Abstract
TEAD proteins are transcription factors that are crucial for development, but also play a role in cancers. Several developmentally and pathologically important genes are upregulated by TEADs. TEADs have a TEA domain that enables them to bind specific DNA elements and a transactivation domain that enables them to interact with coactivators. TEADs on their own are unable to activate transcription and they require the help of coactivators. Several TEAD-interacting coactivators are known and they can be classified into three groups: (1) YAP and its paralog TAZ; (2) Vgll proteins; and (3) p160s. Accordingly, these coactivators also play a role in development and cancers. Recent studies have shown that TEADs and their coactivators aid in the progression of various cancers, including the difficult to treat glioblastoma, liver and ovarian cancers. They facilitate cancer progression through expression of proliferation promoting genes such as c-myc, survivin, Axl, CTGF and Cyr61. There is also a good correlation between high TEAD or its coactivator expression and poor prognosis in various cancers. Given the fact that TEADs and their coactivators need to work together for a functional outcome, disrupting the interaction between them appears to be a viable option for cancer therapy. Structures of TEAD-coactivator complexes have been elucidated and will facilitate drug design and development.
Keywords: YAP, TAZ, Vgll proteins, TEAD, cancer
Introduction
The transcription factor TEAD (TEA/ATTS domain) was first identified as a nuclear protein that had the ability to bind and activate transcription from the SV40 enhancer.1 Later TEAD is also shown to bind human papillomavirus-16 (HPV-16) enhancer and activate the HPV-16 oncogenes E6 and E7.2 TEADs are evolutionarily conserved proteins and TEAD orthologs can be found in all vertebrates, invertebrates and even in single celled eukaryotes. TEADs play a pivotal role in development and TEAD expression is already detected in a 2-cell stage embryo.3 TEADs are required for cardiogenesis,4 myogenesis,5 and for the development of neural crest,6 notochord,7 and trophoectoderm.8 In mammals, there are four genes encoding four homologous members of the TEAD family and they are named TEAD1–4. Each TEAD gene has a distinct but not mutually exclusive expression pattern. Almost all tissues express at least one of the TEAD genes and some express all four of them. The function of the TEAD genes could be deduced from gene inactivation studies performed in mice. TEAD1 null mutation is embryonic lethal and the mutant mice suffer from severe heart defects.4 TEAD1 facilitates the expression of cardiac-specific genes and thereby it has been proposed that TEAD1 is important for the differentiation of cardiac muscle. Intriguingly, in TEAD1 null mutant embryos, only the cardiac muscle growth but not its differentiation is affected. In the case of TEAD2, there have been conflicting reports about its function. In one study, defects in neural tube closure were observed,9 whereas in another study, the TEAD2 null embryos appeared normal.10 There is also some functional redundancy between TEAD1 and TEAD2 because the TEAD1 and TEAD2 double mutant embryos exhibited defects that are more severe than either TEAD1 or TEAD2 single mutant embryos.10 TEAD4 inactivation in mice severely affected the specification of trophectoderm,8 a precursor of placenta. As a result these embryos fail to implant, this appears to be the prime function of TEAD4 because embryos develop normally when function of TEAD4 is disrupted after implantation. Knockout studies have not been reported for TEAD3.
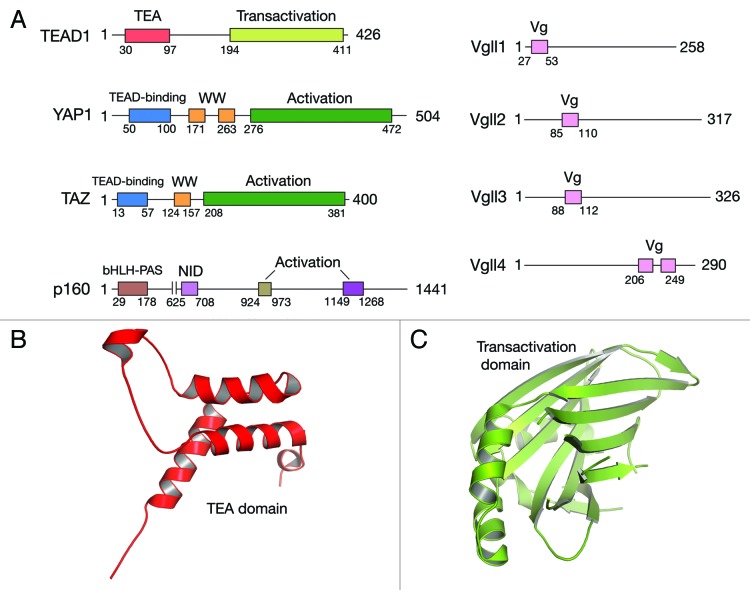
All the four TEAD genes have the same domain architecture (Fig. 1). In the N-terminus, there is a DNA-binding TEA/ATTS domain that adopts a homeodomain fold (Fig. 1B).11 Biochemical and functional studies have revealed that TEA domain binds DNA elements such as 5′-GGAATG-3′ and this element is seen in the SV40 enhancer and in the promoter regions of TEAD target genes.1,11-13 The C-terminal region of TEAD adopts an immunoglobulin-like β-sandwich fold (Fig. 1C).14 This domain recruits transcription coactivators and therefore could be considered as a “transactivation” domain. TEAD on its own is unable to induce gene expression and it was already deduced from initial studies that TEADs require additional factors or coactivators for gene expression.15 Coactivators do not bind DNA; they pair with transcription factors and activate transcription. Coactivators generally have activation domains that facilitate their interaction with the basal transcription or chromatin remodeling machinery. Basal transcription machinery is a set of proteins that are required for the transcription of all genes, whereas the chromatin remodeling machinery decondenses the chromatin and makes it more conducive for the initiation of transcription. Several TEAD interacting coactivators have been identified and they can be classified into three broad groups: (1) YAP and its paralog TAZ; (2) Vgll proteins; and (3) p160 family of nuclear receptor coactivators. Activation domains are seen in YAP/TAZ16 and p160.17 The activation domain in YAP and TAZ may potentially bridge them with the basal transcription and chromatin remodelling machinery in order to activate transcription whereas the activation domains of p160 is shown to recruit proteins that facilitate chromatin remodeling. Vgll proteins on the other hand do not appear to have any activation domains and the molecular mechanism as to how they activate transcription is not known.
Figure 1. Domain architecture of TEAD and its coactivators. (A) TEADs have a N-terminal TEA domain that binds DNA and a C-terminal transactivation domain that binds coactivators. YAP and TAZ have N-terminal TEAD-binding motif, one or two WW domains, which is followed by an activation domain. In Vgll proteins, the only recognizable feature is the Vestigial (Vg) motif and Vgll4 unlike other Vgll proteins, has two Vg motifs. Vg motif facilitates interaction with TEAD. Domain architecture of SRC1, a p160 coactivator, is shown. It interacts with TEAD through its N-terminal bHLH-PAS domain and it interacts with nuclear receptors through its NID (nuclear receptor interacting domain). In the C-terminus p160 has two activation domains. Ribbon diagram representation of TEA (B) and transactivation (C) domains of TEAD are shown. TEA domain adopts a homeodomain fold and the transactivation domain adopts an immunoglobulin-like β- sandwich fold.
TEADs are also involved in human cancers; TEADs are responsible for the overexpression of mesothelin gene, a well-known tumor marker.18 In prostate cancers, high TEAD1 expression correlates with poor clinical outcome.19 There is also up to a 300-fold increase in TEAD1 levels in Kaposi’s sarcoma (www.ebi.ac.uk/gxa).20 Increased TEAD expression is also seen in basal-like breast cancers,21,22 fallopian tube carcinoma23 and germ cell tumors.24 On the other hand, TEAD1 expression is downregulated in other breast cancer types and in renal or bladder cancers (www.oncomine.org). Interestingly, a recent study reconciles this seemingly opposing roles of TEADs in cancers by showing that both up and downregulation of TEAD causes increase in the expression of anti-apoptotic protein Livin.20 Livin expression appears to favor cancer progression. Genetic studies also show that TEADs promote cell proliferation and inhibits apoptosis.10 It is becomingly increasingly clear that TEAD-interacting coactivators are also dysregulated in various cancers. The scenario is more convincing in the case of YAP and TAZ and is emerging in the case of Vgll proteins. Several human cancers are probably caused by their hyperactivity and inhibiting their function appears to be a viable option for future anticancer therapy.
YAP and Its Paralog TAZ
The coactivator YAP (Yes-associated protein) is evolutionarily conserved; with the exception of nematodes, YAP orthologs are found in all metazoans and even in premetazoans.25 TAZ (Transcription coactivator with PDZ-binding motif) on the other hand is seen only in vertebrates.25 YAP and TAZ have a N-terminal TEAD-interacting motif followed by WW domain. Depending on the isoform, YAP has either one or two WW domains26, whereas almost all the isoforms or orthologs of TAZ have a single WW domain. At its C-terminus YAP and TAZ have an activation domain, which is followed by a C-terminal PDZ-binding motif (Fig. 1). To date, eight isoforms of YAP have been identified in humans.26 They were all generated through alternative splicing, which results in either a loss of WW domain or an alteration in the activation domain. Interestingly, alternative splicing also generates a leucine zipper in the activation domain of YAP.26 Leucine zipper could potentially help YAP to homodimerize or to interact with other leucine zipper containing proteins. Leucine zippers are also used by transcription factors to bind DNA.27 But the crucial basic residues that usually contacts DNA in leucine zippers are absent in YAP. This might seriously undermine YAP’s ability to bind DNA. There have been no reports of TAZ isoforms in humans but two TAZ isoforms are reported in mice. Interestingly, there is a TAZ isoform in medaka fish that has two WW domains and the affinity between TAZ and its interacting proteins is increased in the presence of this additional WW domain.28
Evolutionary distance between two orthologs of a gene is calculated based on the number of amino acid or nucleotide substitutions. A strong correlation, correlation coefficient r = 0.85, p = 1.1 ◊ 10−16, is seen in the evolutionary distances of YAP and TEAD orthologs. This indicates that YAP and TEAD genes have coevolved and there is a strong selection pressure for YAP and TEAD to interact with each other.25 Furthermore the residues that are crucial for maintaining YAP-TEAD interaction are conserved throughout evolution. This underscores the functional relationship between YAP and TEAD and the importance of their interaction. YAP, like TEAD, also plays an important role in development. YAP knockout mice embryos do not survive past E8.5.29 In the case of TAZ, the knockout mice survive until adulthood but suffer from polycystic kidney disease and emphysema.30-32 YAP and TAZ are potent promoters of cell proliferation. They pair with TEADs and upregulate the expression of several growth promoting factors. The most important among those are the CCN family secretory proteins CTGF and Cyr6113 and also proteins such as Ki67, receptor tyrosine kinase Axl,33 c-myc and survivin.34 YAP is also involved in the organ size maintenance. The effect of YAP levels on organ size varies among organs; the most sensitive is the liver. YAP overexpression in liver increases liver size by over 5-fold and the normal liver size is restored when the YAP expression is attenuated.34,35 In mouse liver, a sustained YAP overexpression from birth also results in hepatocellular carcinoma. YAP also helps the embryonic stem cells to maintain their stemness and YAP downregulation leads to cell differentiation.36 YAP and TAZ along with TEADs are the prime effectors for the physiologically important Hippo pathway.37 There is also a crosstalk between YAP/TAZ and other signaling pathways such as TGF-β, Wnt, hedgehog and JAK-STAT.38
YAP and TAZ are also bona fide oncogenes and their upregulation is seen in several human cancers. Amplification of YAP-containing 11q22 amplicon is frequently observed in several human tumors. High levels of YAP are observed in human liver tumors and YAP is a key driver of tumorigenesis.39,40 High YAP levels are also seen in about 15% of ovarian cancers and it correlates with poor patient prognosis.41 Similarly, there is also a good correlation between high YAP expression and poor prognosis in non-small cell lung cancer and esophageal squamous cell carcinoma.42,43 YAP overexpression is also seen in medulloblastoma,44 intracranial ependymoma,45 and oral squamous cell carcinoma.46 Additionally Yorkie, the drosophila homolog of YAP promotes the expression of Myc.47 YAP also plays a role in human colorectal cancer progression.35,48 The Hippo insensitive YAP mutant, when overexpressed, promotes proliferation of undifferentiated progenitor cells in the intestine.35 However, it has been shown recently that a restricted overexpression of this mutant in the intestinal epithelium actually causes a reduction in the number of progenitor cells.49 Under these conditions YAP acts as a tumor suppressor and it counters the proliferative Wnt signals.
TAZ increases the expression of several genes that promotes mesenchymal differentiation in malignant glioma.50 As a result the cells lose their epithelial properties such as presence of polarity, intercellular junctions and acquire mesenchymal or stem cell-like properties. Tumors with high TAZ levels are more invasive and metastatic and therefore difficult to treat. TAZ also plays a role in migration, invasion and tumorigenesis of breast cancer cells and is overexpressed in about 20% of human breast cancers.51 TAZ and its downstream transcriptional targets CTGF and Cyr61 also contribute to Taxol resistance in breast cancer cells.52 TAZ also contributes to the oncogenesis of non-small cell lung cancer.53 Furthermore, TAZ-CAMTA1 (calmodulin-binding transcription activator) fusion gene arising due to chromosomal translocation has been implicated to cause epitheloid hemangioendothelioma, a vascular sarcoma.54,55 Apart from this gene fusion, all other YAP and TAZ–mediated tumorigenesis is due to overexpression. Intriguingly we are yet to document activating mutations in YAP and TAZ, although one study showed that TAZ mutation is associated with basal-like breast cancer metastasis.56 YAP and TAZ need to pair with TEADs to upregulate several genes that promote tumorigenesis and therefore molecules that disrupt this interaction are attractive drugs to treat cancers that overexpress YAP and TAZ. Recently, using a screen containing FDA-approved drugs the compound verteporfin has been shown to have the ability to disrupt YAP-TEAD interaction and thereby reduce YAP-induced cell proliferation.57 Verteporfin is able to induce conformational change in YAP, which subsequently prevents its interaction with TEAD. Recently, it has been shown that GPCRs have the ability to activate YAP and TAZ.58,59 Familial and activating mutations in GPCRs have been linked to human cancers.60 It is possible that these cancers are caused due to the activation of YAP and TAZ. In such scenarios it should be possible to use GPCR antagonists to inhibit the activity of YAP and TAZ.
However, in specific circumstances, YAP also acts as a tumor suppressor. Under conditions that cause DNA damage, YAP pairs with the transcription factor p73 and improves its ability to induce apoptosis.61,62 YAP also acts as a tumor suppressor in the intestinal epithelium.49
Vgll Proteins
Vestigial-like (Vgll) proteins are transcription coactivators and they are named after the Drosophila transcription coactivator vestigial (Vg), which is a master regulator of wing development. Only a short motif, roughly 25 amino acids long, is conserved between Vg and Vgll proteins and also among Vgll proteins. Other than this motif, Vg and Vgll proteins bear no sequence similarity. This is why it is difficult to ascertain whether Vgll proteins evolved from Drosophila Vestigial. Vg motif serves a very important function; it facilitates Vgll protein interaction with TEAD and promotes gene expression. There are four Vgll proteins named Vgll 1–4 and they start to appear in vertebrates. Other than the Vg motif the Vgll proteins do not have any recognizable domain (Fig. 1).
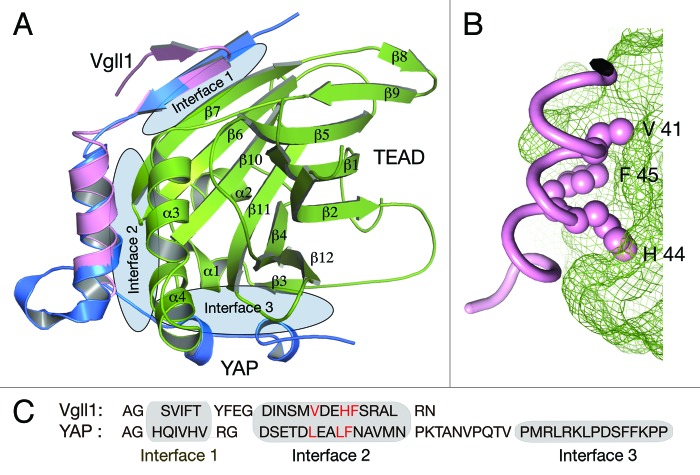
Among the Vgll proteins, Vgll1, also called TONDU, was the first to be identified. In Drosophila, Vgll1 is able to partially rescue Vg loss of function phenotype.63 This is remarkable because other than the Vg motif there is no other sequence similarity between Vg and Vgll1. Interaction with TEAD is crucial for its function and Vgll1 is shown to interact with all four mammalian TEADs.63 In humans it is expressed in the fetal lung, heart, kidney and placenta. In Xenopus its expression is restricted to epidermal cells and the expression levels can be modulated by activin and BMP treatments.64 Vgll1 also promotes cell proliferation, similar to Vg or YAP/TAZ. Even the core complex structure of Vgll1-TEAD is similar to that of YAP-TEAD (Fig. 2A). Given the structural and functional similarity, we proposed that Vgll1 could also play a role in human cancers.65 There is evidence for Vgll1’s role in bladder cancer and its expression is used to classify tumors into good and bad prognosis groups.66 High Vgll1 levels are seen in basal-like breast cancers, but it is not known whether it contributes to cancer progression.22
Figure 2. Structures of YAP/Vgll1-TEAD core complexes. (A) The core complex structures have the transactivation domain of TEAD in complex with either the TEAD-interacting motif of YAP or the Vg motif of Vgll1. YAP interacts with TEAD by forming three interfaces and Vg motif interacts with TEAD by forming two interfaces. The interface 1 and 2 in both the complexes are strikingly similar. (B) A closer look at the interface 2 of Vgll1-TEAD complex. The surface of TEAD is shown as green mesh. Vgll1 helix binds to the hydrophobic groove in TEAD and the crucial region in Vgll1 is the V41xxH44 F45 motif. The side chains of these residues that are shown as pink spheres and they bind to the complementary pockets in TEAD. Similar interaction is also seen in YAP but it has LxxLF motif. (C) The amino acid sequence of the Vg motif of Vgll1 and the TEAD-binding motif of YAP. The LxxLF and the VxxHF motifs are shown in red.
Vgll2, also called VITO-1, is expressed predominantly in the skeletal muscle. Vgll2 plays a role in cell differentiation, as opposed to cell proliferation. It promotes myogenic differentiation by enhancing the expression of muscle-specific genes. A well-known example is the induction of muscle-specific myosin heavy chain expression that is a marker of terminally differentiated muscle.67 Muscle–specific gene expression is regulated by the muscle-specific cytidine-adenosine-thymidine (MCAT) element, 5′-CATTCCT-3′. TEADs are crucial transcription factors for myogenesis and they bind to MCAT elements. MCAT-dependent TEAD activity is enhanced in the presence of Vgll2. TEADs are expressed in almost all tissues, whereas the expression of Vgll2 is largely restricted to skeletal muscle. Therefore it is believed that in the skeletal muscle TEADs require Vgll2 to activate muscle-specific genes. In addition to TEADs, Vgll2 also interacts with myocyte enhancer factor -2 (MEF2) and activates MEF-2 dependent gene expression. MEF2 is a transcription factor that is distinct from TEADs but also plays a crucial role in myogenesis. TEADs and MEF-2 are also shown to interact with each other through their DNA-binding domains, although the functional importance needs to be defined.68 Vgll2 acts as a coactivator for two important myogenic transcription factors, fittingly; knocking down Vgll2 attenuates myogenic differentiation in pluripotent 10T1/2 and C2C12 cell lines.69 Apart from myogenesis Vgll2 has also been shown to play a role in neural crest cell survival and in the development of neural crest derived craniofacial skeleton.70
Vgll3, also called VITO-2, is predominantly expressed in the placenta. Its expression is also seen in the skeletal muscle, heart, kidney, brain and liver. Vgll3 expression is repressed by notch/delta pathway and is activated by myogenic factor-5 and Vgll3 could also play a role in myogenesis.71 Vgll3 also inhibits the differentiation of adipocytes.72 The interaction between Vgll3 and TEADs is not biochemically characterized but the presence of Vg motif suggests that Vgll3 exerts its function through TEAD interaction. Vgll3 also potentially plays a role in cancers, it aids in the progression of soft-tissue sarcoma and also in the development of myxoinflammatory fibroblastic sarcoma.73,74 However, in ovarian cancers Vgll3 could possibly play a tumor suppressor role.75
Vgll4 expression is seen in many tissues. Vgll4, unlike other Vgll proteins, has two partially conserved Vg motifs.76 It uses these motifs to interact with TEADs. Similar to Vgll2, Vgll4 also interacts with MEF2 and uses the same Vg motif for MEF2 interaction.76 It has been proposed that Vgll4 could use its Vg motifs to bridge and reinforce TEAD and MEF2 interaction. The coactivator function of Vgll4 has also been explored; Vgll4 cooperates with TEADs and interferon response factor 2 binding protein 2 (IRF2BP2) and activates the expression of the angiogenic factor VEGFA (vascular endothelial growth factor A).77 However, there is also evidence for Vgll4 suppressing the activity of TEADs in cardiac myocytes.76 It appears that Vgll4 modulates the activity of TEADs in a context-specific manner.
Apart from being a transcription coregulator, Vgll4 also plays a role in regulating signaling in apoptosis. Vgll4 has been shown to bind to inhibitor of apoptosis protein (IAP).78 IAPs, as the name suggests, inhibit apoptosis induced by various stimuli. Vgll4 relocates IAPs from the cytoplasm to the nucleus and thereby retard IAPs ability to inhibit apoptosis. It is not known whether TEADs help in the nuclear retention of Vgll4-IAP complex. Vgll4 also appears to have a potent tumor suppressor function, recently a mutagenic screen identified Vgll4 as one of genes that is inactivated in pancreatic adenocarcinoma.79 There is also a good correlation between low Vgll4 expression and poor prognosis in human pancreatic cancers. Whether the potential tumor suppressor function of Vgll4 is physiologically relevant and whether it is achieved due to its ability to inhibit TEADs or IAPs remain to be investigated.
p160 Family of Nuclear Receptor Coactivators
There is only one report that links p160 and TEADs. p160 coactivators are well studied in the context of nuclear receptor activation but they also have the ability to activate TEAD-dependent transcription.80 Their interaction with TEADs is through their N-terminal basic helix-loop-helix/Per-Arnt-Sim (bHLH-PAS) domain. Although this domain is used to bind DNA in various other proteins, in p160 it is primarily used to mediate protein-protein interaction.
Structures of TEAD—Coactivator Complexes
Structures of the core YAP-TEAD complex and that of core Vgll1-TEAD complex are already determined and they reveal interesting features.65,81,82 The structures contained the TEAD-interacting motif of YAP or the Vg motif of Vgll1 in complex with the transactivation domain of TEAD (Fig. 2A). Vg and YAP motif interaction with TEAD is strikingly similar despite the fact that these motifs differ significantly in their primary sequence (Fig. 2C). YAP/TAZ, Vg/Vgll motif and TEAD domain are highly conserved among all orthologs and therefore these core complex structures are prototypes for all orthologous complexes.
YAP-TEAD1 interaction can be divided into three interfaces and Vg motif-TEAD interaction can be divided into two interfaces. The interface 1 and 2 in both these complexes are remarkably similar. Interface 1 is an antiparallel β-sheet formed between the N-terminal β-strand of the Vg or YAP motif and the β7 of TEAD (Fig. 2A). The interaction is primarily mediated through hydrogen bonds formed between main chain atoms of both strands. As side chains are not involved in this interaction, seemingly different sequences could still interact in a similar fashion. In interface 2, YAP and Vg motif adopts a helical conformation and the helix sits in the hydrophobic groove between α3 and α4 of TEAD. Here the proteins are held together primarily through hydrophobic interactions. The crucial hydrophobic residues in Vgll1 that interacts with the hydrophobic groove in TEAD are V41 H44 and F45 and hereafter we refer to this as VxxHF motif (x: any amino acid). YAP has LxxLF motif instead of VxxHF motif (Fig. 2C). There is also good complementary packing between the residues in VxxHF and LxxLF motifs and the hydrophobic groove in TEAD (Fig. 2B). Therefore in addition to hydrophobicity, structural complementarity is also important for the interaction. Additionally, the residues surrounding these motifs also contribute to the affinity and specificity of interaction. All of these ensure that nonspecific and unproductive interaction between any hydrophobic motif and TEAD does not take place. These can be inferred from the studies done on a similar interaction between the p160 coactivators and nuclear receptors.83 p160 mediates interaction with nuclear receptors through LxxLL motif. Interestingly p160 is also shown to interact with TEAD but this interaction does not appear to involve the LxxLL motif.80
Vg and YAP motif, despite having a different primary sequence, has all the attributes to form interface 1 and 2. However, one prominent difference between these two complexes is the absence of interface 3 in the Vgll1-TEAD complex. Vg motif had only a couple of residues beyond interface 2 in the crystallized Vgll1-TEAD complex. Yet it is likely that Vgll1-TEAD complex might not have interface 3. The rationale for this conclusion comes from the biochemical analysis of YAP/Vgll1 and TEAD interaction.65,81 Vgll1 loses its affinity with TEAD when mutations are made in the crucial interface 2 residues. This is not the case for the YAP interface 2 mutants. YAP is still able to maintain its interaction with TEAD due to the presence of interface 3. The inability of Vgll interface 2 mutant to maintain its interaction with TEAD suggests that Vgll1-TEAD complex lacks interface 3.
In interface 3 YAP adopts a twisted-coil structure and the residues in YAP fit into a groove in TEAD. In this interface, a combination of van der Waals contacts, hydrophobic interactions and hydrogen bonds holds both the proteins together. Sveinsson’s chorioretinal atrophy is a genetic disease that is caused due to Y406H point mutation in TEAD1.84 This residue is in interface 3 and is involved in a hydrogen bond and a hydrophobic contact with YAP residues. A histidine at this position would disrupt these interactions and biochemically it has been shown that this substitution would compromise YAP-TEAD interaction.82 A compromised YAPTEAD interaction could potentially be an underlying reason for this disease. Interface 2 is connected to interface 3 by a linker sequence that has a PXXΦP motif (Φ: any hydrophobic residue). This motif also appears to be important for the YAP-TEAD interaction81 but this linker is absent in TAZ. The lack of linker sequence in TAZ led Sudol et al. to propose that this might facilitate each TEAD molecule to interact with two molecules of TAZ.85 One molecule of TAZ could interact with TEAD through interface 2 and the second TAZ molecule could interact with TEAD through interface 3.
A significant overlap in the interfaces also suggests that Vgll proteins and YAP/TAZ would compete for binding to TEAD and this has also been reported.65 Interaction with TEAD is crucial for YAP, TAZ and Vgll function and any mutation that compromises their interaction with TEAD also affects their function. The structures of these complexes also revealed the possibility that these complexes could be disrupted using staple peptides. Such peptides have the potential to inhibit the activity of TEAD and coactivators and could have utility to treat cancers caused due to their hyperactivity. One example of such an approach is the disruption of the interaction between the transcription factor notch and the MAML coactivator using staple peptides that has the potential to treat leukemias.86 There are also other potential means through which the function of YAP could be inhibited and these have been nicely discussed in the recent review by Sudol et al.85
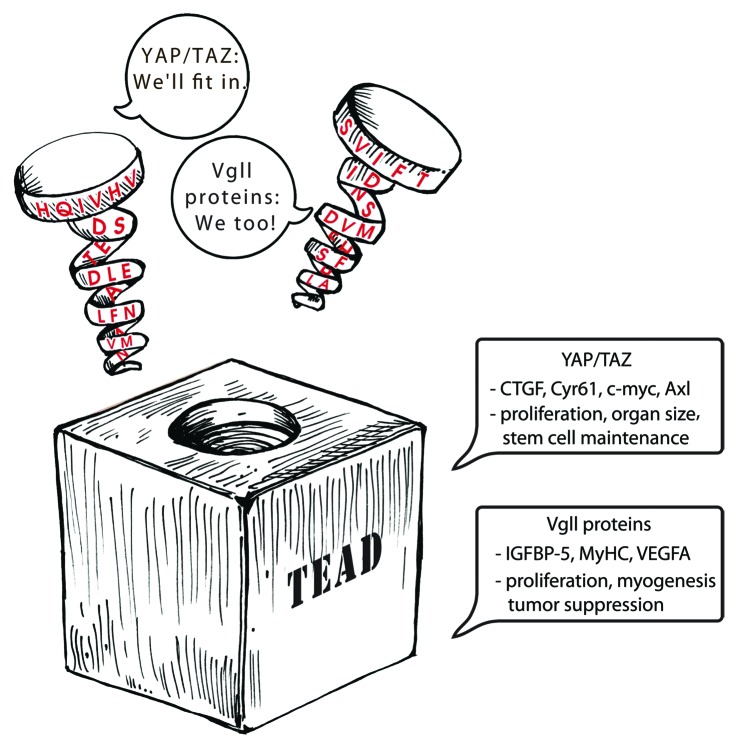
Despite similar structural interaction the gene expression mediated by these coactivators appears to be different (Fig. 3). There is data from limited set of genes that suggests YAP-dependent gene expression is different from that of Vgll-dependent gene expression.65 This is also seen in Drosophila, the YAP ortholog Yorkie upregulates genes such as diap1 and cyclinD, whereas Vg upregulates wing-specific genes. This is remarkable because YAP/TAZ/Yorkie and Vg/Vgll proteins do not have a DNA-binding domain and use the same transcription factor TEAD to mediate gene expression. The specificity in gene expression is unlikely to be solely determined by a transcription factor. The DNA recognition sequence of a transcription factor is often too short or degenerate and could be found everywhere in the genome. In order to enhance or alter specificity transcription factors often pair with other transcription factors or transcription coactivators. In the case of Vg and Vgll proteins, it is already known that they are endowed with the ability to alter the specificity of TEAD87 and this is likely to be the underlying reason for the differential gene expression. Altered specificity could result in differential promoter occupancy and gene expression. Future studies aiming to resolve the molecular basis for the differential expression by different co-activators on the same transcriptional factors will be of great interest and significance.
Figure 3. Cartoon depicting the interaction among YAP/TAZ, Vgll proteins and TEAD. The TEAD-binding region of YAP/TAZ and Vgll proteins are represented as spirals. It adopts a similar structure and fits in the same groove on the surface of TEAD despite having a different primary sequence. YAP/TAZ and Vgll proteins pair with TEAD and upregulate gene expression. CTGF, Cyr61, c-myc and Axl are some of the genes that are upregulated by YAP and TAZ. YAP/TAZ play a significant role in proliferation, organ size and stem cell maintenance. Vgll proteins upregulate the expression genes such as IGFBP-5, myosin heavy chain (MyHC) and VEGFA. They play a role in proliferation, myogenesis and also appear to act as tumor suppressors in certain scenarios.
Conclusion
TEADs require the help of coactivators to activate gene expression. The interaction between TEAD and its coactivators is important for physiologically important processes such as cell differentiation, cell proliferation and stem cell maintenance. However, it also plays a role in human cancers, acting either as cancer drivers or in some cases as tumor suppressors. Inhibiting the interaction between them is a viable option that has therapeutic potential in scenarios where their activity aids cancer progression. The structures of TEAD and coactivator complexes could be put to good use in drug development. Given the similarity in these structures, it is also possible to obtain a compound that has the ability to disrupt several TEAD-coactivator complexes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/23788
References
- 1.Xiao JH, Davidson I, Ferrandon D, Rosales R, Vigneron M, Macchi M, et al. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. EMBO J. 1987;6:3005–13. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishiji T, Lace MJ, Parkkinen S, Anderson RD, Haugen TH, Cripe TP, et al. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. EMBO J. 1992;11:2271–81. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaneko KJ, Cullinan EB, Latham KE, DePamphilis ML. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development. 1997;124:1963–73. doi: 10.1242/dev.124.10.1963. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8:2293–301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida T. MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol. 2008;28:8–17. doi: 10.1161/ATVBAHA.107.155788. [DOI] [PubMed] [Google Scholar]
- 6.Milewski RC, Chi NC, Li J, Brown C, Lu MM, Epstein JA. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development. 2004;131:829–37. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- 7.Sawada A, Nishizaki Y, Sato H, Yada Y, Nakayama R, Yamamoto S, et al. Tead proteins activate the Foxa2 enhancer in the node in cooperation with a second factor. Development. 2005;132:4719–29. doi: 10.1242/dev.02059. [DOI] [PubMed] [Google Scholar]
- 8.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–36. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 9.Kaneko KJ, Kohn MJ, Liu C, DePamphilis ML. Transcription factor TEAD2 is involved in neural tube closure. Genesis. 2007;45:577–87. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–89. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 2006;103:17225–30. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao JH, Davidson I, Ferrandon D, Rosales R, Vigneron M, Macchi M, et al. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. EMBO J. 1987;6:3005–13. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian W, Yu J, Tomchick DR, Pan D, Luo X. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc Natl Acad Sci U S A. 2010;107:7293–8. doi: 10.1073/pnas.1000293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–68. doi: 10.1016/0092-8674(91)90088-G. [DOI] [PubMed] [Google Scholar]
- 16.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–62. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–19. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hucl T, Brody JR, Gallmeier E, Iacobuzio-Donahue CA, Farrance IK, Kern SE. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor dependent MCAT motif. Cancer Res. 2007;67:9055–65. doi: 10.1158/0008-5472.CAN-07-0474. [DOI] [PubMed] [Google Scholar]
- 19.Knight JF, Shepherd CJ, Rizzo S, Brewer D, Jhavar S, Dodson AR, et al. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Cancer. 2008;99:1849–58. doi: 10.1038/sj.bjc.6604774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landin Malt A, Cagliero J, Legent K, Silber J, Zider A, Flagiello D. Alteration of TEAD1 expression levels confers apoptotic resistance through the transcriptional up-regulation of Livin. PLoS One. 2012;7:e45498. doi: 10.1371/journal.pone.0045498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47:490–9. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- 22.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Nowee ME, Snijders AM, Rockx DA, de Wit RM, Kosma VM, Hämäläinen K, et al. DNA profiling of primary serous ovarian and fallopian tube carcinomas with array comparative genomic hybridization and multiplex ligation-dependent probe amplification. J Pathol. 2007;213:46–55. doi: 10.1002/path.2217. [DOI] [PubMed] [Google Scholar]
- 24.Skotheim RI, Autio R, Lind GE, Kraggerud SM, Andrews PW, Monni O, et al. Novel genomic aberrations in testicular germ cell tumors by array-CGH, and associated gene expression changes. Cell Oncol. 2006;28:315–26. doi: 10.1155/2006/219786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilman D, Gat U. The evolutionary history of YAP and the hippo/YAP pathway. Mol Biol Evol. 2011;28:2403–17. doi: 10.1093/molbev/msr065. [DOI] [PubMed] [Google Scholar]
- 26.Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, et al. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene. 2012;509:215–22. doi: 10.1016/j.gene.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–37. doi: 10.1016/S0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 28.Webb C, Upadhyay A, Giuntini F, Eggleston I, Furutani-Seiki M, Ishima R, et al. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway. Biochemistry. 2011;50:3300–9. doi: 10.1021/bi2001888. [DOI] [PubMed] [Google Scholar]
- 29.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol. 2008;294:F542–53. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- 31.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci U S A. 2007;104:1631–6. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, et al. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol. 2007;27:6383–95. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu MZ, Chan SW, Liu AM, Wong KF, Fan ST, Chen J, et al. AXL receptor kinase is a mediator of YAP-dependent oncogenic functions in hepatocellular carcinoma. Oncogene. 2011;30:1229–40. doi: 10.1038/onc.2010.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–60. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 36.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–18. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–93. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varelas X, Wrana JL. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 2012;22:88–96. doi: 10.1016/j.tcb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Zender L, et al. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–85. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, et al. AOCS Study group The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–22. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101:1279–85. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muramatsu T, Imoto I, Matsui T, Kozaki K, Haruki S, Sudol M, et al. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis. 2011;32:389–98. doi: 10.1093/carcin/bgq254. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-L A, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–41. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modena P, Lualdi E, Facchinetti F, Veltman J, Reid JF, Minardi S, et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24:5223–33. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 46.Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–42. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- 47.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19:507–20. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avruch J, Zhou D, Bardeesy N. YAP oncogene overexpression supercharges colon cancer proliferation. Cell Cycle. 2012;11:1090–6. doi: 10.4161/cc.11.6.19453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2012;493:106–10. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–8. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 52.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71:2728–38. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Z, Hao Y, Liu N, Raptis L, Tsao MS, Yang X. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene. 2011;30:2181–6. doi: 10.1038/onc.2010.606. [DOI] [PubMed] [Google Scholar]
- 54.Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- 55.Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, et al. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–53. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–5. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–91. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–43. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 61.Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–73. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 62.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/S1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 63.Vaudin P, Delanoue R, Davidson I, Silber J, Zider A. TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development. 1999;126:4807–16. doi: 10.1242/dev.126.21.4807. [DOI] [PubMed] [Google Scholar]
- 64.Faucheux C, Naye F, Treguer K, Fedou S, Thiebaud P, Theze N. Vestigial like gene family expression in Xenopus: common and divergent features with other vertebrates. Int J Dev Biol. 2012;54:1375–82;. doi: 10.1387/ijdb.103080cf. [DOI] [PubMed] [Google Scholar]
- 65.Pobbati AV, Chan SW, Lee I, Song H, Hong W. Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure. 2012;20:1135–40. doi: 10.1016/j.str.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–55. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 67.Maeda T, Chapman DL, Stewart AF. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J Biol Chem. 2002;277:48889–98. doi: 10.1074/jbc.M206858200. [DOI] [PubMed] [Google Scholar]
- 68.Maeda T, Gupta MP, Stewart AF. TEF-1 and MEF2 transcription factors interact to regulate muscle-specific promoters. Biochem Biophys Res Commun. 2002;294:791–7. doi: 10.1016/S0006-291X(02)00556-9. [DOI] [PubMed] [Google Scholar]
- 69.Günther S, Mielcarek M, Krüger M, Braun T. VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res. 2004;32:791–802. doi: 10.1093/nar/gkh248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson CW, Hernandez-Lagunas L, Feng W, Melvin VS, Williams T, Artinger KB. Vgll2a is required for neural crest cell survival during zebrafish craniofacial development. Dev Biol. 2011;357:269–81. doi: 10.1016/j.ydbio.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mielcarek M, Piotrowska I, Schneider A, Günther S, Braun T. VITO-2, a new SID domain protein, is expressed in the myogenic lineage during early mouse embryonic development. Gene Expr Patterns. 2009;9:129–37. doi: 10.1016/j.gep.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Halperin DS, Pan C, Lusis AJ, Tontonoz P. Vestigial-like 3 (Vgll3) is an Inhibitor of Adipocyte Differentiation. J Lipid Res. 2012;54:473–81. doi: 10.1194/jlr.M032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hélias-Rodzewicz Z, Pérot G, Chibon F, Ferreira C, Lagarde P, Terrier P, et al. YAP1 and VGLL3, encoding two cofactors of TEAD transcription factors, are amplified and overexpressed in a subset of soft tissue sarcomas. Genes Chromosomes Cancer. 2010;49:1161–71. doi: 10.1002/gcc.20825. [DOI] [PubMed] [Google Scholar]
- 74.Hallor KH, Sciot R, Staaf J, Heidenblad M, Rydholm A, Bauer HC, et al. Two genetic pathways, t(1;10) and amplification of 3p11-12, in myxoinflammatory fibroblastic sarcoma, haemosiderotic fibrolipomatous tumour, and morphologically similar lesions. J Pathol. 2009;217:716–27. doi: 10.1002/path.2513. [DOI] [PubMed] [Google Scholar]
- 75.Cody NA, Shen Z, Ripeau JS, Provencher DM, Mes-Masson AM, Chevrette M, et al. Characterization of the 3p12.3-pcen region associated with tumor suppression in a novel ovarian cancer cell line model genetically modified by chromosome 3 fragment transfer. Mol Carcinog. 2009;48:1077–92. doi: 10.1002/mc.20535. [DOI] [PubMed] [Google Scholar]
- 76.Chen HH, Mullett SJ, Stewart AF. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J Biol Chem. 2004;279:30800–6. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- 77.Teng AC, Kuraitis D, Deeke SA, Ahmadi A, Dugan SG, Cheng BL, et al. IRF2BP2 is a skeletal and cardiac muscle-enriched ischemia-inducible activator of VEGFA expression. FASEB J. 2010;24:4825–34. doi: 10.1096/fj.10-167049. [DOI] [PubMed] [Google Scholar]
- 78.Jin HS, Park HS, Shin JH, Kim DH, Jun SH, Lee CJ, et al. A novel inhibitor of apoptosis protein (IAP)-interacting protein, Vestigial-like (Vgl)-4, counteracts apoptosis-inhibitory function of IAPs by nuclear sequestration. Biochem Biophys Res Commun. 2011;412:454–9. doi: 10.1016/j.bbrc.2011.07.117. [DOI] [PubMed] [Google Scholar]
- 79.Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, et al. Australian Pancreatic Cancer Genome Initiative Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109:5934–41. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Belandia B, Parker MG. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem. 2000;275:30801–5. doi: 10.1074/jbc.C000484200. [DOI] [PubMed] [Google Scholar]
- 81.Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, et al. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–40. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–56. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fossdal R, Jonasson F, Kristjansdottir GT, Kong A, Stefansson H, Gosh S, et al. A novel TEAD1 mutation is the causative allele in Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13:975–81. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 85.Sudol M, Shields DC, Farooq A. Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin Cell Dev Biol. 2012;23:827–33. doi: 10.1016/j.semcdb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–8. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halder G, Carroll SB. Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development. 2001;128:3295–305. doi: 10.1242/dev.128.17.3295. [DOI] [PubMed] [Google Scholar]