Abstract
We have previously described the role of red hair (Melanocortin 1 Receptor, MC1R) and blue eye (Oculocutaneous Albinism Type 2, OCA2) gene polymorphisms in modulating risk of cutaneous malignant melanoma (CMM) in a highly sun-exposed population of European descent. A number of recent studies, including genome-wide association studies (GWAS), have identified numerous polymorphisms controlling human hair, eye and skin colour. In this paper, we test a selected set of polymorphisms in pigmentation loci (ASIP, TYR, TYRP1, MC1R, OCA2, IRF4, SLC24A4, SLC45A2) for association with CMM risk in a large Australian population-based case control study. Variants in IRF4 and SLC24A4, despite being strongly associated with pigmentation in our sample, did not modify CMM risk, but the other six did. Three SNPs (rs28777, rs35391, rs16891982) in the MATP gene (SLC45A2) exhibited the strongest crude association with risk, but this was attenuated to approximately the same effect size as that of a MC1R red hair color allele by controlling for ancestry of cases and controls. We also detected significant epistatic interactions between SLC45A2 and OCA2 alleles, and MC1R and ASIP alleles. Overall, these measured variants account for 12% of the familial risk of CMM in our population.
INTRODUCTION
Cutaneous malignant melanoma (CMM) is a common cancer in the fair skinned population of tropical and subtropical Queensland, Australia, with an estimated lifetime risk of 5% (Queensland Cancer Registry, 2008). Within fairer skinned populations, risk varies with degree of paleness, as well as with other pigmentation phenotypes such as hair and eye color, all of which are strongly genetically determined.
We have previously described association between variants in the pigmentation genes encoding the melanocortin-1 receptor (MC1R) (Palmer et al., 2000; Box et al., 2001) and the human melanocyte-specific P-protein (OCA2) with CMM risk (Sturm et al., 2008). A number of recent genome-wide association studies (GWAS) have characterized variants in both known pigmentation pathway (TYR, TYRP1, SLC45A2) (Sulem et al., 2007; Sulem et al., 2008) and novel (IRF4, SLC24A4) (Sulem et al., 2007; Han et al., 2008) genes that influence pigmentation, and one study has reported on associations with CMM (Gudbjartsson et al., 2008). Association to both pigmentation and CMM risk has also been reported with variants in a region of chromosome 20q fairly close to ASIP (Brown et al., 2008; Sulem et al., 2008; Gudbjartsson et al., 2008).
Variants in many of these genes exhibit significant differences in frequency, and underlie the pigmentation differences, between different ethnic groups. It is important therefore, to differentiate between disease associations due to a direct genotype-phenotype relationship, and those to confounding by ancestry.
Here, we replicate and extend association results with CMM to MC1R, OCA2, TYR (OCA1), TYRP1 (OCA3), IRF4, SLC24A4, SLC45A2 (MATP, OCA4), and ASIP and examine the nature of interaction between these loci on disease risk in a large sample from a population at high environmental and genetic risk of melanoma.
RESULTS
There were 1738 CMM cases and 4517 controls. Ancestry and coloring for the sample are shown in Table 1. Differences between cases and controls are in the expected direction, as reported previously for this sample (Palmer et al., 2000), with cases tending to have lighter coloured eyes and hair. Participants were genotyped at up to 161 SNPs across the gene regions of interest (Supplementary Table 1). Table 2 shows case and control allele frequencies and Hardy-Weinberg equilibrium tests for selected SNPs. A number of SNPs exhibited extreme Hardy-Weinberg disequilibrium, but since the chosen SNPs are in pigmentation loci known to be highly informative for colouring and ancestry, it could be shown that the disequilibrium was due to ethnic stratification of the sample rather than to genotyping error (see Supplementary Table 2).
Table 1.
Descriptive statistics for genotyped CMM cases and controls (percentages are of total with data for that phenotype, except for ancestry).
| CMM Cases | Controls | |
|---|---|---|
| All four grandparents of northern European ancestry | 1483 (93.0%) | 3098 (83.3%) |
| Blue/grey eyed | 758 (43.7%) | 1838 (44.7%) |
| Green/hazel eyed | 692 (39.9%) | 1212 (29.5%) |
| Brown/black eyed | 283 (16.3%) | 1059 (25.8%) |
| Blond hair | 419 (24.1%) | 569 (23.7%) |
| Light brown hair | 653 (37.6%) | 1391 (33.9%) |
| Red hair | 193 (11.1%) | 217 (5.3%) |
| Dark brown hair | 402 (23.1%) | 1705 (41.5%) |
| Black hair | 71 (4.1%) | 226 (5.5%) |
| Male | 967 (47.8%) | 2129 (47.2%) |
| Total genotyped individuals | 1738 | 4517 |
Table 2.
Allele frequencies and tests of Hardy-Weinberg Equilibrium for pigmention-associated polymorphisms in Australian CMM cases and controls.
| Chr | Locus/Variant | rs ID | Alleles | Controls | CMM Cases | ||||
|---|---|---|---|---|---|---|---|---|---|
| N | MAFa | HWE-Pb | N | MAFa | HWE-Pb | ||||
| 5 | SLC45A2 i3 c | rs35391 | A (G) | 4107 | 0.0321 | 0.1527 | 1708 | 0.0088 | 1.0000 |
| 5 | SLC45A2 i3 | rs28777 | G (T) | 4111 | 0.0429 | 0.0001 | 1716 | 0.0118 | 1.0000 |
| 5 | SLC45A2*F374L | rs16891982 | G (C) | 4032 | 0.0518 | 0.0000 | 1731 | 0.0133 | 0.0346 |
| 6 | IRF4 i4 | rs12203592 | T (C) | 4113 | 0.2259 | 0.8696 | 1727 | 0.2107 | 0.0264 |
| 9 | TYRP1 | rs1408799 | T (C) | 4110 | 0.3275 | 0.0766 | 1703 | 0.2877 | 0.0980 |
| 9 | TYRP1 i6 | rs2733832 | C (T) | 4098 | 0.4119 | 0.0492 | 1732 | 0.3753 | 0.1514 |
| 11 | TYR i3 | rs10765198 | C (T) | 4103 | 0.3138 | 0.1739 | 1721 | 0.3405 | 0.2930 |
| 11 | TYR*S192Y | rs1042602 | A (C) | 4112 | 0.3486 | 0.7407 | 1728 | 0.3390 | 0.0150 |
| 11 | TYR*R402Q | rs1126809 | A (G) | 4102 | 0.3061 | 0.7688 | 1731 | 0.35 | 0.08 |
| 14 | SLC24A4 | rs12896399 | T (G) | 4102 | 0.4493 | 0.2857 | 1705 | 0.4564 | 0.1512 |
| 15 | OCA2*R419Q | rs1800407 | A (G) | 3996 | 0.0798 | 0.6061 | 1052 | 0.1042 | 1.0000 |
| 15 | HERC2 i86 | rs12913832 | T (C) | 4102 | 0.2315 | 0.0190 | 1747 | 0.2116 | 0.1133 |
| 16 | MC1R*V60L | rs1805005 | T (G) | 2849 | 0.1212 | 0.6061 | 1945 | 0.1181 | 0.2041 |
| 16 | MC1R*D84E | rs1805006 | T (G) | 2851 | 0.0111 | 0.4651 | 1947 | 0.0194 | 0.0008 |
| 16 | MC1R*V92M | rs2228479 | A (G) | 1882 | 0.0961 | 0.0794 | 1286 | 0.0995 | 0.5023 |
| 16 | MC1R*R142H | rs11547464 | A (G) | 1805 | 0.0044 | 0.4878 | 1114 | 0.0198 | 1.0000 |
| 16 | MC1R*R151C | rs1805007 | T (C) | 1876 | 0.1096 | 0.9091 | 1126 | 0.1567 | 0.5223 |
| 16 | MC1R*R160W | rs1805008 | T (C) | 2848 | 0.0697 | 0.6061 | 1947 | 0.1122 | 0.8365 |
| 16 | MC1R*R163Q | rs885479 | A (G) | 1896 | 0.0466 | 0.7407 | 1291 | 0.0476 | 0.6452 |
| 16 | MC1R*D294H | rs1805009 | C (G) | 2851 | 0.0266 | 0.9091 | 1947 | 0.0328 | 1.0000 |
| 20 | ASIP regiond | rs4911442 | C (T) | 4089 | 0.1306 | 0.3003 | 1745 | 0.1848 | 0.2003 |
MAF = minor allele frequency
HWE-P = Hardy-Weinberg Equilibrium test P-value
The letter i denotes intron, so HERC2 i86 denotes the 86th intron of the HERC2 gene
Actually in intron of NCOA6, 500 kbp distal to ASIP
One or more of the SNPs selected in each gene region could be shown to be associated with skin, hair or eye colour (Table 3), and in the subsequent analyses, we only present results for association with CMM for the polymorphisms most penetrant for pigmentary characteristics from each gene. In keeping with the selection criteria, a number of these SNPs were also strongly associated with reported ancestry (see Supplementary Table 2). Among the CMM cases with ancestry information, only 2.7% reported other than 100% Northern European ancestry, as compared to 13.5% of the control sample. To control these effects on CMM association analyses below, we report analyses on the subset of 1438 cases and 3098 controls reporting four grandparents of Northern European ancestry. We found that further inclusion of an ancestry indicator for English, Scottish/Welsh/Irish, and continental Europe as a covariate did not detect finer levels of ethnic confounding on CMM association. Interestingly, rs12203592 (in IRF4) did exhibit significant allele frequency differences between controls from continental Europe, England, and Scotland, Ireland and Wales, but these could be safely ignored given that this locus does not exhibit significant association with CMM (see below).
Table 3.
Results of association analyses of selected SNPs with skin, eye and hair color in individuals with all four grandparents of northern European ancestry.
| Locus/Variant | SNP | Allelic Odds Ratio for pale skin (P-value) | Allelic Odds Ratios for blue eye color (P-value) | Allelic Odds Ratios for light hair (P-value)a |
|---|---|---|---|---|
| SLC45A2 i3 | rs35391 | 3.42 (<1.0e-5) | 2.60 (0.001) | 5.34 (<1.0e-5) |
| SLC45A2 i3 | rs28777 | 4.31 (<1.0e-5) | 2.56 (0.017) | 6.30 (<1.0e-5) |
| SLC45A2*F374L | rs16891982 | 3.80 (<1.0e-5) | 2.50 (<1.0e-5) | 6.72 (<1.0e-5) |
| IRF4 i4 | rs12203592 | 1.63 (<1.0e-5) | 1.28 (0.0017) | 2.22 (<1.0e-5) |
| TYRP1 | rs1408799 | 1.06 (0.2985) | 1.22 (0.018) | 1.22 (<1.0e-5) |
| TYRP1 i6 | rs2733832 | 1.05 (0.3333) | 1.22 (0.007) | 1.22 (<1.0e-5) |
| TYR i3 | rs10765198 | 1.25 (0.0015) | 1.33 (<1.0e-5) | 1.01 (0.77) |
| TYR*S192Y | rs1042602 | 1.06 (0.11) | 1.04 (0.57) | 1.13 (0.13) |
| TYR*R402Q | rs1126809 | 1.27 (<1.0e-5) | 1.43 (<1.0e-5) | |
| SLC24A4 | rs12896399 | 1.17 (0.061) | 1.85 (<1.0e-5) | 1.45 (<1.0e-5) |
| OCA2*R419Q | rs1800407 | 1.10 (0.59) | 2.37 (<1.0e-5) | 1.21 (0.14) |
| HERC2 i86 | rs12913832 | 1.74 (<1.0e-5) | 30.77 (<1.0e-5) | 2.95 (<1.0e-5) |
| MC1R*V60L | rs1805005 | 1.01 (0.85) | 1.00 (0.80) | 1.13 (0.008) |
| MC1R*D84E | rs1805006 | 5.19 (<1.0e-5) | 1.23 (0.61) | 1.74 (0.041) |
| MC1R*V92M | rs2228479 | 1.38 (0.024) | 1.06 (0.61) | 1.17 (0.016) |
| MC1R*R142H | rs11547464 | 1.31 (0.54) | 1.03 (0.71) | 1.28 (0.74) |
| MC1R*R151C | rs1805007 | 2.57 (<1.0e-5) | 1.09 (0.94) | 2.13 (<1.0e-5) |
| MC1R*R160W | rs1805008 | 1.72 (<1.0e-5) | 1.12 (0.17) | 1.82 (<1.0e-5) |
| MC1R*R163Q | rs885479 | 1.10 (0.53) | 1.21 (0.15) | 1.12 (0.014) |
| MC1R*D294H | rs1805009 | 2.88 (<1.0e-5) | 1.16 (0.74) | 1.68 (<1.0e-5) |
| MC1R “R”b | 3.70 (<1.0e-5) | 1.09 (0.43) | 2.30 (<1.0e-5) | |
| MC1R “r” c | 1.84 | 1.10 | 1.26 | |
| ASIP region | rs4911442 | 1.43 (<1.0e-5) | 1.01 (0.87) | 1.28 (<1.0e-5) |
Light hair = blond, light brown, red.
High penetrance red hair color MC1R haplotype: variant at D84E, R142H, R151C, R160W, or D294H.
Low penetrance red hair color MC1R haplotype: variant at V60L, V92M, or R163Q.
In univariate analyses, MC1R, ASIP, TYRP1, (IRF4) and SLC45A2 SNPs were the markers most strongly associated with CMM risk in terms of crude risk (Table 4). Given the star-shaped genealogy of MC1R red-hair associated haplotypes (ie all these coding variants are in complete negative LD; in our population we have observed only three haplotypes with more than one coding variant present), we have recoded MC1R haplotypes as “R” (high red hair penetrance: rs1805006 (D84E), rs11547464 (R142H), rs1805007 (R151C), rs1805008 (R160W), rs1805009 (D294H) and “r” (low red hair penetrance: rs1805005, rs2228479, rs885479) (Sturm et al., 2003) for this and subsequent analyses. On restricting the case-control analysis only to individuals reporting all four grandparents to be of Northern European ancestry, the significant predictors of CMM risk were four “R” SNPs in MC1R (rs1805007, rs1805008, rs1805006 and rs11547464); the three SLC45A2 SNPs, rs1126809 in TYR, and rs4911442 in the region of ASIP. In the case of SLC45A2, the three SNPs were in very strong disequilibrium, and the noncoding SNPs rs35391 and rs28777 were not significant melanoma risk predictors in multivariate logistic regression models after adjusting for rs16891982.
Table 4.
Results of association analyses of individual pigmentation-associated SNPs and risk of CMM.
| Locus/Variant | SNP | Allelic Odds Ratio for CMM (P-value) | |||
|---|---|---|---|---|---|
| All cases and controls | Subset: 100% Northern European ancestry | Same subset, adjusted for hair, eye and skin colora | Same subset, adjusted for MC1R genotype | ||
| SLC45A2 i3 | rs35391 | 3.71 (<1.0e-4)d | 2.49 (0.0014) | 1.53 (0.11) | 1.68 (0.16) |
| SLC45A2 i3 | rs28777 | 3.75 (<1.0e-4) | 2.37 (0.0012) | 1.68 (0.045) | 2.28 (0.0028) |
| SLC45A2*F374L | rs16891982 | 3.44 (<1.0e-4) | 2.86 (<1.0e-5) | 1.68 (0.0001) | 1.68 (0.0004) |
| IRF4 i4 | rs12203592 | 1.04 (0.32) | 1.18 (0.0075) | 1.05 (0.45) | 1.03 (0.77) |
| TYRP1 | rs1408799 | 1.20 (0.0007) | 1.14 (0.0117) | 1.166 (0.014) | 1.06 (0.56) |
| TYRP1 i6 | rs2733832 | 1.16 (0.0018) | 1.12 (0.0243) | 1.121 (0.025) | 1.06 (0.33) |
| TYR*R402Q | rs1126809 | 1.20 (0.002) | 1.13 (0.001) | 1.19 (0.0059) | 1.37 (0.0004) |
| TYR*S192Y | rs1042602 | 1.05 (0.30) | 1.10 (0.10) | 1.14 (0.040) | 1.17 (0.041) |
| SLC24A4 | rs12896399 | 1.11 (0.24) | 1.12 (0.37) | 1.07 (0.30) | 1.09 (0.24) |
| OCA2*R419Q | rs1800407 | 1.31 (0.004) | 1.18 (0.20) | 1.23 (0.096) | 1.35 (0.022) |
| HERC2 i86 | rs12913832 | 1.15 (0.042) | 1.06 (0.51) | 1.20 (0.078) | 1.21 (0.048) |
| MC1R*V60L | rs1805005 | 1.01 (0.80) | 1.02 (0.87) | 1.02 (0.83) | - |
| MC1R*D84E | rs1805006 | 1.61 (0.013) | 2.15 (0.004) | 1.58 (0.10) | - |
| MC1R*V92M | rs2228479 | 1.07 (0.53) | 1.12 (0.42) | 1.08 (0.62) | - |
| MC1R*R142H | rs11547464 | 2.25 (0.055) | 3.33 (0.014) | 3.85 (0.011) | - |
| MC1R*R151C | rs1805007 | 1.58 (7.0e-4) | 1.39 (0.0042) | 1.00 (0.99) | - |
| MC1R*R160W | rs1805008 | 1.68 (<1.0e-4) | 1.53 (<1e-4) | 1.29 (0.092) | - |
| MC1R*R163Q | rs885479 | 1.15 (0.54) | 1.04 (0.99) | 1.04 (0.83) | - |
| MC1R*D294H | rs1805009 | 1.42 (0.019) | 1.27 (0.87) | 1.01 (0.87) | - |
| MC1R “R”b | 1.65 (<1e-5) | 1.56 (<1e-5) | 1.12 (0.1481) | ||
| MC1R “r” c | 1.10 | 1.08 | 0.95 | ||
| ASIP region | rs4911442 | 1.49 (<1e-5) | 1.46 (<1e-5) | 1.32 (8.0e-4) | 1.39 (0.0037) |
Hair color encoded as 4 category categorical variable (fair, light brown, red, dark). Eye color as 3 category ordinal trait. Skin color as 3 category ordinal trait.
High penetrance red hair color MC1R haplotype: variant at D84E, R142H, R151C, R160W, or D294H.
Low penetrance red hair color MC1R haplotype: variant at V60L, V92M, or R163Q.
Bolding denotes P value < 10−3 (equivalent to P<0.04 after a Bonferroni correction for forty tests).
When we further adjusted for hair, eye and skin colour, several SNPs exhibited a persistent association, suggesting they act as mediating variables (Table 4) between ancestry and melanoma risk. The TYR coding SNP rs1126809 remained important, but rs12913832 in OCA2 (a regulatory SNP actually located in an intron of the adjacent HERC2 gene, but the main determinant of blue eye colour, Sturm et al., 2008) and the TYRP1 SNPs were less impressive as CMM risk factors. The effect of OCA2*R419Q was relatively small in its own right. When we combine the strongest associated SNPs from TYR, SLC45A2, OCA2, MC1R and ASIP in a multivariate logistic regression (Table 5), this gives a Nagelkerke R2 index=0.059. For comparison, a regression including eye, skin and hair color obtained R2=0.048, and supplementing this with the genotype data increased the R2 to 0.08. Because logistic regression R2 are difficult to interpret (Nagelkerke,1991), we also present locus-specific sibling recurrence risk ratios (λS). Under the usual multiplicative model assumption, these can be multiplied to give an total contribution of 1.14 to the sibling recurrence risk. We can calculate the equivalent contribution of measured pigmentation phenotype (skin, hair and eye colour) at 1.37, so that the current SNPs explain approximately 43% of the effects of pigmentation on familial risk (log(1.15)/log(1.37)), and about 15% of the total observed sibling risk λS=2.49 estimated previously in our population (Do et al, 2004).
Table 5.
Multivariate analysis of CMM versus 5 polymorphic pigmentation loci, and pigmentation phenotype in individuals of 100% Northern European ancestry (1062 cases, 1262 controls).
| Variant | Allelic Odds Ratio (no covariates) | Allelic Odds Ratios (pigmentation as covariate) | Locus-specific Sibling Recurrence Risk Ratio5 (Attributable Risk6) |
|---|---|---|---|
| MC1R “r” | 1.10 (0.94–1.27) | 1.09 (0.94–1.27) | 1.047 (19.4%) |
| MC1R “R” | 1.74 (1.49–2.01) | 1.46 (1.24–1.72) | |
| rs16891982*C SLC45A2 |
2.47 (1.51–4.07) | 2.04 (1.27–3.40) | 1.058 (90.6%) |
| rs1800407*A OCA2*R419Q |
1.32 (1.09–1.61) | 1.34 (1.10–1.64) | 1.008 (5.1%) |
| rs1126809*A TYR*Q402R |
1.24 (1.09–1.41) | 1.18 (1.04–1.34) | 1.010 (8.0%) |
| rs4911442*C ASIP |
1.44 (1.23–1.69) | 1.40 (1.19–1.65) | 1.016 (10.8%) |
| Hair colour1 | - | 0.88, 1.50, 0.72, 0.59 (P=1.8e-5)4 | 1.374 |
| Skin Colour2 | - | 0.98, 0.63 (P=0.007)4 | |
| Eye Colour3 | - | 1.22, 1.05 (P=0.10)4 | |
| Total Model R2 | 5.9% | 8.3% |
Five categories (fair, light brown, red, dark brown, black); reference category, fair.
Three categories (light, medium, dark); reference category, light.
Three categories (blue/grey, green/hazel, brown/black); reference category, blue/grey.
P-value from sequential likelihood ratio test.
Assuming a lifetime melanoma risk of 5.2% in the Queensland population
Proportional reduction in disease rate if entire population carried lowest risk genotype.
In analyses including pairwise interactions, we observed an interaction only between OCA2*R419Q and rs1126809 (P=0.01, Supplementary Table 3), though there was a suggestive pattern of interaction between MC1R and ASIP genotypes and CMM which mirrored a highly significant interaction between these loci in their effects of red hair colour (Table 6).
Table 6.
Interaction between rs4911442 (near ASIP) and MC1R effects on red hair color, skin color and CMM risk.
| Joint Genotype | Cutaneous Melanoma | Red Hair | |||||
|---|---|---|---|---|---|---|---|
| MC1R | rs4911442 | Cases | N | Odds Ratio | Red | Total | Penetrance |
| R/R | C/C | 1 | 2 | 5.00 | 1 | 2 | 0.500 |
| C/T | 17 | 34 | 5.00 | 30 | 38 | 0.789 | |
| T/T | 45 | 94 | 4.59 | 73 | 108 | 0.676 | |
| R/r | C/C | 3 | 11 | 1.88 | 5 | 12 | 0.417 |
| C/T | 27 | 60 | 4.09 | 12 | 74 | 0.162 | |
| T/T | 73 | 226 | 2.55 | 13 | 250 | 0.052 | |
| R/w | C/C | 2 | 5 | 3.33 | 0 | 6 | 0.000 |
| C/T | 43 | 112 | 3.12 | 8 | 140 | 0.057 | |
| T/T | 99 | 343 | 2.03 | 13 | 412 | 0.032 | |
| r/r | C/C | 3 | 5 | 7.50 | 0 | 5 | 0.000 |
| C/T | 20 | 35 | 6.67 | 2 | 39 | 0.051 | |
| T/T | 19 | 83 | 1.48 | 0 | 100 | 0.000 | |
| r/w | C/C | 2 | 16 | 0.71 | 0 | 19 | 0.000 |
| C/T | 33 | 127 | 1.76 | 3 | 148 | 0.020 | |
| T/T | 74 | 323 | 1.49 | 2 | 417 | 0.005 | |
| w/w | C/C | 2 | 2 | 24.81 | 0 | 3 | 0.000 |
| C/T | 24 | 101 | 1.56 | 0 | 135 | 0.000 | |
| T/T | 52 | 312 | 1.00 | 0 | 412 | 0.000 | |
As we noted elsewhere (Brown et al., 2008), a number of SNPs spread over the region around ASIP exhibit association to CMM, and the analyses above have concentrated on rs4911442 only because it is genotyped in more controls than the other peak SNPs. The pattern of linkage disequilibrium around ASIP is quite complex, with “hotspots” just distal to the gene and 70 kbp proximally (see Figure 1), and the pattern of individual SNP association to CMM and to red hair color is bimodal, dipping rather than rising over ASIP.
Figure 1.
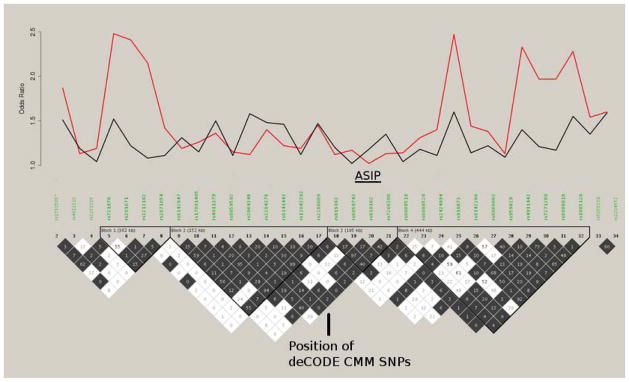
Odds Ratios for red hair color (red line) and cutaneous malignant melanoma (black line) and pairwise linkage disequilibrium (r2, with colors generated using the “4 gamete rule”) versus SNPs around ASIP.
Examining haplotypes comprised of 31 SNPs spanning from rs17305657 to rs1885120 (a distance of 1.77 Mbp) revealed that the commonest haplotype (present in 4% of controls) is significantly increased in CMM cases (7.3%, OR=1.89; see Supplementary Table 4). This haplotype is tagged by the SNPs that are most strongly individually associated with CMM (including rs4911442). When analysis is restricted just to six tagging SNPs, there are eight haplotypes present at greater than 1% study frequency, one (78% of control samples) being significantly increased in controls, and three (3.8%, 3.9%, 0.6%) increased in cases (global X2=47.2, df=14, P=2×10−5). These latter three share only the distal three markers in common, so the most likely location for the susceptibility locus lies downstream to ASIP.
Using a publicly available US population GWAS dataset (Simon-Sanchez et al., 2007), we were able to show that the peak CMM SNP set described by Gudbjartsson et al. (2008) was in strong linkage disequilibrium with rs4911442, despite being 600 kbp distant (P=4×10−13). Specifically, an individual carrying the G-A risk haplotype defined by Gudbjartsson et al. (2008) had a 50% probability of carrying the rs4911442*C on that haplotype, while the other 3 haplotypes were associated with rs4911442*T in 99% of cases.
DISCUSSION
This paper attempts to address two topics. The first is to confirm and quantify the overall contribution of currently recognized pigmentation loci to variation in risk of cutaneous melanoma. Secondly, we expand the evidence of the recently recognized association between SNPs near ASIP and CMM, and attempt to reconcile results from different studies (Gudbjartsson et al. 2008; Brown et al., 2008).
We have replicated association of SNPs at our chosen loci MC1R, OCA2, ASIP, SLC45A2, SLC24A4, TYR, TYRP1, and IRF4 with skin, hair and eye color, and shown that most of these SNPs are also significant predictors of CMM risk in our population. Because some SNPs are not genotyped in all subjects, the dataset suitable for multivariate analysis is reduced, so the confidence intervals around simultaneously adjusted effect sizes are broader than for (most of) the univariate analyses. The pattern is broadly consistent with multiplicative epistasis., with only weak evidence for non-multiplicative interactions The combination of SNPs in the five associated loci (MC1R, SLC45A2, OCA2, TYR, ASIP) explains roughly one-third to one-half of the variation in risk due to observed pigmentation phenotype. Pigmentation phenotype itself explains approximately one-third of the familial aggregation of melanoma. By contrast, the high penetrance CDKN2A mutations segregating in families with multiple cases of melanoma explain only 0.1% of recurrence risk in the total population. This is because they are extremely rare, involved in 0.2% of all Queensland melanoma cases (Aitken et al, 1999), with an allele frequency ~5×10−6. Attributable risk is another measure of the importance of a risk locus, but is not completely satisfactory in that genotypes that are rare in our population may offer the lowest absolute reduction in risk (for example, the rare protective allele in SLC45A2).
This is only the third study reporting an association between SLC45A2 and CMM. Fernandez and coworkers (Fernandez et al., 2008) genotyped the coding polymorphism rs16891982 (F374L) in a collection of Spanish CMM cases and controls, while Guedj et al. (2008) report results from a French case-control study. In keeping with our findings, the minor (L374) allele was protective (7% frequency in Spanish cases and 16% frequency in Spanish controls; 4% and 10% in the French sample). This protective allele is associated with olive and dark skin, and so is least common in northern European populations (1.7% in the Utah-derived CEU HapMap sample), more common in southern Europeans, but is fixed in African and most Asian populations (Yuasa et al., 2006). In our Australian sample, examination of the 3 SNP haplotypes, as well as logistic regression analysis showed that rs16891982 is the SNP most strongly associated with melanoma risk in our population.
The variant in the tyrosinase gene most strongly associated with CMM risk by Gudbjartsson and coworkers (2008) was rs1126809 (R402Q). We found significant association with rs1126809, and any association to the intronic SNP rs10765198 disappeared in multivariate models where rs1126809 was included as a covariate. The Q402 variant form of tyrosinase has 25% of the catalytic activity of the R402 form at 37°C (Tripathi et al., 1991), and causes autosomal recessive ocular albinism when co-occurring with more penetrant OCA mutations in compound heterozygote form (Fukai et al., 1995; Hutton et al., 2008). It is therefore a strong candidate for the causative risk variant in this gene. Like Gudbjartsson and coworkers (2008), we did not detect any association between rs1042602 (TYR*S192Y) with CMM in our Australian sample, even though those authors did detect an association with freckling in their Scandinavian sample.
In the case of OCA2, we found that the SNP (rs12913832) most strongly associated with blue eye colour in our sample (Sturm et al., 2008) was not associated with CMM, especially after adjusting for ancestry. Rather, the less common coding mutation R419Q (rs1800407) was a significant risk factor, notably in the multivariate analysis adjusting for other pigmentation loci. This association has also been seen in a Spanish dataset (Fernandez et al., 2009), with similar effect sizes. In the French case-control study of Jannot et al. (2005), R419Q was not a significant risk factor for melanoma, but the nearby R305W and neighbouring intronic SNPs were. The R305W and R419Q variants are in complete linkage disequilibrium in our population. The OCA2 variants screened by Gudbjartsson et al. (2008) were not associated with CMM.
The TYRP1 (OCA3) variants we genotyped have been previously shown to be associated with eye and hair colour (Sulem et al. 2008), and with CMM risk (Gudbjartsson et al., 2008). We confirmed these associations and effect size estimates. These variants differ significantly in frequency between the main HapMap ethnic samples, and we could demonstrate a gradient with proportion Northern European ancestry paralleling hair and eye colour. We did not detect specific interactions between TYR and TYRP1 in their effects on melanoma risk or pigmentation phenotypes.
The two recent reports of association of SNPs in the region of ASIP to CMM risk (Gudbjartsson et al., 2008; Brown et al., 2008) implicated widely separated SNPs. We observed that the association of CMM to the ASIP region closely paralleled that with red hair color, and seems to be due largely to a single long (approximately 1.8 Mb) haplotype that straddles a number of haplotype block boundaries. This is likely to represent the effects of selection along with a relatively recent origin, as has been observed around other pigmentation loci (Barreiro et al., 2008), given that this particular haplotype is also associated with lighter hair and skin color.
Given the role of Agouti signalling protein (ASIP) as an antagonist of the melanocortin pathway, it seems biologically plausible that there would be an interaction between MC1R genotype and ASIP genotype. For red hair color, this was markedly so, with the rs4911442*C allele being roughly equivalent to an “r” MC1R haplotype. This is seen most easily seen comparing the penetrances of the joint “R/w; C/T” genotype to the “R/r; T/T” genotype, and in the linear dose-response relationship between number of rs4911442*C alleles on a “R/r” background (Table 8). In the same fashion, MC1R “R” haplotypes were dominant to rs4911442*C. Therefore ASIP is a red hair color locus, in the same way as MC1R. In keeping with this, rs4911442 genotype was a significant predictor of facial freckling (P=0.0001), as noted by others (Gudbjartsson et al., 2008). A similar pattern was observed for CMM, but did not reach formal statistical significance. The association signal for red hair color and CMM across chromosome 20 did track quite strongly, so we expect that the epistatic pattern seen for these two loci in respect to hair color would also be demonstrable for CMM given a larger sample size.
An interesting negative finding in this study is the absence of an association between IRF4 genotype and melanoma risk, despite the very strong evidence for association with this locus and skin coloring in this and other studies (Han et al., 2008; Sulem et al., 2008). This was also noted by Gudbjartsson et al. (2008), and is in some ways the obverse of our previous observation (Palmer et al., 2000) that some pigmentation loci such as MC1R seem to affect CMM risk by pathways other than that mediated by measurable changes in skin coloring or tanning ability. Here, we observe an effect on coloring which one would expect to significantly influence response of the organism to exposure to ultraviolet light, but appears neutral with respect to CMM risk. In the case of OCA2, we and others have previously described how a single SNP rs12913832 strongly predicts eye color (Sturm et al., 2008; Kayser et al., 2008; Eiberg et al., 2008), but has little or no effect on CMM risk. We do see an effect of another OCA2 polymorphism rs1800407 on CMM risk, but this is less impressive in terms of statistical significance.
A peculiarity of these analyses is that most of these pigmentation loci markedly differ in allele frequencies between different ethnic groups, in a way that parallels the differences in CMM rates between these groups. The simplest hypothesis, given that we have shown that all the selected variants are associated with pigmentation, is that these variants underlie ethnic variation in CMM risk. To definitively exclude ethnic confounding as a cause of the observed association between ancestry informative variants and disease risk, we would need either a cohort design, where we could show that the rate of CMM in carriers of all low risk variants is comparable to that seen in ethnic groups of known low “intrinsic” risk, or a family based design where we could also demonstrate linkage of these variants. Unfortunately, the age of onset of CMM in our sample and sampling design means that we cannot pursue the latter strategy, although we can demonstrate significant transmission-disequilibrium test results for pigmentation phenotypes such as skin colouring.
In conclusion, the variants in pigmentation genes we describe explain approximately 40% of the variation in familial risk of melanoma ascribable to phenotypes such as skin, hair and eye color. In the cases of TYR, SLC45A2, OCA2 and MC1R, it is likely that the major causative risk variants have been identified, but this is still open for ASIP, and other minor risk variants at all loci await characterization.
MATERIALS AND METHODS
Study participants
CMM cases were a stratified sample of all cases of CMM diagnosed in the state of Queensland in the period 1982–1990, as described in detail elsewhere (Baxter et al., 2008). These individuals were originally studied 1991–1994, but were recontacted and interviewed in 2002–2004.
The controls for the present analysis come from the Brisbane Twin Nevus Study. As described in detail elsewhere (Sturm et al., 2008), adolescent twins, their siblings and parents have been recruited over sixteen years into an ongoing study of genetic and environmental factors contributing to the development of pigmented nevi and other risk factors for skin cancer. The proband twins are recruited at age twelve years via schools around Brisbane, Australia, and followed up at age fourteen. All controls are screened to be unaffected by CMM. The sample is overwhelmingly (>95%) of northern European origin (mainly Anglo-Celtic). All cases and controls gave informed consent to participation in this study, and the study protocol was approved by appropriate institutional review boards.
Ancestry was measured via questions to either the individual or a parent about the country of birth and ancestry of each of the grandparents of the individual. Since most subjects were of European ancestry, we have constructed three ancestry scores based on the proportion of grandparents of Northern European (British, Scandinavian, Danish, Dutch, German, French), Southern European (Spanish, Italian, Greek) and Eastern European ancestry. In addition, the Northern indicator was supplemented by “Celtic” (Ireland, Scotland and Wales), English and Continental indicators, to test whether there were further substructure effects. Ancestry data is available for all cases and controls included in these analyses.
Genotyping and statistical analysis
We selected SNPs for association testing based on association with pigmentation phenotypes in our own and other studies (Palmer et al., 2000; Sturm et al., 2008; Sulem et al., 2007;, Han et al., 2008). In the case of SNPs near ASIP (Brown et al., 2008), we had observed maximal association to CMM at SNPs away from ASIP itself, so the two SNPs reported to be most strongly associated to CMM in the study of Gudbjartsson et al. (2008) were not genotyped. We did not genotype within SLC24A5 (“Golden”), given that the variant associated with human skin colour is almost monomorphic in whites. SNPs were typed using iPLEX™ Gold chemistry on a MALDI-TOF Mass Spectrometer (Sequenom Inc, San Diego). PCR reactions were carried out in 2.5 ul in standard 384-well plates. PCR was performed with 10ng genomic DNA, 0.5 unit of Taq polymerase (HotStarTaq, Qiagen, Valencia, CA), 500 umol of each dNTP, and 100 nmol of each PCR primer. PCR thermal cycling in an ABI-9700 instrument was 15 min at 94°C, followed by 45 cycles of 20 sec at 94°C, 30 sec at 56°C, 60 sec at 72°C. To the completed PCR reaction, 1 μl containing 0.15 units Shrimp Alkaline Phosphatase was added and the reaction incubated for 30 min at 37°C followed by inactivation for 5 min at 85°C. After adjusting the concentrations of extension primers to equilibrate signal-to-noise ratios, the post-PCR primer extension reaction of the iPLEX assay was performed in a final 5 ul volume extension reaction containing 0.1 ul of termination mix, 0.02 ul of DNA polymerase (Sequenom, San Diego, CA), and 600 nM to 1200 nM extension primers. A two step 200 short cycles program was used for the iPLEX reaction: initial denaturation was 30 sec at 94°C followed by 5 cycles of 5 sec at 52°C and 5 sec at 80°C. An additional 40 annealing and extension cycles were then looped back to 5 sec at 94°C, 5 sec at 52°C and 5 sec at 80°C. The final extension was carried out at 72°C for three minutes and the sample was cooled to 20°C. The iPLEX reaction products were desalted by diluting samples with 15 ul of water and adding 3 ul of resin, centrifuged to remove resin. The products spotted on a SpectroChip (Sequenom Inc, San Diego), processed and analysed in a Compact Mass Spectrometer by MassARRAY Workstation (version 3.3) software (Sequenom Inc, San Diego). Since genotyping has been carried out in various subsets of the data, the number of genotyped cases and controls varies from marker to marker. Specifically, some markers are genotyped in the entire collection, while others have been genotyped only in a nested sample of unrelated cases and controls.
Since some cases (105 individuals) are from 48 multiplex families and all our controls come from twin families, the association analyses were carried out using Sib-pair 1.0 (Duffy et al., 2008). The association analysis in Sib-pair implements a logistic regression penetrance analysis of measured genotypes within pedigrees that may include monozygotic twins, and uses Monte Carlo (gene-dropping) simulation to obtain P-values adjusted for the relationships of cases and controls. Other analyses have been carried out in the R statistical analysis environment (R Core Development Team, 2008), using especially the haplo.stats package (Sinnwell et al., 2007), and also using Haploview (Barrett et al., 2005) to generate plots and long range haplotypes. We express the overall contribution of the measured loci to risk of melanoma in logistic regression analyses as the Nagelkerke R2 (Nagelkerke 1991), and have also calculated the locus-specific sibling recurrence risk (James, 1971; Risch, 1990) predicted for that locus under an assumed lifetime risk of 5.2% (Queensland Cancer Registry, 2008). This further assumes a proportional hazards model is appropriate, but is quite robust to the assumed lifetime risk.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRC 241944, 389875, 389891) and the National Cancer Institute (CA88363). RAS, DLD, NKH and GWM are NHMRC Senior Research Fellows. We thank Anjali Henders, Megan Campbell, Shane Thomas and Mitchell Stark for assistance with sample preparation and genotyping. An anonymous reviewer helped greatly with comments.
Footnotes
This study used data from the SNP Database at the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds/).
Conflict of Interest
The authors have no conflict of interest arising from the publication of this paper.
References
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15; doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baxter AJ, Hughes MC, Kvaskoff M, Siskind V, Shekar S, Aitken JF, et al. The Queensland Study of Melanoma: Environmental and Genetic Associations (Q-MEGA); Study design, baseline characteristics, and repeatability of phenotype and sun exposure measures. Twin Res Hum Genet. 2008;11:183–196. doi: 10.1375/twin.11.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box NF, Duffy DL, Chen W, Stark M, Martin NG, Sturm RA, et al. MC1R genotype modifies risk of melanoma in families segregating CDKN2A mutations. Am J Hum Genet. 2001;69:765–773. doi: 10.1086/323412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do K-A, Aitken JF, Green AC, Martin NG. Analysis of melanoma onset: assessing familial aggregation by using estimating equations and fitting variance components via Bayesian random effects models. Twin Res. 2004;7(1):98–113. doi: 10.1375/13690520460741480. [DOI] [PubMed] [Google Scholar]
- Duffy DL. [accessed 7th July 2008];Sib-pair [Computer Program]. Version 1.0b. URL: http://www.qimr.edu.au/davidD#sib-pair.
- Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 2008;123:177–87. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- Fernandez LP, Milne RL, Pita G, Avilés JA, Lázaro P, Benítez J, et al. SLC45A2: a novel malignant melanoma-associated gene. Hum Mutat. 2008;29:1161–7. doi: 10.1002/humu.20804. [DOI] [PubMed] [Google Scholar]
- Fernandez LP, Milne RL, Pita G. Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp Dermatol. 2009 Mar 3; doi: 10.1111/j.1600-0625.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Fukai K, Holmes SA, Lucchese NJ, et al. Autosomal recessive ocular albinism associated with a functionally significant tyrosinase gene polymorphism. Nat Genet. 1995;9:92–95. doi: 10.1038/ng0195-92. [DOI] [PubMed] [Google Scholar]
- Guedj M, Bourillon A, Combadières C, Rodero M, Dieudé P, Descamps V, et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum Mutat. 2008;29:1154–60. doi: 10.1002/humu.20823. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Sulem P, Stacey SN, Goldstein AM, Rafnar T, Sigurgeirsson B, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008 May 18; doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008 May 16;4(5):e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton SM, Spritz RA. A Comprehensive Genetic Study of Autosomal Recessive Ocular Albinism in Caucasian Patients. Invest Ophthalmol Vis Sci. 2008;49:868–872. doi: 10.1167/iovs.07-0791. [DOI] [PubMed] [Google Scholar]
- Jannot AS, Meziani R, Bertrand G, et al. Allele variations in the OCA2 gene (pink-eyed-dilution locus) are associated with genetic susceptibility to melanoma. Eur J Hum Genet. 2005;13(8):913–920. doi: 10.1038/sj.ejhg.5201415. [DOI] [PubMed] [Google Scholar]
- James JW. Frequency in relatives for an all-or-none trait. Ann Hum Genet. 1971;35:47–49. doi: 10.1111/j.1469-1809.1956.tb01377.x. [DOI] [PubMed] [Google Scholar]
- Sinnwell Jason P, Schaid Daniel J, Yu Zhaoxia. haplo.stats: Statistical Analysis of Haplotypes with Traits and Covariates when Linkage Phase is Ambiguous. R package version 1.3.0. 2007 http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm.
- Kayser M, Liu F, Janssens AC, et al. Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J Hum Genet. 2008 Feb;82(2):411–23. doi: 10.1016/j.ajhg.2007.10.003. Epub 2008 Jan 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78:691–692. [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O’Gorman LE, Green AC, et al. MC1R polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet. 2000;66:176–186. doi: 10.1086/302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queensland Cancer Registry. Cancer in Queensland: Incidence and Mortality 1982–2005. Queensland: The Cancer Council Queensland; 2008. [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing Version 271. R Foundation for Statistical Computing; Vienna, Austria: 2008. URL http://www.R-project.org. [Google Scholar]
- Risch N. Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet. 1990;46:222–228. [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Box NF, Newton RA, Shepherd AG, Chen W, et al. Genetic association and cellular function of MC1R variant alleles in human pigmentation. Ann New York Acad Sci 2003. 2003;994:348–358. doi: 10.1111/j.1749-6632.2003.tb03199.x. [DOI] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Zhao ZZ, Leite FP, Stark MS, Hayward NK, et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 2008 Feb;82(2):424–31. doi: 10.1016/j.ajhg.2007.11.005. Epub 2008 Jan 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008 May 18; doi: 10.1038/ng.160. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39(12):1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Tripathi RK, Giebel LB, Strunk KM, Spritz RA. A polymorphism of the human tyrosinase gene is associated with temperature-sensitive enzymatic activity. Gene Expression. 1991;1:103–110. [PMC free article] [PubMed] [Google Scholar]
- Yuasa I, Umetsu K, Harihara S, Kido A, Miyoshi A, Saitou N, et al. Distribution of the F374 allele of the SLC45A2 (MATP) gene and founder-haplotype analysis. Ann Hum Genet. 2006;70:802–11. doi: 10.1111/j.1469-1809.2006.00261.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


