Abstract
Background
Gonad differentiation is one of the most important developmental events in vertebrates. Some heat shock proteins are associated with gonad development. Heat shock protein 70 (Hsp70) in the teleost fish and its roles in sex differentiation are poorly understood.
Methods and Findings
We have identified a testis-enriched heat shock protein Hspa8b2 in the swamp eel using Western blot analysis and Mass Spectrometry (MS). Fourteen Hsp70 family genes were further identified in this species based on transcriptome information. The phylogenetic tree of Hsp70 family was constructed using the Maximum Likelihood method and their expression patterns in the swamp eel gonads were analyzed by reverse transcription-polymerase chain reaction (RT-PCR).
Conclusion
There are fourteen gene members in the Hsp70 family in the swamp eel genome. Hsp70 family, particularly Hspa8, has expanded in the species. One of the family members Hspa8b2 is predominantly expressed in testis of the swamp eel.
Introduction
Gonad differentiation is one of the most important developmental events in vertebrates, and the underlying molecular mechanism is always a debate focus. A number of key genes involved in sex determination have been identified in mammals, such as Sry (sex-determining region on Y chromosome), which encodes a DNA-binding protein that acts dominantly to trigger differentiation of testes from undifferential gonads [1], [2]. Sox9 can induce testis development in the XX transgenic mice (Mus musculus), a condition of complete absence of Sry [3], [4] and it may be a direct downstream target of Sry to initiate the male development. However, Sox9 is dispensable during subsequent embryonic and postnatal testis development, only leads to late-onset sterility at about 5 months [5], [6]. Sox8 may play a role in maintenance of the integrity of the basal lamina [7]. Together, sex determination in mammals is regulated by two groups of factors: Sox (Sry, Sox8, Sox9 and Sox10) and the Rspo1/Wnt/beta-catenin [8], [9], [10], [11]. In addition, insulin and Igf1 receptors are essential for testis determination and/or differentiation in mice [12], [13], and Dmrt1 is evolutionarily conserved and play a prominent role in regulation of testicular differentiation and gametogenesis in vertebrates [6], [14]. In fish, Dmy/Dmrt1Y, which is a duplicated copy of Dmrt1 on the Y chromosome, is required for male sex determination in the teleost fish medaka (Oryzias latipes) [15], [16]. Recently, four novel sex determining genes in the teleost fish species were identified, and they were amhy in the Patagonian pejerrey (Odontesthes hatcheri), Amhr2 in fugu (Takifugu rubripes), sdY in rainbow trout (Oncorhynchus mykiss) and Gsdf in Oryzias luzonensis (a relative of medaka) [17], [18], [19], [20]. Together, sex determination and differentiation in fish are complex because of diverse range of species and common molecular mechanisms need to be studied.
Some heat shock proteins are closely related with gonad development and spermatogenesis. Down regulation of Hsp10 will result in apoptosis in testis, which provided new aspects for understanding the mechanisms of germ cell apoptosis [21]. Hspa2 (also named Hsp70-2) is a heat shock protein involved in maintenance of the nucleolus and centrosome integrity in cancer cells subjected to heat shock and protecting cells against cytotoxic stress [22], [23], [24], [25]. In addition, Hspa2 plays an important role during meiosis in mouse [26], [27], even post-meiosis [28]. And male mice lacking Hspa2 were infertile while females fertile [26]. HSPA2 has a similar expression pattern in the male germ cells of human and mouse, and is also associated with sperm morphology and concentration [29], [30], [31], [32]. We have previously identified Hsp10 in the swamp eel and found that down regulation of Hsp10 is consistent with high apoptosis during the gonadal transformation. However, we are still lack of understanding of the roles and mechanisms of heat shock proteins in sexual differentiation.
In this study, we have cloned a testis-enriched Hsp70 member, and further identified all members of Hsp70 family in the swamp eel (Monopterus albus), a freshwater fish with a characteristic of natural sex reversal [33]. These results in the Hsp70 family genes might shed light on Hsp70 family evolution and their roles in gonadal development.
Materials and Methods
Animals and Reagents
The swamp eels were obtained from markets in the Wuhan area of China. Their gonads were confirmed by microscopic analysis of sections. Mice (Kunmingbai) were purchased from Wuhan Center for Disease Prevention and Control of Hubei in China. The polyclonal antibodies: anti-human NF-kB1 p50 (NLS) and anti-β-actin were purchased from Santa Cruz Biotechnology (CA, USA); the secondary antibody conjugated with AP was purchased from Vector (Buringame, CA, USA). The animals were treated in accordance with the guiding principles for biomedical research involving animals of Ethics and Animal Welfare Committee of College of Life Sciences of Wuhan University and the committee.
Western Blot Analysis
Western blots were performed as the routine protocols [21]. We extracted proteins from freshly obtained tissues with buffer containing 50 mM Tris-Cl (pH 7.5), 140 mM NaCl, 1% Nonidet P-40, 2mM EDTA and complete inhibitor cocktail. Then we analyzed whole extract by Glycine-SDS-PAGE and transferred them onto 0.45 µm PVDF membrane (Hybond-P, Amersham Pharmacia Biotech, Sweden). The membranes were blocked with 5% low fat milk powder in TBST (20mM Tris-HCL pH7.5, 150mM NaCl, 0.1% Tween 20) and incubated with primary antibody at 4°C over night, and then with AP labeled secondary antibody for 1 hour at 25°C. The immunoreactive signals were revealed by NBT/BCIP regents.
Immunoprecipitation (IP) and Mass Spectrometry (MS) Analysis
Proteins from the swamp eel and mouse gonads were extracted in IP buffer containing 50 mM Tris-Cl (pH 7.5), 140 mM NaCl, 1% Nonidet P-40, 2mM EDTA and complete inhibitor cocktail. The lysates were centrifuged at 10,000 g for 30 min at 4°C and precleared with Protein G PLUS-Agarose (Santa Cruz Biotech, USA). The precleared lysates (1 mg of protein) were incubated with 1 µg of anti-NF-kB1 p50 (NLS) antibody for 12 hrs at 4°C. Samples were washed three times with NP-40 lysis buffer, resuspended in 5X protein sample buffer, and placed for 5 min in boiling water, then placed in ice at once. A part of samples were analyzed by Western blot analysis with anti-NF-kB1 p50 (NLS) antibody. Most of samples from IP were analyzed by SDS-PAGE, and stained by Coomassie brilliant blue. Specific bands were cut from gels and put into 1.5mL EP tubes for MS analysis. The gels were washed twice with Milli-Q water for 15 min, and then washed three times in 25 mM NH4HCO3 and 50% CH3CN for 30 min with vortexing. The gels were then dehydrated in 100% CH3CN for 10 min with vortexing and allowed to air-dry for 1 h. Suspension of 1.5 mM trypsin (Promega, Madison, WI, USA) in 25 mM NH4HCO3 was added to gels, then digested at 37°C overnight. The 1.5ml EP tubes were then gently centrifuged, and the supernatant was removed for the MS analysis. Peptide mixtures were analyzed by the MALDI-TOF/TOF MS (Bruker-Daltonics AutoFlex TOF-TOF LIFT Mass Spectrometer, Bruker-Daltonics). Proteins were identified using the Mascot software (http://www.matrixscience.com) and SWISS-PROT, NCBInr database. Valid identification required a protein score greater than 65 when peptide mixtures were analyzed using the MALDI-TOF/TOF MS.
Cloning of Hsp70 Family Genes
Based on the amino acid sequences from MS analysis, the transcriptome data (unpublished) of the swamp eel were searched using BLASTp to find all related genes. PCR primers were designed to clone these genes. Primer sequences and amplification conditions were shown in Table 1. PCR was performed in a 20 µl reaction mixture containing 100ng DNA, 10mM Tris-HCl pH 8.3, 1.5mM MgCl2, 50mM KCl, 200 µM dNTP, 0.2 µM each primer, and 1 U Taq DNA polymerase. Amplified products were electrophoresed in a 0.8% agarose gel. The PCR products were cloned into T-vector and sequenced.
Table 1. The primers for sequencing.
| Primer name | Primer sequences (5′-3′) | PCR |
| Hspa1a | ||
| Hspa1a(F1) | ATGTCTGCAGCTAAAGGTGT | 94°C, 30 s; 55°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa1a(R1) | TCAATCTACCTCCTCAATAG | |
| Hspa1b | ||
| Hspa1b(F1) | ATGTCCTCAGCTAAAGGAAT | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa1b(R1) | TTAGTCCACTTCCTCAATAG | |
| Hspa4a | ||
| Hspa4a(F1) | ATGGCAGTTGTCGGATTTGA | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa4a(R1) | TTAATCAAGGTCCATGTCAG | |
| Hspa4b | ||
| Hspa4b(F1) | ATGTCAGTGGTGGGATTTGA | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa4b(R1) | TTAGTCAATGTTCATTTCAG | |
| Hspa4L | ||
| Hspa4L(F1) | ATGTCAGTGGTAGGCATTGA | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa4L(R1) | CTCCATCTCTTTCGTGCCAG | |
| Hspa5 | ||
| Hspa5(F1) | ATGAAGCTGTTATGGGTTGT | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa5(R1) | CTACAACTCATCCTTCTCAT | |
| Hspa8a1 | ||
| Hspa8a1(F1) | ATGTCTAAAGGACCAGCAG | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa8a1(R1) | TTAGTCGACCTCTTCAATGG | |
| Hspa8a2 | ||
| Hspa8a2(F1) | ATGTCAGTGGTAGGCATTGA | 94°C, 30 s; 57°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa8a2(R1) | TCAGTCGACTTCCTCGATGG | |
| Hspa8b1 | ||
| Hspa8b1(F1) | ATGTCCAAGGGACCAGCAGT | 94°C, 30 s; 61°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa8b1(R1) | TTAGTCAACCTCCTCAATGG | |
| Hspa8b2 | ||
| Hspa8b2(F1) | ATGTCTAAAGGACCAGCAGT | 94°C, 30 s; 57°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa8b2(R1) | CTGGTATAGCTTGGAGATGA | |
| Hspa9 | ||
| Hspa9(F1) | GGTTTCCAGCCAGATGTTCT | 94°C, 30 s; 57°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa9(R1) | TTAATCGTCATGACATATTG | |
| Hspa12a | ||
| Hspa12a(F1) | ATGGCTAACCCTTCTCCAGC | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa12a(R1) | TTAATGGCTCAGGAAGTCAA | |
| Hspa12b | ||
| Hspa12b(F1) | GACAGTCCATCAGCCCCGTC | 94°C, 30 s; 59°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa12b(R1) | TCAGTTAGACAGGAAGTCTA | |
| Hspa14 | ||
| Hspa14(F1) | ATGGCTGCGATTGGAGTCCA | 94°C, 30 s; 61°C, 30 s; 72°C, 2 min; 35cycles |
| Hspa14(R1) | TTATGAAGCAGCCGCTATGG |
Phylogenetic Analysis
We searched homologous Hsp70 genes of other vertebrates, including human, mouse, rat (Rattus norvegicus), camel (Camelus dromedarius), cattle (Bos taurus), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), frog (Xenopus laevis), zebrafish (Danio rerio), medaka, pufferfish (Tetraodon nigroviridis), stickleback (Gasterosteus aculeatus), fugu (Takifugu rubripes) and tilapia (Orepchromis niloticus) in NCBI and Ensembl databases. Hsp70 family members have many synonyms, including Hspa1a (other aliases: Hsp70-1a, Hsp70I, Hsp70-1, Hsp72 and Hspa1), Hspa1b (other aliases: Hsp70-1, Hsp68 and Hsp70.1), Hspa1L (other aliases: Hsp70-1L, Hsp70T, Hsp70-hom and hum70t), Hspa2 (other aliases: Hsp70-3 and Hsp70-2), Hspa4 (other aliases: Hsp70RY, APG-2, Hsph2 and HS24/P52), Hspa4L (other aliases: Osp94 and APG-1), Hspa5 (other aliases: Bip, Grp78 and Mif2), Hspa6 (other aliases: Hsp70B), Hspa8 (other aliases: Hsc54, Hsc71, Hsc70, Hsp71, Hsp73, Hspa10, Lap1 and Nip71), Hspa9 (other aliases: CSA, Grp-75, Hspa9b, Grp75, Mot, Mot2, mtHsp75 and Pbp74), Hspa12 and Hspa14 (other aliases: Hsp70-4 and Hsp70L1). We constructed phylogenetic tree using the Maximum Likelihood method in 100 bootstrap replicates (PHYLIP, version 3.68) (protein ID showed in Table S1).
Sequence and Domain Analysis of Hsp70 Proteins
Hsp70 protein sequences and domains of the swamp eel were analyzed using Interproscan software. The protein sequences were aligned with the mammalian Hsp70 proteins. A complete protein alignment was generated by ClustalX V2.0 and Genedoc 2.7.0 (protein ID in Table S1), and the positives and identities were analyzed by Vector NT 12.0.
RT-PCR Analysis
Total RNAs were isolated from adult tissues with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cDNAs were reverse transcribed from the RNAs using the MMLV system (Promega, Madison, WI, USA) with 0.5 µg of oligo (dT)18 and 5 µg of total RNAs in a 25 µl reaction. PCR was performed in a 20 µl reaction mixture containing 100ng DNA, 10mM Tris-HCl pH 8.3, 1.5mM MgCl2, 50mM KCl, 200 µM dNTP, 0.2 µM each primer, and 1 U Taq DNA polymerase, and cDNA templates from testis, ovotestis and ovary of the swamp eel. Primers designed for semi-quantity PCR and amplification conditions were showed in Table 2. Amplified products were electrophoresed in a 2% agarose gel.
Table 2. Primers for semi-quantitative RT-PCR analysis.
| Primer name | Primer sequences(5′–3′) | PCR |
| Hspa1a | ||
| Hspa1a(F2) | TCCCAGCGACAGGCGACTAA | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa1a(R2) | CTGGCTGATGTCCTTCTTGTGC | |
| Hspa1b | ||
| Hspa1b(F2) | ACGAAATTGTCCTGGTTGGC | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa1b(R2) | GGTATTGCGTTTGATTAGTGGTGT | |
| Hspa4a | ||
| Hspa4a(F2) | CAGTTGAAATAGTGGGTGGAG | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa4a(R2) | AGGAGATGGGATAAGGAA | |
| Hspa4b | ||
| Hspa4b(F2) | CAGTGACTTTAGCGTATGG | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa4b(R2) | CTCGCAGAAGTGGTTGA | |
| Hspa4L | ||
| Hspa4L(F2) | TCCCTTTCCCATTACTCTTCG | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa4L(R2) | CACGCACTTTGACCTTCACTTT | |
| Hspa5 | ||
| Hspa5(F2) | ACCAGCCTACTGTCACTATTA | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa5(R2) | TTGTTCTTGTTGCCTGTG | |
| Hspa8a1 | ||
| Hspa8a1(F2) | ATGGATAAAGGGCAGATTC | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa8a1(R2) | CCAGGGACAGAGGAGTGA | |
| Hspa8a2 | ||
| Hspa8a2(F2) | ACTCCTCTGTCCCTGGGTATTG | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa8a2(R2) | CCTTGCTGAGACGACCCTTG | |
| Hspa8b1 | ||
| Hspa8b1(F2) | GGGGTGCTCATTCAGGTGTT | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa8b1(R2) | TCTGCTTCCTGAACCATACGC | |
| Hspa8b2 | ||
| Hspa8b2(F2) | GGAGGCTGAGCAAGGAGGAA | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa8b2(R2) | CTGTCCAGCCAGGCAATAACT | |
| Hspa9 | ||
| Hspa9(F2) | TGTGGGCATACCTGCTAAACG | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa9(R2) | TTGGTAGCCTGAGAATCAAAGAAA | |
| Hspa12a | ||
| Hspa12a(F2) | CTCCAACCACAATCCTGCTGAC | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa12a(R2) | GGCTTTGACTCGCTTCCCATT | |
| Hspa12b | ||
| Hspa12b(F2) | GCAGCCTGGGTGGACCTAAC | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa12b(R2) | TGGTGATGGTGGGTTGGAAGA | |
| Hspa14 | ||
| Hspa14(F2) | TGAGCGTGACAGTGCTACAGG | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hspa14(R2) | TCAAAGTCAATGCCGTCGTG | |
| Hprt | ||
| Hprt(F) | GAACAGTGACCGCTCCATCC | 94°C, 30 s; 58°C, 30 s; 72°C, 30 s; 30cycles |
| Hprt(R) | TCTTCATCGTCTTTCCCGTGTC |
Results
Identification of a Testis-enriched Hsp70 Gene in the Swamp Eel
Western blot analysis using anti-NF-kB1 antibody in HeLa cells showed normal two bands [50 kD (P50) and 105 kD (P105)], while only a testis-enriched band of 70 kD in the swamp eel was observed (Figure 1a). Further immunoprecipitation using anti-NF-kB1 antibody clearly showed the object unknown protein in testis samples of both the swamp eel and mouse (Figure 1b). The MALDI-TOF-TOF MS analysis was used to identify the proteins of both the swamp eel and mouse. The results indicated that the unknown testis-enriched protein was similar to Hsp70 family member Hspa2 or Hspa8 (Table 3), which had a conserved domain Hspa1–2_6–8-like_NBD (nucleotide-binding domain).
Figure 1. Western blot analysis of unknown testis-enriched protein in mouse and swamp eel.
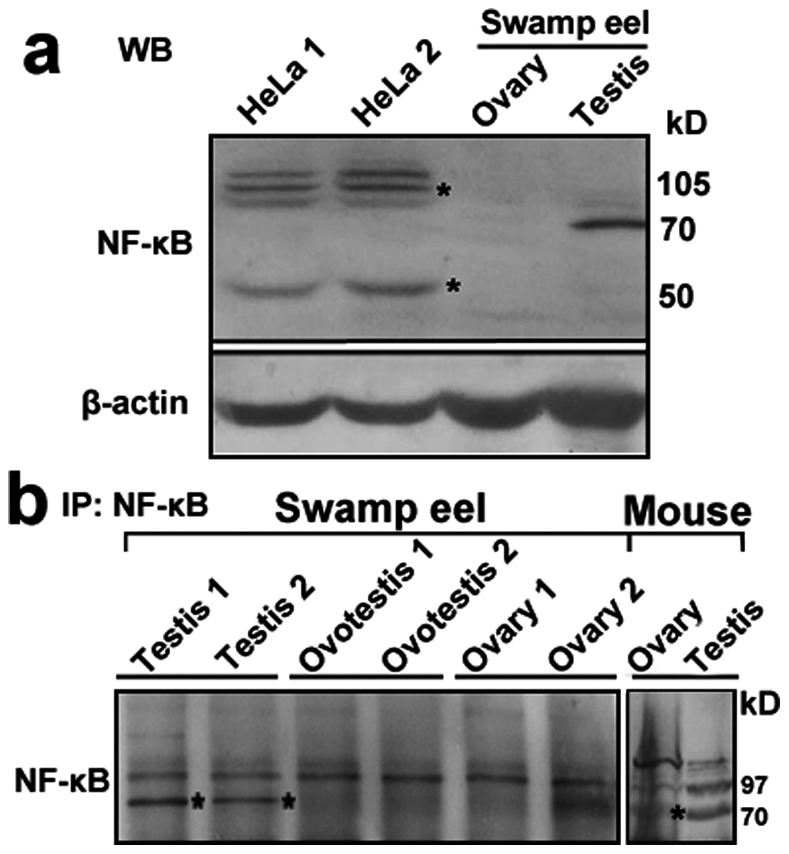
a. Western blot analysis using anti-NF-kB1 antibody in HeLa cells and the swamp eels showed two bands: 50kD (P50) and 105kD (P105) (stars denote the bands) in HeLa cells (two repeat samples), while only a dominant 70 KD band in testis of the swamp eel. β-actin protein was used as an internal control. Molecular weight sizes for proteins were shown on the right. b. Immunoprecipitation using anti-NF-kB1 antibody enriched the unknown testis-enriched protein in the swamp eel and mouse. Testis samples showed more obvious band of 70 kD in the swamp eel and mouse. Stars denote the band.
Table 3. MS results of the unknown proteins from mouse and the swamp eel.
| Species # Protein groups | Amino Acid Sequences | GenBank Accession No. | Identified Protein Names | Coverage | MASCOT Scores |
| Mouse | |||||
| #1–1 | K.DAGTITGLNVLR.I | NP_001002012 | Heat shock protein 2 (Hspa2) [Mus musculus] | 17.54% | 805.6 |
| K.LDKGQIQEIVLVGGSTR.I | |||||
| K.LLQDFFNGK.E | |||||
| K.NAVESYTYNIK.Q | |||||
| K.NQVAMNPTNTIFDAK.R | |||||
| K.VQSAVITVPAYFNDSQR.Q | |||||
| R.IINEPTAAAIAYGLDK.G | |||||
| R.IINEPTAAAIAYGLDKK.V | |||||
| R.TTPSYVAFTDTER.L | |||||
| #2–1 | K.DAGTIAGLNVLR.I | NP_112442 | Heat shock cognate 71 (Hspa8) [Mus musculus] | 10.68% | 1339.8 |
| K.LLQDFFNGK.E | |||||
| K.TVTNAVVTVPAYFNDSQR.Q | |||||
| R.IINEPTAAAIAYGLDK.G | |||||
| R.IINEPTAAAIAYGLDKK.V | |||||
| R.TTPSYVAFTDTER.L | |||||
| #3–1 | K.APQVSTPTLVEAAR.N | NP_033784 | Serum albumin precursor [Mus musculus] | 6.91% | 690.1 |
| #4–1 | R.NHTLQKWHDK.T | XP_993489.1 | similar to Protein AATF [Mus musculus] | 1.88% | 453.8 |
| Swamp eel | |||||
| #1–1 | K.TVLTQEALISVK.G | XP_003976390.1 | PREDICTED: protein slowmo homolog 2-like [Takifugu rubripes] | 6.35% | 963 |
| #2–1 | K.YKLIKLGMSK.V | CAG00227.1 | unnamed protein product [Tetraodon nigroviridis] | 2.89% | 849.6 |
| #3–1 | K.FTASGGEGMLSILKK.S | CAG11484.1 | unnamed protein product [Tetraodon nigroviridis] | 2.15% | 374 |
| #4–1 | K.HQRELENLQEEKER.L | CAF91893.1 | unnamed protein product [Tetraodon nigroviridis] | 0.8% | 897.9 |
| #5–1 | K.LENVLLDENLNIK.I | XP_003444058.1 | PREDICTED: NUAK family SNF1-like kinase 1 [Oreochromis niloticus] | 2.02% | 357.1 |
| #6–1 | K.TVNNAVITVPAYFNDSQR.Q | NP_001098270.1 | Heat shock cognate 71 (Hspa8) [Oryzias latipes] | 2.62% | 185.5 |
| #7–1 | R.ISITLVSVPLIVR.Y | XP_003973504.1 | PREDICTED: protein FAM210B-like [Takifugu rubripes] | 4.44% | 483.8 |
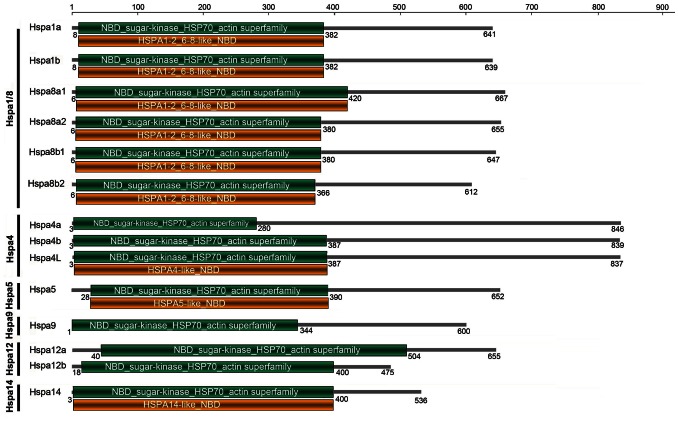
To further characterize the testis-enriched gene, we cloned 14 members of Hsp70 family from the swamp eel (Table S1 and Figure 2). Conserved domain analysis showed that Hspa1a/b and Hspa8a/b had the same Hspa1–2_6–8-like_NBD domain (Figure 2). Further sequence analysis showed that there were four copies of Hspa8 (–8a1/−8a2 and –8b1/−8b2), three copies of Hspa4 (–4a/−4b and –4L), two copies of Hspa1 (–1a/−1b) and Hspa12 (–12a/−12b), one copy of Hspa5, Hspa9 and Hspa14 in the swamp eel.
Figure 2. Conserved domain analysis of Hsp70 family members in swamp eel.
All the 14 proteins had NBD_sugar-kinase_HSP70_actin superfamily domain (green bars), and some genes had their own conserved domain (yellow bars). Hspa1a/b and Hspa8a1/a2/b1/b2 had a common domain (HSPA1–2_6–8-like_NBD). The numbers on the bars denote amino acid positions. GenBank accession numbers were shown in Table S1.
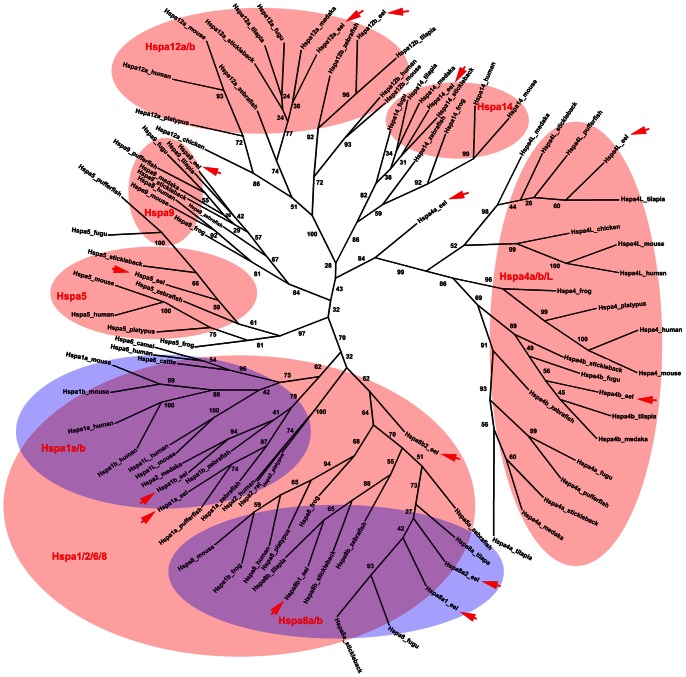
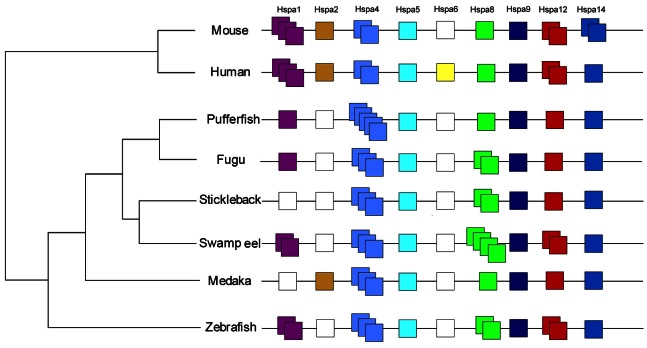
Phylogenetic and Copy Number Analysis of Hsp70 Family Genes in Vertebrates
As Hsp70 family members have many synonyms, we aligned all Hsp70 members of vertebrates and analyzed their clusters using the Maximum Likelihood method. A phylogenetic tree was finally constructed (Figure 3). Each Hsp70 gene in the swamp eel could be grouped into a cluster of Hsp70 family. The evolutionary analysis of these Hsp70 family genes in vertebrates showed that more copies of Hsp70 members were observed in fishes than mammals (Figure 4), especially 3–5 copies of Hspa4 in the fish species. Because of a high level of homology between Hspa8 and Hspa2, we further compared their sequence homology to determine the branching of the Hspa8 and Hspa2 genes. The protein sequence alignments of the swamp eel Hspa8 (–8a1/−8a2 and –8b1/−8b2) with the mammalian Hspa8 and Hspa2 indicated that the swamp eel Hspa8b2 was more similar to Hspa8 (positive and identity of Hspa8b2 with Hspa8 of human and mouse: 100% and 86.7%; Hspa8b2 with Hspa2 of human and rat: 98.9% and 82.5%, respectively) (Figure S1 a, b, c). Thus four copies of Hspa8 genes (named as Hspa8a1/−a2 and Hspa8b1/−b2) in the swamp eel were further confirmed. Hspa2 and Hspa6 may be lost in the teleost fish species (Figure 4), and previously named Hspa2 in medaka may be Hspa1b (Figure 3).
Figure 3. Phylogenetic tree of Hsp70 family in vertebrates.
The phylogenetic tree was constructed using the Maximum Likelihood method based on the amino acids of Hsp70 members from human, mouse, rat, camel, cattle, platypus, chicken, frog, zebrafish, pufferfish, fugu, stickleback, medaka, tilapia and the swamp eel. The protein sequences were from NCBI and Ensembl database (protein ID in Table S1). 14 proteins of the swamp eel had been grouped into 7 cluster of Hsp70 family (red arrows). The numbers at the nodes indicate bootstrap values.
Figure 4. Copy number variation analysis of Hsp70 family genes from various vertebrates.
Hspa5 and Hspa9 had one copy in these species, while copy numbers of the other genes ranged from 0 to 5. Hspa4 and Hspa8 had duplicated in fishes, while Hspa2 and Hspa6 may be lost in fishes (previously named Hspa2 in medaka would be Hspa1). Squares with different colors represent different genes and copies, while white squares denote the genes, which have not been detected in these species.
Expression Analysis of Hsp70 Family Genes
We further analyzed expression pattern of Hsp70 family genes in three types of gonads in the swamp eel by RT-PCR. Most of the Hsp70 genes were expressed equally in different types of gonads except Hspa4L, Hspa5, Hspa8b2 and Hspa9. Hspa9 was expressed faintly in gonads. Only Hspa8b2 and Hspa5 were upregulated from ovary, ovotestis to testis (Figure 5). Hspa8b2 gene has Hspa1–2_6–8-like_NBD conserved domain, which is similar to the protein identified by the MS analysis. Together with phylogenetic analysis, these results indicated that the testis-enriched gene may be Hspa8b2 in the swamp eel, instead of Hspa2.
Figure 5. RT-PCR analysis of Hsp70 gene expression in gonad samples of the swamp eel.
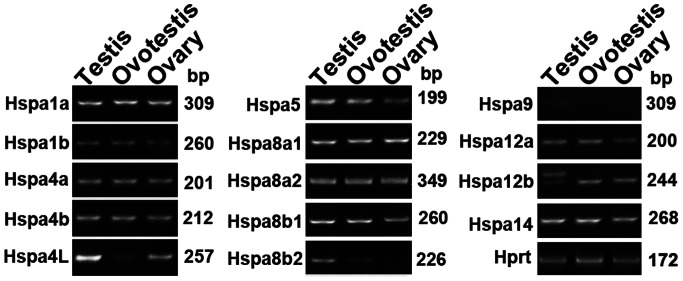
Most of the Hsp70 family genes were expressed equally in different types of gonads except Hspa4L, Hspa5, Hspa8b2, Hspa9 and Hspa12a. Hspa8b2 was expressed differentially among testis, ovotestis and ovary. Hprt was used as an internal control. Length sizes of amplified products were shown on the right.
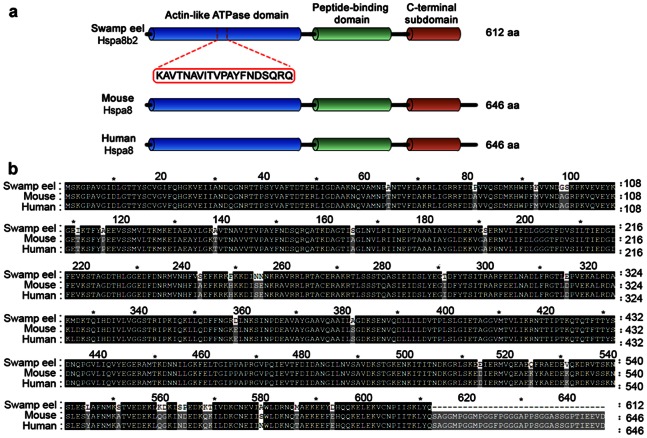
Hspa8 is Evolutionarily Conserved with Three Conserved Domains
Protein sequence of Hspa8b2 in the swamp eel was further aligned with the mammalian Hspa8, which showed a high level of sequence homology. Conserved domains were further analyzed using Interproscan software. Three conserved domains were identified, which were actin-like ATPase domain, peptide-binding domain and C-terminal subdomain (Figure 6a). In addition, in comparison with those of mouse and human, the protein size of the swamp eel Hspa8b2 was shorted of 34 amino acids, and the positive and identity of the alignments were 100% and 86.7% respectively (Figure 6a, b).
Figure 6. Sequence analysis of Hspa8b2 protein of the swamp eel.
a. Cartoon showed a linear representation of the swamp eel Hspa8b2 in comparison with Hspa8 of mouse and human. Different colors showed three conserved domains. Protein sequence in red square showed the corresponding sequence, which is similar to Hspa8 identified by MS analysis. b. A complete protein alignment of the swamp eel Hspa8b2 with Hspa8 of mouse and human using ClustalX V2.0 and Genedoc 2.7.0 (protein ID in Table S1), and the positive and identity of the alignments were 100% and 86.7% respectively. White letters on black background indicated identical amino acids.
Discussion
Hsp70 family genes play crucial roles in protecting cells against heat and other stresses in most animal species. In the present study, we have identified 14 members of Hsp70 family in the swamp eel. In comparison with this, there are about 10 members in the Hsp70 family in other fish species including zebrafish, medaka, pufferfish, threespine stickleback, pufferfish and fugu. Particularly Hspa8 has expended to 4 members in the swamp eel genome, while other fishes have 1–2 copies. The expansion of Hsp70 family members in the swamp eel may reflect its adaptation to physiological change in sex reversal and water/air-breathing of the species. The expansion to 88 members of the Hsp70 family in the Pacific oyster Crassostrea gigas genome is a typical example to adapt to sessile life in the highly stressful intertidal zone [34].
As a multigene family, Hsp70 genes have evolved by duplications, subfunctionalization, nonfunctionalization or even loss. The birth-and-death model may explain well the evolution mechanism of the Hsp70 family. New genes are created by gene duplication and some duplicated genes stay in the genome for a long time, whereas others are inactivated or deleted from the genome [35], [36]. Gene duplication is one of the main forces acting on the evolution of organisms, which creates new genes for adaptation evolution in favor of natural selection.
Hsp70 genes have many functions including protein transportation between cellular compartments, protein folding, degradation of unstable and misfolded proteins, and control of regulatory proteins [37], [38], [39]. Some Hsp70 family proteins can prevent caspase-independent cell death [40], [41], while most Hsp70 genes play crucial roles in protecting cells against heat and other stresses [34]. Hsp70 family members in cells have different but closely related gene products: the stress-inducible form such as Hspa1a, Hspa1b and Hspa6, and the constitutively expressed form, such as Hspa1L, Hspa2, Hspa5, Hspa8 and Hspa9 [42]. Some Hsp70 family members have tissue-special functions. For example, Hspa1a/b proteins can be released from cells and act as messengers and play a role in the immune system [43], [44]. Hspa1L and Hspa2 are sperm proteins which are important for sperm functions [45], [46]. Hspa8 is important in viral assembly in cells, independently of its chaperone function [47]. Some heat shock proteins have functions in gonad development, for example, Hsp10 and Hspa2 [21], [30]. Here, we have further identified testis-enriched Hspa8b2 in the swamp eel. Phylogenetic analysis, sequence characteristic and the expression pattern support this gene as a candidate for gonad development/spermatogenesis. This finding derives from Western blot analysis of gonadal samples using NF-kB1 antibody. In fact, it has been predicted that NF-kB protein could interact selectively and non-covalently with heat shock protein 70, which was based on Gene Ontology (Function GO:0031072) [48]. Further studies of roles and functions of Hspa8b2 will provide comprehensive understanding of the molecular mechanisms of gonadal transformation from ovary to testis via ovotestis in the special fish species, the swamp eel.
Supporting Information
Protein alignments of the swamp eel Hspa8b2 with Hspa2 and Hspa8 of mammals. a. Complete protein alignments of Hspa8a1/a2/b1/b2 of the swamp eel with Hspa8 of human and mouse. b. Complete protein alignments of Hspa8a1/a2/b1/b2 of the swamp eel with Hspa2 of human and rat. c. Complete protein alignments of Hspa8b2 of the swamp eel with Hspa2 of human and rat. White letters on black background indicated identical amino acids, and these genes accession numbers were showed in Table S1.
(TIF)
List of protein sequences used for sequence analysis.
(DOC)
Funding Statement
This work was supported by the National Natural Science Foundation of China, the National Key Basic Research project, National Key Technologies R&D Program and the Chinese 111 project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, et al. (1990) Genetic evidence equating SRY and the testis-determining factor. Nature 348: 448–450. [DOI] [PubMed] [Google Scholar]
- 2. Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R (1991) Male development of chromosomally female mice transgenic for Sry. Nature 351: 117–121. [DOI] [PubMed] [Google Scholar]
- 3. Wagner T, Wirth J, Meyer J, Zabel B, Held M, et al. (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79: 1111–1120. [DOI] [PubMed] [Google Scholar]
- 4. Kikuchi K, Hamaguchi S (2013) Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn 242: 339–53. [DOI] [PubMed] [Google Scholar]
- 5. Barrionuevo F, Scherer G (2010) SOX E genes: SOX9 and SOX8 in mammalian testis development. Int J Biochem Cell Biol 42: 433–436. [DOI] [PubMed] [Google Scholar]
- 6. Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D (2000) Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev 14: 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Georg I, Barrionuevo F, Wiech T, Scherer G (2012) Sox9 and Sox8 are required for basal lamina integrity of testis cords and for suppression of FOXL2 during embryonic testis development in mice. Biol Reprod 87: 99. [DOI] [PubMed] [Google Scholar]
- 8. Wilhelm D (2007) R-spondin1–discovery of the long-missing, mammalian female-determining gene? Bioessays 29: 314–318. [DOI] [PubMed] [Google Scholar]
- 9. Wei Q, Yokota C, Semenov MV, Doble B, Woodgett J, et al. (2007) R-spondin1 is a high affinity ligand for LRP6 and induces LRP6 phosphorylation and beta-catenin signaling. J Biol Chem 282: 15903–15911. [DOI] [PubMed] [Google Scholar]
- 10. Tomizuka K, Horikoshi K, Kitada R, Sugawara Y, Iba Y, et al. (2008) R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum Mol Genet 17: 1278–1291. [DOI] [PubMed] [Google Scholar]
- 11. Lavery R, Chassot AA, Pauper E, Gregoire EP, Klopfenstein M, et al. (2012) Testicular differentiation occurs in absence of R-spondin1 and Sox9 in mouse sex reversals. PLoS Genet 8: e1003170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pitetti JL, Calvel P, Romero Y, Conne B, Truong V, et al. (2013) Insulin and IGF1 receptors are essential for XX and XY gonadal differentiation and adrenal development in mice. PLoS Genet 9: e1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nef S, Verma-Kurvari S, Merenmies J, Vassalli JD, Efstratiadis A, et al. (2003) Testis determination requires insulin receptor family function in mice. Nature 426: 291–295. [DOI] [PubMed] [Google Scholar]
- 14. Zarkower D (2013) DMRT genes in vertebrate gametogenesis. Curr Top Dev Biol 102: 327–356. [DOI] [PubMed] [Google Scholar]
- 15. Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, et al. (2002) DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- 16. Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, et al. (2002) A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci USA 99: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, et al. (2012) A Y-linked anti-mullerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci U S A 109: 2955–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, et al. (2012) Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, et al. (2012) A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet 8: e1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, et al. (2012) An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Current Biology 22: 1423–1428. [DOI] [PubMed] [Google Scholar]
- 21. He Y, Shang X, Sun J, Zhang L, Zhao W, et al. (2010) Gonadal apoptosis during sex reversal of the rice field eel: implications for an evolutionarily conserved role of the molecular chaperone heat shock protein 10. J Exp Zool B Mol Dev Evol 314: 257–266. [DOI] [PubMed] [Google Scholar]
- 22. Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, et al. (2005) Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev 19: 570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daugaard; M, Jaattela; M, Rohde M (2005) Hsp70–2 is required for tumor cell growth and survival. cell cycle 4: 877–880. [DOI] [PubMed] [Google Scholar]
- 24. Scieglinska D, Piglowski W, Mazurek A, Malusecka E, Zebracka J, et al. (2008) The HspA2 protein localizes in nucleoli and centrosomes of heat shocked cancer cells. J Cell Biochem 104: 2193–2206. [DOI] [PubMed] [Google Scholar]
- 25. Filipczak PT, Piglowski W, Glowala-Kosinska M, Krawczyk Z, Scieglinska D (2012) HSPA2 overexpression protects V79 fibroblasts against bortezomib-induced apoptosis. Biochem Cell Biol 90: 224–231. [DOI] [PubMed] [Google Scholar]
- 26. Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, et al. (1996) Targeted gene disruption of Hsp70–2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc Natl Acad Sci U S A 93: 3264–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dix DJ, Allen JW, Collins BW, Poorman-Allen P, Mori C, et al. (1997) HSP70–2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development 124: 4595–4603. [DOI] [PubMed] [Google Scholar]
- 28. Govin J, Caron C, Escoffier E, Ferro M, Kuhn L, et al. (2006) Post-meiotic shifts in HSPA2/HSP70.2 chaperone activity during mouse spermatogenesis. J Biol Chem 281: 37888–37892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krawczyk Z, Wisniewski J, Biesiada E (1987) A hsp70-related gene is constitutively highly expressed in testis of rat and mouse. Mol Biol Rep 12: 27–34. [DOI] [PubMed] [Google Scholar]
- 30. Zakeri ZF, Wolgemuth DJ (1987) Developmental-stage-specific expression of the hsp70 gene family during differentiation of the mammalian male germ line. Mol Cell Biol 7: 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Motiei M, Tavalaee M, Rabiei F, Hajihosseini R, Nasr-Esfahani MH (2013) Evaluation of HSPA2 in fertile and infertile individuals. Andrologia 45: 66–72. [DOI] [PubMed] [Google Scholar]
- 32. Son WY, Hwang SH, Han CT, Lee JH, Kim S, et al. (1999) Specific expression of heat shock protein HspA2 in human male germ cells. Mol Hum Reprod 5: 1122–1126. [DOI] [PubMed] [Google Scholar]
- 33. Cheng H, Guo Y, Yu Q, Zhou R (2003) The rice field eel as a model system for vertebrate sexual development. Cytogenet Genome Res 101: 274–277. [DOI] [PubMed] [Google Scholar]
- 34. Zhang G, Fang X, Guo X, Li L, Luo R, et al. (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490: 49–54. [DOI] [PubMed] [Google Scholar]
- 35. Nei M (1969) Gene duplication and nucleotide substitution in evolution. Nature 221: 40–42. [DOI] [PubMed] [Google Scholar]
- 36.Nei M, Hughes A (1992) Balanced polymorphism and evolution by the birth-and-death process in the MHC loci. In: Tsuji K, Aizawa M, Sasazuki T, editors. 11th Histocompatibility Workshop and Conference. Oxford, UK: Oxford Univ.
- 37. Pelham HR (1984) Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J 3: 3095–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bukau B, Deuerling E, Pfund C, Craig EA (2000) Getting newly synthesized proteins into shape. Cell 101: 119–122. [DOI] [PubMed] [Google Scholar]
- 39. Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parcellier A, Gurbuxani S, Schmitt E, Solary E, Garrido C (2003) Heat shock proteins, cellular chaperones that modulate mitochondrial cell death pathways. Biochem Biophys Res Commun 304: 505–512. [DOI] [PubMed] [Google Scholar]
- 41. Ravagnan L, Gurbuxani S, Susin SA, Maisse C, Daugas E, et al. (2001) Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat Cell Biol 3: 839–843. [DOI] [PubMed] [Google Scholar]
- 42. Daugaard M, Rohde M, Jaattela M (2007) The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett 581: 3702–3710. [DOI] [PubMed] [Google Scholar]
- 43. Udono H, Srivastava PK (1993) Heat shock protein 70-associated peptides elicit specific cancer immunity. J Exp Med 178: 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jolesch A, Elmer K, Bendz H, Issels RD, Noessner E (2012) Hsp70, a messenger from hyperthermia for the immune system. Eur J Cell Biol 91: 48–52. [DOI] [PubMed] [Google Scholar]
- 45. Ito Y, Ando A, Ando H, Ando J, Saijoh Y, et al. (1998) Genomic structure of the spermatid-specific hsp70 homolog gene located in the class III region of the major histocompatibility complex of mouse and man. J Biochem 124: 347–353. [DOI] [PubMed] [Google Scholar]
- 46. Eddy EM (1999) Role of heat shock protein HSP70–2 in spermatogenesis. Rev Reprod 4: 23–30. [DOI] [PubMed] [Google Scholar]
- 47. Susanne Kaufer, Caroline M Coffey, Parker JSL (2011) The cellular chaperone Hsc70 is specifically recruited to reovirus viral factories independently of its chaperone function. Journal of Virology 86: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein alignments of the swamp eel Hspa8b2 with Hspa2 and Hspa8 of mammals. a. Complete protein alignments of Hspa8a1/a2/b1/b2 of the swamp eel with Hspa8 of human and mouse. b. Complete protein alignments of Hspa8a1/a2/b1/b2 of the swamp eel with Hspa2 of human and rat. c. Complete protein alignments of Hspa8b2 of the swamp eel with Hspa2 of human and rat. White letters on black background indicated identical amino acids, and these genes accession numbers were showed in Table S1.
(TIF)
List of protein sequences used for sequence analysis.
(DOC)