in a multicellular organism |
14 |
7.07 |
6.43E-06 |
4.69 |
0.004 |
||
| CC |
Acrosomal vesicle |
10 |
5.05 |
1.06E-10 |
25.52 |
1.76E-08 |
|
| Secretory granule |
10 |
5.05 |
1.11E-06 |
9.26 |
1.84E-04 |
||
| 4: Late spermatids |
BP |
Sexual reproduction |
16 |
7.24 |
4.98E-08 |
6.01 |
2.95E-05 |
| Spermatogenesis |
12 |
5.43 |
1.27E-06 |
6.83 |
7.51E-04 |
||
| Male gamete generation |
12 |
5.43 |
1.27E-06 |
6.83 |
7.51E-04 |
||
| Gamete generation |
13 |
5.88 |
2.51E-06 |
5.70 |
0.001 |
||
| Reproductive process in a multicellular organism |
14 |
6.33 |
3.97E-06 |
4.97 |
0.002 |
||
| Multicellular organism reproduction |
14 |
6.33 |
3.97E-06 |
4.97 |
0.002 |
||
| CC |
Cytoplasm |
73 |
33.03 |
1.65E-05 |
1.49 |
0.002 |
|
| 5: Somatic cells | BP |
Oxidation reduction |
28 |
15.56 |
2.32E-09 |
3.87 |
3.34E-06 |
| Lipid metabolic process |
28 |
15.56 |
4.93E-09 |
3.74 |
7.10E-06 |
||
| Response to oxidative stress |
10 |
5.56 |
3.62E-07 |
10.68 |
5.22E-04 |
||
| Lipid biosynthetic process |
16 |
8.89 |
4.00E-07 |
5.21 |
5.76E-04 |
||
| Cellular ketone metabolic process |
20 |
11.11 |
1.23E-06 |
3.79 |
0.002 |
||
| Oxoacid metabolic process |
19 |
10.56 |
3.61E-06 |
3.69 |
0.005 |
||
| Carboxylic acid metabolic process |
19 |
10.56 |
3.61E-06 |
3.69 |
0.005 |
||
| Organic acid metabolic process |
19 |
10.56 |
3.71E-06 |
3.68 |
0.005 |
||
| Cellular lipid metabolic process |
19 |
10.56 |
3.93E-06 |
3.66 |
0.006 |
||
| Response to hydrogen peroxide |
6 |
3.33 |
4.71E-06 |
23.23 |
0.007 |
||
| CC |
Cytoplasm |
114 |
63.33 |
4.81E-13 |
1.68 |
1.11E-10 |
|
| Cytoplasmic part |
85 |
47.22 |
1.37E-11 |
1.94 |
3.14E-09 |
||
| Mitochondrion |
37 |
20.56 |
6.64E-08 |
2.67 |
1.53E-05 |
||
| Intracellular part |
128 |
71.11 |
5.25E-06 |
1.27 |
0.001 |
||
| Intracellular |
130 |
72.22 |
2.52E-05 |
1.23 |
0.006 |
||
| MF | Oxidoreductase activity |
29 |
16.11 |
2.67E-10 |
4.12 |
1.02E-07 |
|
| Oxidoreductase activity, acting on CH-OH group of donors |
13 |
7.22 |
2.96E-09 |
10.68 |
1.13E-06 |
||
| Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 6.67 | 6.67 | 1.19E-08 | 10.91 | 4.56E-06 |
BP: Biological process; CC: Cellular component; MF: Molecular function.
IHC suggested irradiation-induced Leydig cell hyperplasia
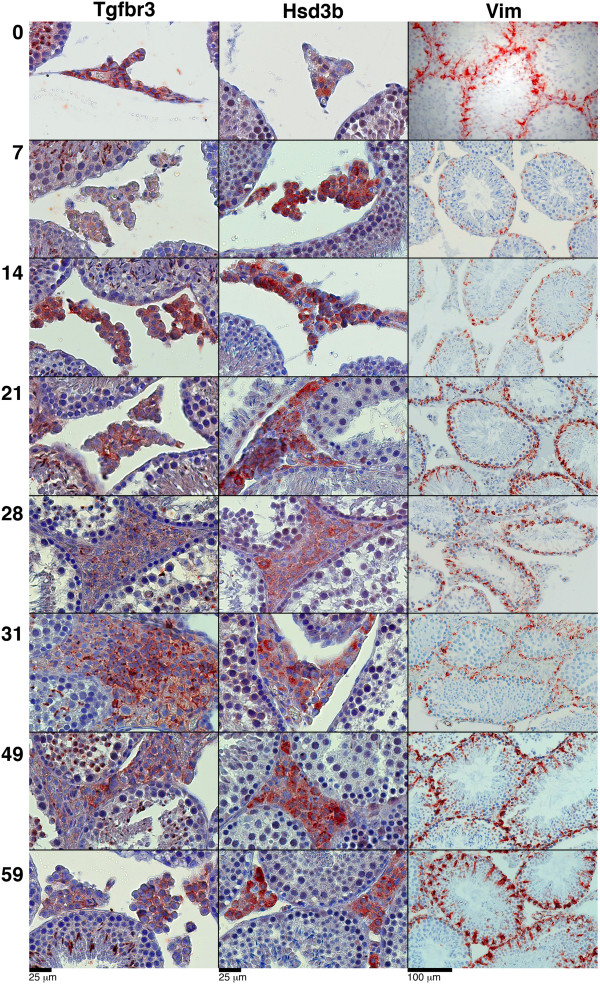
Large expression changes were observed in the somatic cell cluster. We expected this to be caused mainly by changes in cellularity, but we decided to use IHC to investigate whether histological changes contributed to the observed changes in expression. We stained sections of the irradiated testis tissue with the following somatic cell-specific markers: two Leydig cell markers: Tgfbr3 and Hsd3b; a PTM cell marker: SMA; and a Sertoli cell marker: Vimentin (Figure 3).
Figure 3.
IHC staining of the testis tissue pi with Leydig and Sertoli cell markers. Tgfbr3 and Hsd3b were used as Leydig cell markers and Vimentin (Vim) was used as a Sertoli cell marker. Indications of Leydig cell hyperplasia are observed at pi day 21–31, whereas the Sertoli cells do not seem affected. A detailed description of the pictures is found in the text.
In control material (day 0; Figure 3), Tgfbr3 was expressed in the cell membrane of the majority of Leydig cells. Already at pi day 7 the Leydig cell membrane reaction appeared disturbed, incomplete or became cytoplasmic. In addition, the shape of the Leydig cells became more round and the close configuration apparent in the control material seemed to be lost. At pi day 21 some membrane reaction reappeared, but was lost again day 28–31 where hyperplasia of the Leydig cells was pronounced. During pi day 49–59 some of the Leydig cell membrane reactions was restored, but Tgfbr3 was only expressed at the membrane in about half of the Leydig cells at pi day 59 while the rest either showed a cytoplasmic reaction or no reaction at all.
Hsd3b was expressed in the Leydig cell cytoplasm in the control (day 0; Figure 3). Only a few Leydig cells reacted strongly, whereas the majority reacted moderately to faint or were negative. Again, already from pi day 7, this picture changed with the vast majority of the Leydig cells now reacting strongly to the antibody and although the intensity of the reaction gradually declined, it was still elevated at pi day 59. Staining with Hsd3b indicated an increase in Leydig cell numbers at pi day 21–28, but the number gradually returned to almost pre-irradiation levels at pi day 59.
Vimentin showed a specific staining in Sertoli cells in the control material (day 0; Figure 3). The staining remained in Sertoli cells throughout recovery, but the intensity and amount of staining seemed to decrease as specific germ cells disappeared. Already from pi day 7 where only spermatogonia are absent from the testis, the amount of staining was reduced, and it remained reduced until pi day 49 where the intensity and amount of staining was back to control levels. In addition, the classical extension of Sertoli cell cytoplasm towards the centre of the tubuli was mainly observed at day 0 and after pi day 49. Although the staining was reduced as distinct germ cell populations were missing, we did not find any indications of cell death among the Sertoli cells (deduced from the Vimentin staining). Neither did we find any histological changes pi in the PTM cell population by staining with SMA (results not shown).
Nevertheless, IHC staining of the testis tissue pi with the Leydig cell markers Tgfbr3 and Hsd3b suggested that some Leydig cell hyperplasia occurred after low dose irradiation. To confirm the hyperplasia pi, we quantified the ratio of the areas occupied by Leydig cells relative to the areas covered by other testicular cells. We found that the area of Leydig cells increased after irradiation and gradually decreased towards pre-irradiation levels as the testis was repopulated by germ cells (Figure 4). This further underlined that the number of Leydig cells seemed to be affected by low dose irradiation that may have caused Leydig cell hyperplasia.
Figure 4.
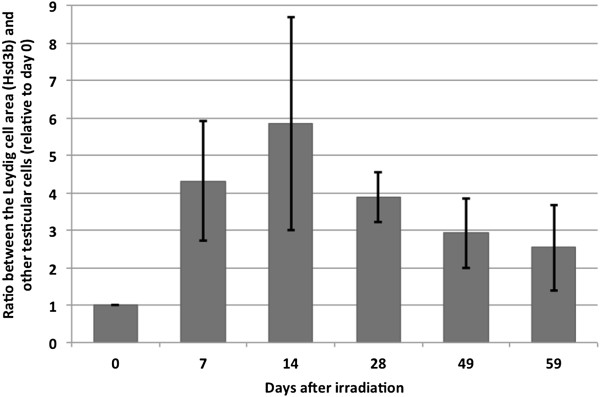
In silico determination of Leydig cells relative to other testicular cells pi. Areas covered by Leydig cells highly increased following irradiation compared to areas covered by other testicular cells. The peak is observed at pi day 14 where the ratio decreased again.
Discussion
Cell-specific transcript clusters
We identified five unique transcript clusters with an unsupervised cluster analysis of the transcripts that changed in expression during recovery from irradiation. The expression changes observed in this study were caused by a combination of changes in cellularity and cells changing transcription pattern due to their differentiation towards spermatids and spermatozoa - both events change the total RNA pool. It further complicates the separation of the clusters that cells have a mixed cellular expression of many of the genes that are specific for spermatogenesis e.g. a relatively low expression that initiates in spermatogonia that is followed by a large increase in expression in spermatocytes (for example Deleted in azoospermia-like (Dazl) [22]).
We identified two spermatid clusters, one associated with early spermatids and a novel cluster associated with late spermatids. We anticipate that the transcripts in the early spermatid cluster initially were expressed in late pachytene or diplotene spermatocytes just before they entered meiosis, but with their main expression in early spermatids. It has previously been hypothesized that DNA is inaccessible to transcription during meiosis but also that a large group of mRNAs needed in early spermatids are expressed in the late spermatocyte stages [16,23]. Transcription is reinitiated in early spermatids [16] resulting in a massive wave of transcriptional activity immediately after meiosis before the chromatin is condensed and transcription is silenced during spermiogenesis [24,25]. Thus, the late spermatid cluster that we identified likely includes transcripts that are initially expressed during this wave of transcription in early spermatids.
The somatic cell cluster included markers specific for Leydig, PTM and Sertoli cells, suggesting that this cluster contained transcripts expressed by all the somatic cells in the testis. The somatic cells further comprise the microvasculature, lymph vessels, nerve fibres and connective tissue [26] and transcripts expressed by these cells are properly also present in the somatic cell cluster. From inspection of the expression profile of the somatic cell cluster it seemed that the somatic cells underwent large expression changes pi. However, the majority of the somatic cells did not seem to be affected by the irradiation and the gene expression changes detected in the somatic cell cluster were most likely caused by changes in cellularity. Since pachytene spermatocytes physically are the largest and most RNA-rich germ cells in the testis [10,26] their disappearing leads to the apparent expressional upregulation of genes from the somatic cell cluster where especially RNA from Sertoli cells comprise a larger part of the RNA from the testis.
We tried to separate more than five cell clusters from this data set. Yet, some of the original clusters just separated into subclusters with very similar expression patterns. The matured spermatozoa have a highly condensed transcriptionally inactive chromatin [27] and it is therefore reasonable that we did not identify any cluster(s) originating from the later germ cell stages.
Gene set enrichment analysis
We performed GSEA of the subsets of the transcripts in each cluster to gain knowledge of their protein function. The analyses supported the association of the clusters with the cell types. However, the data set was complex and other cells than those specifically associated to the cluster may contribute to the pool of transcripts in each cluster.
The transcripts in the spermatogonia cluster were enriched for transcripts involved in mRNA processing, spliceosome and nucleus, in agreement with the transcriptional regulation involved during the numerous divisions and differentiations undergone by spermatogonia [28]. The transcripts in the spermatocyte cluster were enriched in male gamete generation, spermatogenesis and cell cycle process, which fit well with the meiotic processes in spermatocytes. Both spermatid clusters were enriched in spermatogenesis. The early spermatid cluster was additionally enriched in acrosomal vesicle, which agrees with the acrosome being formed during the maturation of early to late spermatids and further to spermatozoa [29]. The transcripts in the late spermatid cluster were in addition enriched in serine-type peptidase activity and especially one serine protease, acrosin, has been identified as an important enzyme in the acrosome [30].
The very diverse functions of the somatic cells were also observed in the GSEA where the enriched categories among others included lipid metabolic process, response to oxidative stress and lipid biosynthetic process. Since cholesterol is the basis for steroidogenesis in Leydig cells, lipid metabolism is essential for steroid production [31-33]. Both the many cell divisions in the germinal tissue and Leydig cell steroidogenesis require large amounts of oxygen. The testis is poorly vascularized and therefore vulnerable to oxidative stress and consequently have several defence mechanisms where Sertoli cells superoxide dismutase is important [34]. Thus, the results from the GSEA on the transcripts in the somatic cell cluster represented both Leydig and Sertoli cell functions.
Low dose irradiation may cause Leydig cell hyperplasia
IHC validation of the two Leydig cell markers, Tgfbr3 and Hsd3b, showed that low dose irradiation affected the appearance of the Leydig cells with indications of Leydig cell hyperplasia around pi day 21–31. Previously published studies reported that human Leydig cells are preserved even after high dose irradiation up to 20 Gy, but that the endocrine function of the cells are affected also at lower doses [35-38]. In this study, the testes were irradiated with 1 Gy in a single dose, which is a low dose even in mice compared to the clinical doses of 16–20 Gy used to treat human testicular cancer [39].
Leydig cell hyperplasia has also been observed in men with fertility problems and testicular cancer [40]. The proliferation might arise due to a decreased testosterone production by the Leydig cells pi, which will induce increased LH production that may stimulate proliferation of pre-Leydig cells and their differentiation to Leydig cells. This is also the clinical observation upon irradiation of human testis harbouring carcinoma in situ[41] and in rats with Sertoli cell-only testes [42]. However, it is not clear if a dose of 1 Gy leads to reduced testosterone production and increased LH level, although higher irradiation doses lead to changes in testosterone and LH levels [43]. Also, the presence of Follicle-stimulating hormone (FSH) receptors in Leydig cells could suggest that the proliferation could be a response to decreased inhibin production caused by the absence of germ cells, which leads to increased FSH levels that may induce proliferation of pre-Leydig cells [44]. Further, the GSEA showed that the transcripts in the somatic cell cluster were highly enriched in lipid and steroid biosynthetic processes (see Additional file 11: Table S10). This enrichment can either be caused by an increased activity of the remaining Leydig cells or by an increased number of Leydig cells. From our results it seems to be caused by an increased number of Leydig cells. Alternatively, the reduced number of germ cells in the tubuli will likely result in a reduced size of the tubuli, and this might alter the ratio between tubuli and interstitial areas.
IHC staining of Vimentin specific to Sertoli cells did not show any histologically changes during recovery from low dose irradiation. Earlier studies found Sertoli cell death following irradiation, but these were all studies that used higher irradiation doses [45,46]. SMA staining of PTM cells did not suggest any histological changes in the PTM cells.
Conclusion
In this study, we investigated the molecular and cellular changes following re-establishment of spermatogenesis pi by analysis of the testicular transcriptome. We identified five clusters each representing a profile of a specific type of cell(s) during recovery from irradiation. We associated the five clusters with: spermatogonia, spermatocytes, early spermatids, late spermatids and somatic cells, respectively. We observed large expression changes in the somatic cell cluster during the time series. Therefore, we investigated if changes in histology, in addition to changes in cellularity, contributed to this observation. IHC staining of Leydig cell-specific markers suggested Leydig cell hyperplasia at pi day 21–31, which was further supported by in silico quantification of the relative area occupied by Leydig cells.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HL designed the study; MT conducted irradiation experiments; MDD and KCB performed the microarray experiments; KCB and HBN performed the data analysis; JEN performed the immunohistochemistry; KA performed the in silico determination of Leydig cells; KCB, HBN, SB, KA and HL interpreted the results; all authors contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
An example of testis tissue sections stained with the Leydig cell-specific Hsd3b (red) and counter stained with Meyers haematoxylin (blue). Red stain was assigned a red colour, blue stain a blue colour and the background assigned white. In silico separation of the colour space using a Baysian algorithm separated blue from red stain and hence Leydig cells from all other cells. This figure shows the layer of assigned colours with different transparency on top of the original image. Sections from day 0 and pi day 7, 14, 28, 49 and 59 were used for in silico quantification of the Leydig cells versus the other testis cells.
Gene names and identifiers of the 109 transcripts assigned cluster 1 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 164 transcripts assigned cluster 2 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 246 transcripts assigned to cluster 3 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 273 transcripts assigned cluster 4 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 196 transcripts assigned cluster 5 based on the cluster analysis of the subset of 988 transcripts.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 1 - spermatogonia.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 2 - spermatocytes.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 3 – early spermatids.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 4 – late spermatids.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 5 – somatic cells.
Contributor Information
Kirstine C Belling, Email: belling@cbs.dtu.dk.
Masami Tanaka, Email: masami-t@jc.u-aizu.ac.jp.
Marlene Danner Dalgaard, Email: marlene.dalgaard@gmail.dk.
John Erik Nielsen, Email: john.erik.nielsen@regionh.dk.
Henrik Bjørn Nielsen, Email: hbjorn@cbs.dtu.dk.
Søren Brunak, Email: brunak@cbs.dtu.dk.
Kristian Almstrup, Email: kristian@almstrup.net.
Henrik Leffers, Email: henrik@leffers.co.dk.
Acknowledgements
We thank Ana Ricci Nielsen, Betina F. Nielsen and Brian V. Hansen for skilful technical assistance, and Jose MG Izarzugaza for valuable discussion and revision of the manuscript. This work was supported by the Villum Kann-Rasmussen foundation, the European Commission, and the Japanese Science and Technology Agency CREST program.
References
- Jahnukainen K, Ehmcke J, Hou M, Schlatt S. Testicular function and fertility preservation in male cancer patients. Best Pract Res Clin Endocrinol Metab. 2011;25:287–302. doi: 10.1016/j.beem.2010.09.007. [DOI] [PubMed] [Google Scholar]
- van der Meer Y, Huiskamp R, Davids JA, van der Tweel I, de Rooij DG. The sensitivity of quiescent and proliferating mouse spermatogonial stem cells to X irradiation. Radiat Res. 1992;130:289–295. doi: 10.2307/3578373. [DOI] [PubMed] [Google Scholar]
- van der Meer Y, Huiskamp R, Davids JA, van der Tweel I, de Rooij DG. The sensitivity to X rays of mouse spermatogonia that are committed to differentiate and of differentiating spermatogonia. Radiat Res. 1992;130:296–302. doi: 10.2307/3578374. [DOI] [PubMed] [Google Scholar]
- Oakberg EF. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am J Anat. 1956;99:507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Sonne SB, Almstrup K, Dalgaard M, Juncker AS, Edsgard D, Ruban L, Harrison NJ, Schwager C, Abdollahi A, Huber PE. et al. Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res. 2009;69:5241–5250. doi: 10.1158/0008-5472.CAN-08-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen MS, Gundgaard MG, Daugaard G. Treatment options for carcinoma in situ testis. Int J Androl. 2011;34:e32–e36. doi: 10.1111/j.1365-2605.2011.01178.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shao S, Shetty G, Meistrich ML. Donor Sertoli cells transplanted into irradiated rat testes stimulate partial recovery of endogenous spermatogenesis. Reproduction. 2009;137:497–508. doi: 10.1530/REP-08-0120. [DOI] [PubMed] [Google Scholar]
- Delic JI, Hendry JH, Morris ID, Shalet SM. Leydig cell function in the pubertal rat following local testicular irradiation. Radiother Oncol. 1986;5:29–37. doi: 10.1016/S0167-8140(86)80006-8. [DOI] [PubMed] [Google Scholar]
- Delic JI, Hendry JH, Morris ID, Shalet SM. Serum androgen binding protein and follicle stimulating hormone as indices of Sertoli cell function in the irradiated testis. Br J Cancer Suppl. 1986;7:105–107. [PMC free article] [PubMed] [Google Scholar]
- Shah FJ, Tanaka M, Nielsen JE, Iwamoto T, Kobayashi S, Skakkebaek NE, Leffers H, Almstrup K. Gene expression profiles of mouse spermatogenesis during recovery from irradiation. Reprod Biol Endocrinol. 2009;7:130. doi: 10.1186/1477-7827-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivell R, Spiess AN. Analysing differential gene expression in the testis. Berlin: Springer; 2002. [Google Scholar]
- The R project. http://www.r-project.org.
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Almstrup K, Nielsen JE, Hansen MA, Tanaka M, Skakkebaek NE, Leffers H. Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biol Reprod. 2004;70:1751–1761. doi: 10.1095/biolreprod.103.026575. [DOI] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Acharya KK, Chandrashekar DS, Chitturi N, Shah H, Malhotra V, Sreelakshmi KS, Deepti H, Bajpai A, Davuluri S, Bora P, Rao L. A novel tissue-specific meta-analysis approach for gene expression predictions, initiated with a mammalian gene expression testis database. BMC Genomics. 2010;11:467. doi: 10.1186/1471-2164-11-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- da Huang W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Wright WW, Dicandeloro P, Wilson E, Kopf GS, Jelinsky SA. Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells. Proc Natl Acad Sci U S A. 2008;105:8315–8320. doi: 10.1073/pnas.0709854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijo RA, Dorfman DM, Slee R, Renshaw AA, Loughlin KR, Cooke H, Page DC. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- Hansen MA, Nielsen JE, Tanaka M, Almstrup K, Skakkebaek NE, Leffers H. Identification and expression profiling of 10 novel spermatid expressed CYPT genes. Mol Reprod Dev. 2006;73:568–579. doi: 10.1002/mrd.20463. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development. 1995;121:2373–2383. doi: 10.1242/dev.121.8.2373. [DOI] [PubMed] [Google Scholar]
- Holstein AF, Schulze W, Davidoff M. Understanding spermatogenesis is a prerequisite for treatment. Reprod Biol Endocrinol. 2003;1:107. doi: 10.1186/1477-7827-1-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- Godmann M, Auger V, Ferraroni-Aguiar V, Di Sauro A, Sette C, Behr R, Kimmins S. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol Reprod. 2007;77:754–764. doi: 10.1095/biolreprod.107.062265. [DOI] [PubMed] [Google Scholar]
- Hermo L, Pelletier RM, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: changes in spermatid organelles associated with development of spermatozoa. Microsc Res Tech. 2010;73:279–319. doi: 10.1002/jemt.20787. [DOI] [PubMed] [Google Scholar]
- Baba T, Azuma S, Kashiwabara S, Toyoda Y. Sperm from mice carrying a targeted mutation of the acrosin gene can penetrate the oocyte zona pellucida and effect fertilization. J Biol Chem. 1994;269:31845–31849. [PubMed] [Google Scholar]
- de Catalfo GE H, de Alaniz MJ, Marra CA. Influence of commercial dietary oils on lipid composition and testosterone production in interstitial cells isolated from rat testis. Lipids. 2009;44:345–357. doi: 10.1007/s11745-008-3277-z. [DOI] [PubMed] [Google Scholar]
- Premalatha R, Jubendradass R, Rani SJ, Srikumar K, Mathur PP. A phytooxysterol, 28-homobrassinolide modulates Rat testicular steroidogenesis in normal and diabetic rats. Reprod Sci. 2013;20:589–596. doi: 10.1177/1933719112459241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocco DM, Clark BJ. Regulation of the acute production of steroids in steroidogenic cells. Endocr Rev. 1996;17:221–244. doi: 10.1210/edrv-17-3-221. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Roman SD. Antioxidant systems and oxidative stress in the testes. Adv exper med biol. 2008;636:154–171. doi: 10.1007/978-0-387-09597-4_9. [DOI] [PubMed] [Google Scholar]
- Colpi GM, Contalbi GF, Nerva F, Sagone P, Piediferro G. Testicular function following chemo-radiotherapy. Eur J Obstet Gynecol Reprod Biol. 2004;113(Suppl 1):S2–S6. doi: 10.1016/j.ejogrb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Giwercman A, von der Maase H, Berthelsen JG, Rorth M, Bertelsen A, Skakkebaek NE. Localized irradiation of testes with carcinoma in situ: effects on Leydig cell function and eradication of malignant germ cells in 20 patients. J Clin Endocrinol Metab. 1991;73:596–603. doi: 10.1210/jcem-73-3-596. [DOI] [PubMed] [Google Scholar]
- Petersen PM, Giwercman A, Daugaard G, Rorth M, Petersen JH, Skakkeaek NE, Hansen SW, von der Maase H. Effect of graded testicular doses of radiotherapy in patients treated for carcinoma-in-situ in the testis. J Clin Oncol. 2002;20:1537–1543. doi: 10.1200/JCO.20.6.1537. [DOI] [PubMed] [Google Scholar]
- Shapiro E, Kinsella TJ, Makuch RW, Fraass BA, Glatstein E, Rosenberg SA, Sherins RJ. Effects of fractionated irradiation of endocrine aspects of testicular function. J Clin Oncol. 1985;3:1232–1239. doi: 10.1200/JCO.1985.3.9.1232. [DOI] [PubMed] [Google Scholar]
- Mortensen MS, Gundgaard MG, Daugaard G. Treatment options for carcinoma in situ testis. Int J Androl. 2011;34:e32–36. doi: 10.1111/j.1365-2605.2011.01178.x. [DOI] [PubMed] [Google Scholar]
- Holm M, Rajpert-De Meyts E, Andersson AM, Skakkebaek NE. Leydig cell micronodules are a common finding in testicular biopsies from men with impaired spermatogenesis and are associated with decreased testosterone/LH ratio. J Pathol. 2003;199:378–386. doi: 10.1002/path.1309. [DOI] [PubMed] [Google Scholar]
- Petersen PM, Daugaard G, Rorth M, Skakkebaek NE. Endocrine function in patients treated for carcinoma in situ in the testis with irradiation. APMIS. 2003;111:93–98. doi: 10.1034/j.1600-0463.2003.11101131.x. discussion 98–99. [DOI] [PubMed] [Google Scholar]
- Rich KA, Kerr JB, de Kretser DM. Evidence for Leydig cell dysfunction in rats with seminiferous tubule damage. Mol Cell Endocrinol. 1979;13:123–135. doi: 10.1016/0303-7207(79)90013-3. [DOI] [PubMed] [Google Scholar]
- Bang AK, Petersen JH, Petersen PM, Andersson AM, Daugaard G, Jorgensen N. Testosterone production is better preserved after 16 than 20 Gray irradiation treatment against testicular carcinoma in situ cells. Int J Radiat Oncol Biol Phys. 2009;75:672–676. doi: 10.1016/j.ijrobp.2008.11.057. [DOI] [PubMed] [Google Scholar]
- Sarkar PS, Paul S, Han J, Reddy S. Six5 is required for spermatogenic cell survival and spermiogenesis. Hum Mol Genet. 2004;13:1421–1431. doi: 10.1093/hmg/ddh161. [DOI] [PubMed] [Google Scholar]
- Guitton N, Brouazin-Jousseaume V, Dupaix A, Jegou B, Chenal C. Radiation effect on rat Sertoli cell function in vitro and in vivo. Int J Radiat Biol. 1999;75:327–333. doi: 10.1080/095530099140500. [DOI] [PubMed] [Google Scholar]
- Kochar NK, Bateman AJ. Post-irradiation changes in Sertoli cells. J Reprod Fertil. 1969;18:265–273. doi: 10.1530/jrf.0.0180265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An example of testis tissue sections stained with the Leydig cell-specific Hsd3b (red) and counter stained with Meyers haematoxylin (blue). Red stain was assigned a red colour, blue stain a blue colour and the background assigned white. In silico separation of the colour space using a Baysian algorithm separated blue from red stain and hence Leydig cells from all other cells. This figure shows the layer of assigned colours with different transparency on top of the original image. Sections from day 0 and pi day 7, 14, 28, 49 and 59 were used for in silico quantification of the Leydig cells versus the other testis cells.
Gene names and identifiers of the 109 transcripts assigned cluster 1 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 164 transcripts assigned cluster 2 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 246 transcripts assigned to cluster 3 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 273 transcripts assigned cluster 4 based on the cluster analysis of the subset of 988 transcripts.
Gene names and identifiers of the 196 transcripts assigned cluster 5 based on the cluster analysis of the subset of 988 transcripts.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 1 - spermatogonia.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 2 - spermatocytes.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 3 – early spermatids.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 4 – late spermatids.
All Gene Ontology categories from the gene set enrichment analysis performed using DAVID on cluster 5 – somatic cells.