Abstract
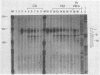
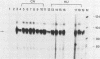
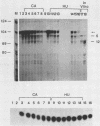
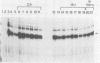
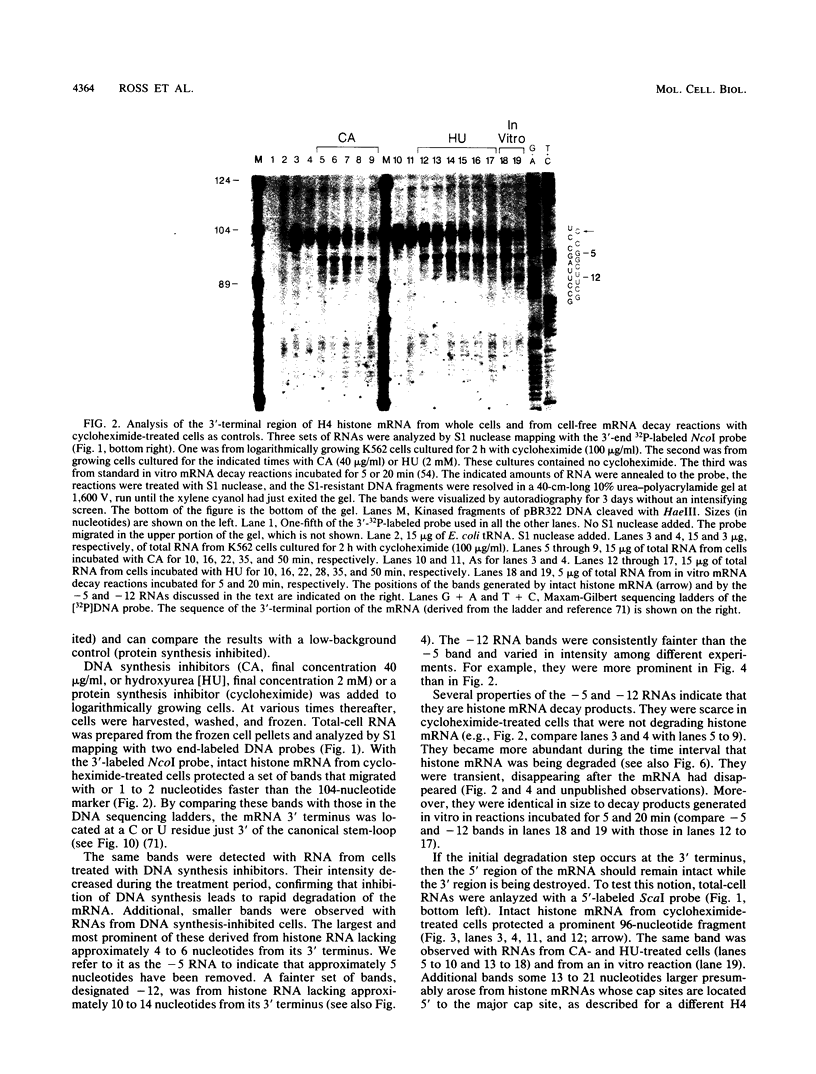
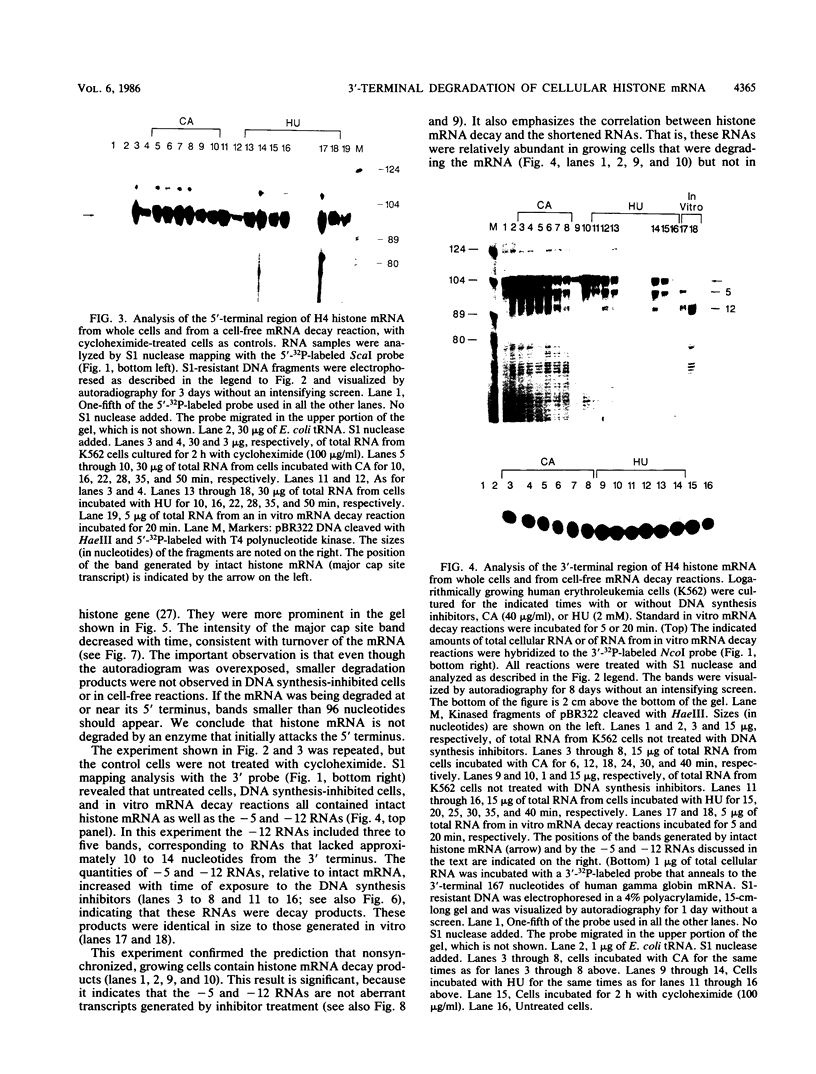
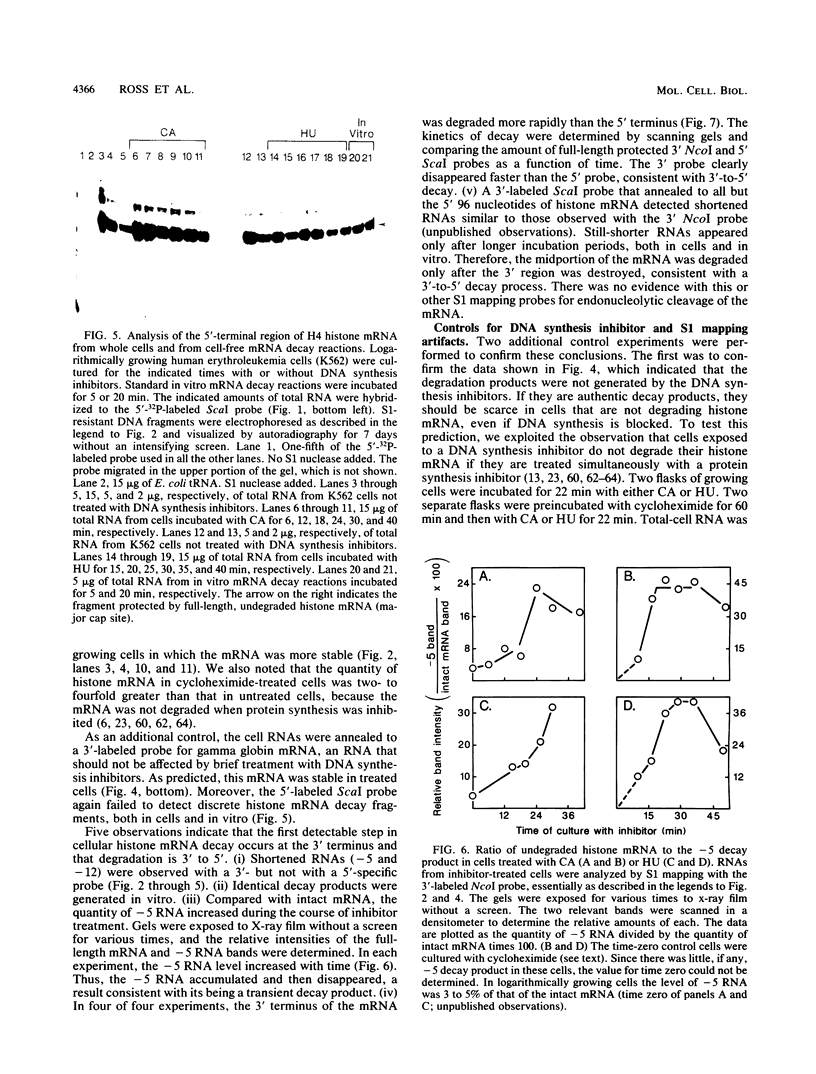
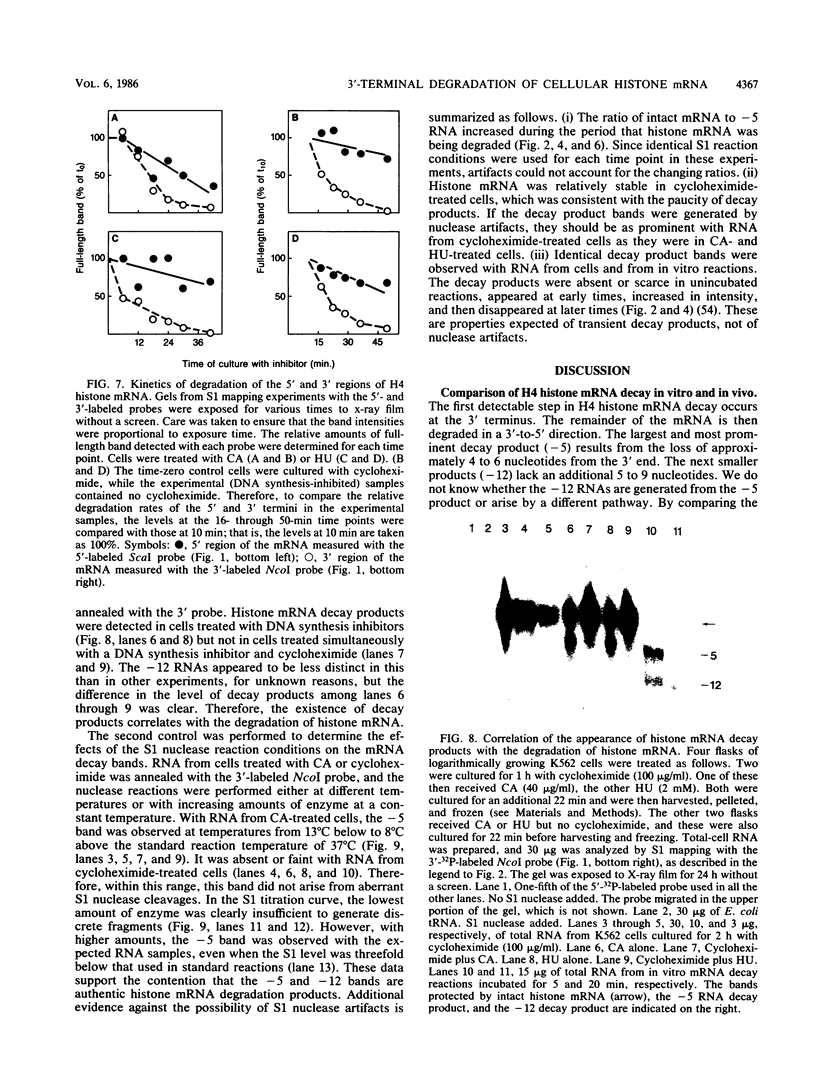
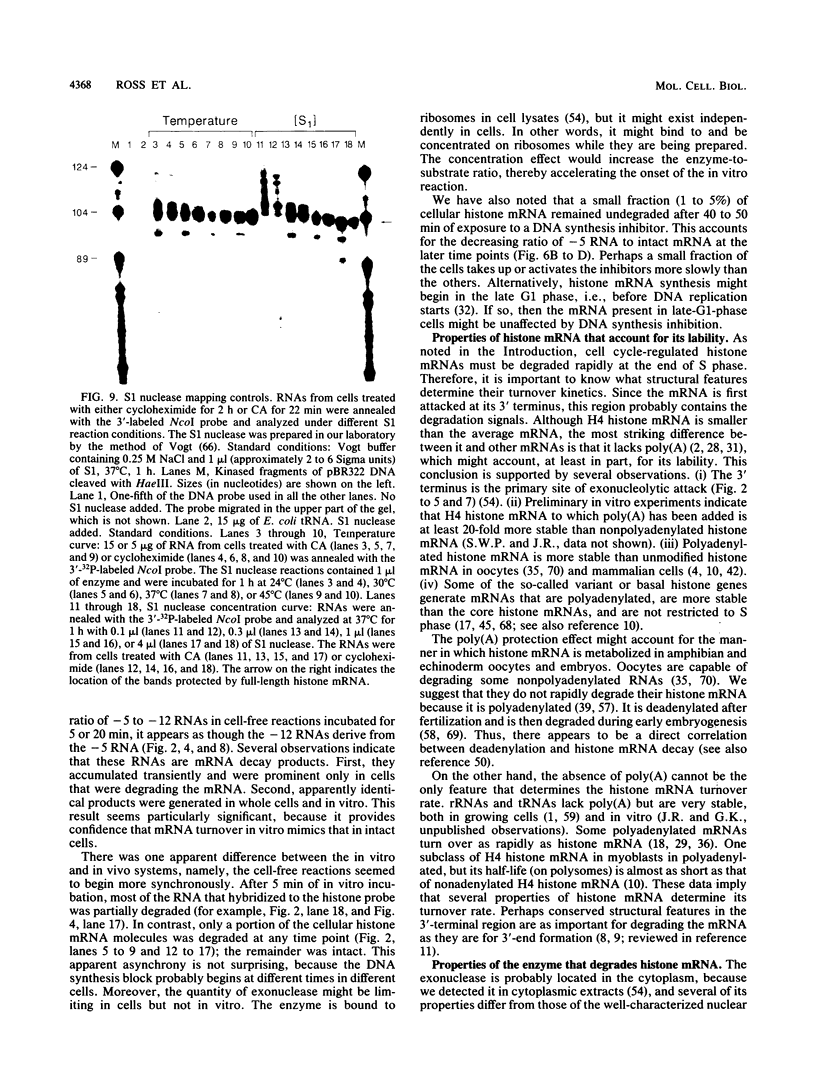
The first detectable step in the degradation of human H4 histone mRNA occurs at the 3' terminus in a cell-free mRNA decay system (J. Ross and G. Kobs, J. Mol. Biol. 188:579-593, 1986). Most or all of the remainder of the mRNA is then degraded in a 3'-to-5' direction. The experiments described here were designed to determine whether a similar degradation pathway is followed in whole cells. Two sets of short-lived histone mRNA decay products were detected in logarithmically growing erythroleukemia (K562) cells. These products, designated the -5 and -12 RNAs, were generated by the loss of approximately 4 to 6 and 11 to 13 nucleotides, respectively, from the 3' terminus of histone mRNA. The same decay products were observed after a brief incubation in vitro. They were in low abundance or absent from cells that were not degrading histone mRNA. In contrast, they were readily detectable in cells that degraded the mRNA at an accelerated rate, i.e., in cells cultured with a DNA synthesis inhibitor, either cytosine arabinoside or hydroxyurea. During the initial stages of the decay process, as the 3' terminus of the mRNA was being degraded, the 5'-terminal region remained intact. These results indicate that the first detectable step in human H4 histone mRNA decay occurs at the 3' terminus and that degradation proceeds 3' to 5', both in cells and in cell-free reactions.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adesnik M., Darnell J. E. Biogenesis and characterization of histone messenger RNA in HeLa cells. J Mol Biol. 1972 Jun 28;67(3):397–406. doi: 10.1016/0022-2836(72)90458-5. [DOI] [PubMed] [Google Scholar]
- Alterman R. B., Ganguly S., Schulze D. H., Marzluff W. F., Schildkraut C. L., Skoultchi A. I. Cell cycle regulation of mouse H3 histone mRNA metabolism. Mol Cell Biol. 1984 Jan;4(1):123–132. doi: 10.1128/mcb.4.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alterman R. B., Sprecher C., Graves R., Marzluff W. F., Skoultchi A. I. Regulated expression of a chimeric histone gene introduced into mouse fibroblasts. Mol Cell Biol. 1985 Sep;5(9):2316–2324. doi: 10.1128/mcb.5.9.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artishevsky A., Grafsky A., Lee A. S. Isolation of a mammalian sequence capable of conferring cell cycle regulation to a heterologous gene. Science. 1985 Nov 29;230(4729):1061–1063. doi: 10.1126/science.4059922. [DOI] [PubMed] [Google Scholar]
- Baumbach L. L., Marashi F., Plumb M., Stein G., Stein J. Inhibition of DNA replication coordinately reduces cellular levels of core and H1 histone mRNAs: requirement for protein synthesis. Biochemistry. 1984 Apr 10;23(8):1618–1625. doi: 10.1021/bi00303a006. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Schümperli D., Sconzo G., Birnstiel M. L. 3' editing of mRNAs: sequence requirements and involvement of a 60-nucleotide RNA in maturation of histone mRNA precursors. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1057–1061. doi: 10.1073/pnas.81.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird R. C., Jacobs F. A., Stein G., Stein J., Sells B. H. A unique subspecies of histone H4 mRNA from rat myoblasts contains poly(A). Proc Natl Acad Sci U S A. 1985 Oct;82(20):6760–6764. doi: 10.1073/pnas.82.20.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Borun T. W., Gabrielli F., Ajiro K., Zweidler A., Baglioni C. Further evidence of transcriptional and translational control of histone messenger RNA during the HeLa S3 cycle. Cell. 1975 Jan;4(1):59–67. doi: 10.1016/0092-8674(75)90134-8. [DOI] [PubMed] [Google Scholar]
- Butler W. B., Mueller G. C. Control of histone synthesis in HeLa cells. Biochim Biophys Acta. 1973 Feb 4;294(1):481–496. doi: 10.1016/0005-2787(73)90104-4. [DOI] [PubMed] [Google Scholar]
- Cabrera C. V., Lee J. J., Ellison J. W., Britten R. J., Davidson E. H. Regulation of cytoplasmic mRNA prevalence in sea urchin embryos. Rates of appearance and turnover for specific sequences. J Mol Biol. 1984 Mar 25;174(1):85–111. doi: 10.1016/0022-2836(84)90366-8. [DOI] [PubMed] [Google Scholar]
- Capasso O., Heintz N. Regulated expression of mammalian histone H4 genes in vivo requires a trans-acting transcription factor. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5622–5626. doi: 10.1073/pnas.82.17.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M., Schibler U. Accumulation of rare and moderately abundant mRNAs in mouse L-cells is mainly post-transcriptionally regulated. J Mol Biol. 1984 Oct 5;178(4):869–880. doi: 10.1016/0022-2836(84)90316-4. [DOI] [PubMed] [Google Scholar]
- Dalton S., Coleman J. R., Wells J. R. Transcription of the histone H5 gene is not S-phase regulated. Mol Cell Biol. 1986 Feb;6(2):601–606. doi: 10.1128/mcb.6.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A., Ley T. J., Humphries R. K., Fordis M., Schechter A. N. Inducible transcription of five globin genes in K562 human leukemia cells. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detke S., Lichtler A., Phillips I., Stein J., Stein G. Reassessment of histone gene expression during cell cycle in human cells by using homologous H4 histone cDNA. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4995–4999. doi: 10.1073/pnas.76.10.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler D. C., Eales S. J. Purification and properties of a novel nucleolar exoribonuclease capable of degrading both single-stranded and double-stranded RNA. Biochemistry. 1985 Jan 29;24(3):686–691. doi: 10.1021/bi00324a022. [DOI] [PubMed] [Google Scholar]
- Gallwitz D. Kinetics of inactivation of histone mRNA in the cytoplasm after inhibition of DNA replication in synchronised HeLa cells. Nature. 1975 Sep 18;257(5523):247–248. doi: 10.1038/257247a0. [DOI] [PubMed] [Google Scholar]
- Gallwitz D., Mueller G. C. Histone synthesis in vitro on HeLa cell microsomes. The nature of the coupling to deoxyribonucleic acid synthesis. J Biol Chem. 1969 Nov 10;244(21):5947–5952. [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984 Feb;4(2):351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L., Schlaffer I., Wright K., Moreno M. L., Berand D., Hager G., Stein J., Stein G. Cell cycle-dependent expression of a stable episomal human histone gene in a mouse cell. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2315–2319. doi: 10.1073/pnas.83.8.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J. R., Perry R. P. Relative occurrence of polyadenylic acid sequences in messenger and heterogeneous nuclear RNA of L cells as determined by poly (U)-hydroxylapatite chromatography. J Mol Biol. 1972 Dec 14;72(1):91–98. doi: 10.1016/0022-2836(72)90070-8. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Zernik M., Roeder R. G. The structure of the human histone genes: clustered but not tandemly repeated. Cell. 1981 Jun;24(3):661–668. doi: 10.1016/0092-8674(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Osley M. A., Ludwig T. R., 2nd, McLaughlin C. S. Cell-cycle regulation of yeast histone mRNA. Cell. 1981 May;24(2):367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Hereford L., Bromley S., Osley M. A. Periodic transcription of yeast histone genes. Cell. 1982 Aug;30(1):305–310. doi: 10.1016/0092-8674(82)90036-8. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Bonner J. Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry. 1973 Jun 5;12(12):2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- Huez G., Marbaix G., Gallwitz D., Weinberg E., Devos R., Hubert E., Cleuter Y. Functional stabilisation of HeLa cell histone messenger RNAs injected into Xenopus oocytes by 3'-OH polyadenylation. Nature. 1978 Feb 9;271(5645):572–573. doi: 10.1038/271572a0. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Cooper J. A., Hunter T., Verma I. M. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 1984 Dec 20;312(5996):711–716. doi: 10.1038/312711a0. [DOI] [PubMed] [Google Scholar]
- Lasater L. S., Eichler D. C. Isolation and properties of a single-strand 5'----3' exoribonuclease from Ehrlich ascites tumor cell nucleoli. Biochemistry. 1984 Sep 11;23(19):4367–4373. doi: 10.1021/bi00314a019. [DOI] [PubMed] [Google Scholar]
- Lazarus H. M., Sporin M. B. Purification and properties of a nuclear exoribonuclease from Ehrlich ascites tumor cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1386–1393. doi: 10.1073/pnas.57.5.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson R. G., Marcu K. B. On the existence of polyadenylated histone mRNA in Xenopus laevis oocytes. Cell. 1976 Oct;9(2):311–322. doi: 10.1016/0092-8674(76)90121-5. [DOI] [PubMed] [Google Scholar]
- Lim L. W., Kennell D. Evidence for random endonucleolytic cleavages between messages in decay of Escherichia coli trp mRNA. J Mol Biol. 1980 Aug 5;141(2):227–233. doi: 10.1016/0022-2836(80)90388-5. [DOI] [PubMed] [Google Scholar]
- Lozzio C. B., Lozzio B. B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975 Mar;45(3):321–334. [PubMed] [Google Scholar]
- Lüscher B., Stauber C., Schindler R., Schümperli D. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3'-terminal part of the gene. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4389–4393. doi: 10.1073/pnas.82.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D., Hartwell L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986 Jan 17;44(1):43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- Mercer J. F., Wake S. A. An analysis of the rate of metallothionein mRNA poly(A)-shortening using RNA blot hybridization. Nucleic Acids Res. 1985 Nov 25;13(22):7929–7943. doi: 10.1093/nar/13.22.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molgaard H. V., Perucho M., Ruiz-Carrillo A. Histone H5 messenger RNA is polyadenylated. Nature. 1980 Jan 31;283(5746):502–504. doi: 10.1038/283502a0. [DOI] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R. Histone genes: not so simple after all. Cell. 1984 Oct;38(3):624–626. doi: 10.1016/0092-8674(84)90256-3. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Gould J., Kim S., Kane M. Y., Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986 May 23;45(4):537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Hereford L. M. Yeast histone genes show dosage compensation. Cell. 1981 May;24(2):377–384. doi: 10.1016/0092-8674(81)90327-5. [DOI] [PubMed] [Google Scholar]
- Palatnik C. M., Wilkins C., Jacobson A. Translational control during early Dictyostelium development: possible involvement of poly(A) sequences. Cell. 1984 Apr;36(4):1017–1025. doi: 10.1016/0092-8674(84)90051-5. [DOI] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Prescott D. M. The syntheses of total macronuclear protein, histone, and DNA during the cell cycle in Euplotes eurystomus. J Cell Biol. 1966 Oct;31(1):1–9. doi: 10.1083/jcb.31.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E., Borun T. W. The cytoplasmic synthesis of histones in hela cells and its temporal relationship to DNA replication. Proc Natl Acad Sci U S A. 1967 Feb;57(2):409–416. doi: 10.1073/pnas.57.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Kobs G. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3' terminus and proceeds 3' to 5'. J Mol Biol. 1986 Apr 20;188(4):579–593. doi: 10.1016/s0022-2836(86)80008-0. [DOI] [PubMed] [Google Scholar]
- Ross J., Pizarro A. Human beta and delta globin messenger RNAs turn over at different rates. J Mol Biol. 1983 Jul 5;167(3):607–617. doi: 10.1016/s0022-2836(83)80101-6. [DOI] [PubMed] [Google Scholar]
- Ross J., Sullivan T. D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985 Nov;66(5):1149–1154. [PubMed] [Google Scholar]
- Ruderman J. V., Pardue M. L. Cell-free translation analysis of messenger RNA in echinoderm and amphibian early development. Dev Biol. 1977 Oct 1;60(1):48–68. doi: 10.1016/0012-1606(77)90109-9. [DOI] [PubMed] [Google Scholar]
- Ruderman J. V., Woodland H. R., Sturgess E. A. Modulations of histone messenger RNA during the early development of Xenopus laevis. Dev Biol. 1979 Jul;71(1):71–82. doi: 10.1016/0012-1606(79)90083-6. [DOI] [PubMed] [Google Scholar]
- Sameshima M., Liebhaber S. A., Schlessinger D. Dual pathways for ribonucleic acid turnover in WI-38 but not in I-cell human diploid fibroblasts. Mol Cell Biol. 1981 Jan;1(1):75–81. doi: 10.1128/mcb.1.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Heintz N., Roeder R. G. Regulation of human histone gene expression during the HeLa cell cycle requires protein synthesis. Mol Cell Biol. 1984 Dec;4(12):2723–2734. doi: 10.1128/mcb.4.12.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding J., Kajiwara K., Mueller G. C. The metabolism of basic proteins in HeLa cell nuclei. Proc Natl Acad Sci U S A. 1966 Nov;56(5):1535–1542. doi: 10.1073/pnas.56.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl H., Gallwitz D. Fate of histone messenger RNA in synchronized HeLa cells in the absence of initiation of protein synthesis. Eur J Biochem. 1977 Jan;72(2):385–392. doi: 10.1111/j.1432-1033.1977.tb11263.x. [DOI] [PubMed] [Google Scholar]
- Stimac E., Groppi V. E., Coffino P. Increased histone mRNA levels during inhibition of protein synthesis. Biochem Biophys Res Commun. 1983 Jul 18;114(1):131–137. doi: 10.1016/0006-291x(83)91604-2. [DOI] [PubMed] [Google Scholar]
- Stimac E., Groppi V. E., Jr, Coffino P. Inhibition of protein synthesis stabilizes histone mRNA. Mol Cell Biol. 1984 Oct;4(10):2082–2090. doi: 10.1128/mcb.4.10.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai S., Borun T. W., Muchmore J., Lieberman I. Concurrent synthesis of histone and deoxyribonucleic acid in liver after partial hepatectomy. Nature. 1968 Aug 24;219(5156):860–861. doi: 10.1038/219860a0. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci U S A. 1985 May;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H. R., Flynn J. M., Wyllie A. J. Utilization of stored mRNA in Xenopus embryos and its replacement by newly synthesized transcripts: histone H1 synthesis using interspecies hybrids. Cell. 1979 Sep;18(1):165–171. doi: 10.1016/0092-8674(79)90365-9. [DOI] [PubMed] [Google Scholar]
- Woodland H. R., Wilt F. H. The functional stability of sea urchin histone mRNA injected into oocytes of Xenopus laevis. Dev Biol. 1980 Mar;75(1):199–213. doi: 10.1016/0012-1606(80)90155-4. [DOI] [PubMed] [Google Scholar]
- Zhong R., Roeder R. G., Heintz N. The primary structure and expression of four cloned human histone genes. Nucleic Acids Res. 1983 Nov 11;11(21):7409–7425. doi: 10.1093/nar/11.21.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]