In this study, Mendelian randomization was used to study genes known to modify iron levels, and the effect of iron on Parkinson's disease (PD) risk was estimated. Based on estimates of the genetic effects on both iron and PD obtained from the largest sample meta-analyzed to date, the findings suggest that increased iron levels in the blood are associated with a 3% reduction in the risk of Parkinson's disease for every 10 µg/dL increase in iron. The results of this analysis have potentially important implications for future research into the prevention of Parkinson's disease.
Please see later in the article for the Editors' Summary
Abstract
Background
Although levels of iron are known to be increased in the brains of patients with Parkinson disease (PD), epidemiological evidence on a possible effect of iron blood levels on PD risk is inconclusive, with effects reported in opposite directions. Epidemiological studies suffer from problems of confounding and reverse causation, and mendelian randomization (MR) represents an alternative approach to provide unconfounded estimates of the effects of biomarkers on disease. We performed a MR study where genes known to modify iron levels were used as instruments to estimate the effect of iron on PD risk, based on estimates of the genetic effects on both iron and PD obtained from the largest sample meta-analyzed to date.
Methods and Findings
We used as instrumental variables three genetic variants influencing iron levels, HFE rs1800562, HFE rs1799945, and TMPRSS6 rs855791. Estimates of their effect on serum iron were based on a recent genome-wide meta-analysis of 21,567 individuals, while estimates of their effect on PD risk were obtained through meta-analysis of genome-wide and candidate gene studies with 20,809 PD cases and 88,892 controls. Separate MR estimates of the effect of iron on PD were obtained for each variant and pooled by meta-analysis. We investigated heterogeneity across the three estimates as an indication of possible pleiotropy and found no evidence of it. The combined MR estimate showed a statistically significant protective effect of iron, with a relative risk reduction for PD of 3% (95% CI 1%–6%; p = 0.001) per 10 µg/dl increase in serum iron.
Conclusions
Our study suggests that increased iron levels are causally associated with a decreased risk of developing PD. Further studies are needed to understand the pathophysiological mechanism of action of serum iron on PD risk before recommendations can be made.
Please see later in the article for the Editors' Summary
Editors' Summary
Background
Parkinson disease is a degenerative disorder of the central nervous system caused by the death of dopamine-generating cells in the substania nigra, a region of the midbrain. The earliest symptoms are usually movement-related and include tremor, slow movements, and difficulty walking, and later cognitive and behavioral problems may arise, with dementia commonly occurring in the advanced stages of the disease. Parkinson disease affects around ten million people world-wide and incidence increases with age, with men more affected than women. To date, the causes of Parkinson disease remain unknown although a combination of genetic and environmental factors is thought to play a role. Identifying possible modifiable risks is an important step in the possible prevention of Parkinson disease.
Why Was This Study Done?
Previous studies have shown a possible association between lower blood levels of iron in people with Parkinson disease compared with controls, although the quality of these studies makes this finding difficult to interpret. So in this study, the researchers used a mendelian randomization approach to investigate whether there was any evidence of an effect of blood iron levels on the risk of Parkinson disease and if so to further explore the direction and scale of any link. Mendelian randomization is a method of using measured variation in genes of known function to examine the causal effect of a modifiable exposure on disease in situations where it is inappropriate to perform a randomized controlled trial.
What Did the Researchers Do and Find?
The researchers estimated the effect of blood iron levels on the risk of Parkinson disease using three polymorphisms in two genes, HFE and TMPRSS6. For each polymorphism, they performed a meta-analysis combining the results of studies investigating the genetic effect on iron levels, which included almost 22,000 people from Europe and Australia, and a meta-analysis of studies investigating the genetic effect on the risk of Parkinson disease, which included a total of 20,809 people with Parkinson disease and 88,892 controls from Europe and North America. They then performed three separate mendelian randomization analyses to estimate the effect of iron on Parkinson disease for the three polymorphisms. By combining the three estimates, they obtained a statistically significant odds ratio of 0.97 for Parkinson disease per 10 µg/dl increase in iron, corresponding to a 3% reduction in the risk of Parkinson disease for every 10 µg/dl increase in blood iron. Since genotype influences on blood iron levels represent differences that generally persist throughout adult life, the combined mendelian randomization estimate reflects an effect of iron over the course of a lifetime.
What Do These Findings Mean?
These findings suggest that increased iron levels in the blood are associated with a 3% reduction in the risk of Parkinson disease for every 10 µg/dl increase in iron. This finding is important as it suggests that increased blood iron levels may have a protective effect against Parkinson disease, although the underlying mechanism remains unclear. Furthermore, although mendelian randomization is an increasingly used approach to address the issue of classical confounding, there may be remaining confounding factors specific of mendelian randomization that may influence the interpretation of this study. Nevertheless, the results of this analysis have potentially important implications for future research into the prevention of Parkinson disease. Further studies on the underlying mechanisms are needed before any specific treatment recommendations can be proposed.
Additional Information
Please access these Web sites via the online version of this summary at http://dx.doi.org/10.1371/journal.pmed.1001462.
The National Institutes of Neurological Disorder and Stroke, MedlinePlus, and NHS Choices have several pages with comprehensive information on Parkinson disease
Wikipedia gives an explanation of mendelian randomization (note that Wikipedia is a free online encyclopedia that anyone can edit; available in several languages)
Introduction
Iron is involved in fundamental biochemical activities, such as oxygen delivery, mitochondrial respiration, and DNA synthesis in almost all cell types. In the brain, iron is a cofactor for a large number of enzymes, including key enzymes of neurotransmitter biosynthesis, such as the tyrosine hydroxylase, which represents the rate-limiting enzyme of dopamine synthesis [1]. However, iron is also potentially toxic as an excess of free iron contributes to the generation of reactive oxygen species and can favor oxidative tissue damage [1]. In the brains of patients with Parkinson disease (PD), increased levels of iron in the substantia nigra (SN) and the lateral globus pallidus have been observed, and yet the mechanisms responsible for this phenomenon are not completely understood [2],[3]. PD is characterized by the rather selective loss of dopaminergic neurons [4] and the presence of α-synuclein-enriched Lewy body inclusions in the SN [5], and several studies have demonstrated that free iron in the SN can enhance the aggregation of α-synuclein and may thus promote the formation of Lewy bodies [1].
Limited epidemiological evidence on the relationship between peripheral blood levels of iron and PD risk is available. A recent meta-analysis of ten studies, with a total of 520 PD cases and 711 controls, showed a trend for lower serum iron levels in PD patients compared with controls, although the difference in iron levels was not statistically significant (standardized mean difference: −0.45; 95% CI −0.98 to 0.08; p = 0.09) [6]. However, the very large degree of heterogeneity observed across studies (I2: 93%; p<0.0001) makes it difficult to interpret these findings.
A major limitation of observational studies is the difficulty in distinguishing between causal and spurious associations due to problems of confounding and reverse causation. Mendelian randomization (MR) is an approach based on the use of genes as instrumental variables, which has been proposed to assess causality and provide estimates of the effect of modifiable intermediate phenotypes on disease unaffected by classical confounding or reverse causation, whenever randomized clinical trials are not feasible [7]. Genes are randomly allocated at conception, so that genetic effects on the intermediate phenotype cannot be affected by classical confounding, such as lifestyle factors, or reverse causation, as in the situation where the phenotype level is influenced by the presence of the disease [8]. For this reason, demonstration that a genetic polymorphism known to modify the phenotype level also modifies the disease risk represents indirect evidence of a causal association between phenotype and disease.
The MR estimate of the effect of the intermediate phenotype on the disease is derived from the estimates of the associations of the polymorphism with both intermediate phenotype and disease. MR, as any other instrumental variable approach, has low statistical power and therefore requires very large sample sizes [9]. The recent availability of large collections of genome-wide data on intermediate phenotypes, such as blood biomarkers, and disease traits within international consortia represents a great opportunity to exploit the potentials of this approach, and indeed MR studies have become increasingly popular over the last few years.
The validity of the MR approach relies on the crucial assumption that the polymorphism acts on the disease only through the intermediate phenotype of interest and not through others (assumption of no pleiotropy) [8]. Evaluating the possibility of pleiotropic effects of the polymorphism is therefore fundamental when using MR, and yet pleiotropy can only be excluded with confidence if the function of the gene and its polymorphisms is completely known, which is rarely the case. This problem can be addressed by using multiple instruments (polymorphisms in multiple genes influencing the same intermediate phenotype), since in the absence of pleiotropy, similar MR estimates should be obtained regardless of the instrument used, so that differences across MR estimates beyond what can be expected by chance can indicate the presence of pleiotropy [10].
In this study, we provide evidence on the presence, direction, and magnitude of a causal effect of serum iron levels on PD risk by performing a MR study, based on iron data in 21,567 individuals from the general population and PD data from 20,809 PD cases and 88,892 controls. We used three polymorphisms as instruments in order both to increase statistical power by combining their MR estimates and to investigate the possible presence of pleiotropy.
Methods
Mendelian Randomization Approach
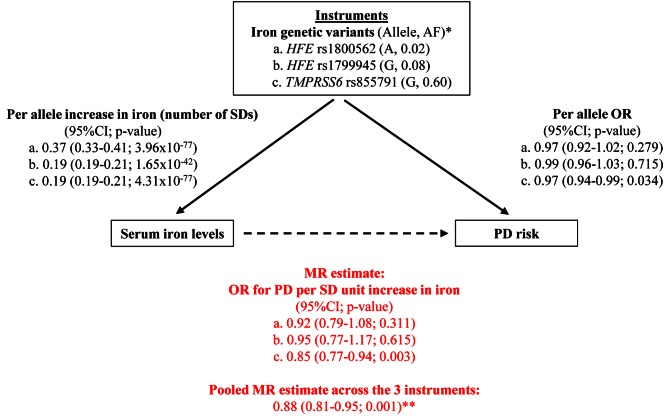
The selection of the genes modifying iron levels to be used as instruments in our MR study was based on published results showing that polymorphisms in the hemochromatosis (HFE, ENSG00000010704) gene and the transmembrane protease 6 (TMPRSS6, ENSG00000187045) gene have the strongest effects on serum iron in the general population of European ancestry [11]. The choice of the polymorphisms within these two genes was based on the findings of a recent large meta-analysis of genome-wide association (GWA) studies on iron levels in the general population (unpublished data). We selected the polymorphisms with the strongest statistical evidence, two for the HFE gene, rs1800562 (C282Y) and rs1799945 (H63D), which are not in linkage disequilibrium (HapMap CEU r2<0.01) and therefore represent independent signals of association, and one for the TMPRSS6 gene, rs855791 (V736A) (Figure 1).
Figure 1. Graphical representation of the MR approach, with all estimates used to derive the final MR estimate.
*Reported is the allele that increases iron levels, together with its frequency (AF). **This corresponds approximately to an OR per unit µg/dl increase in iron of 0.997 (95%CI 0.994–0.999), that is 0.3% (0.1%–0.6%) relative reduction in PD risk per 1 µg/dl increase in iron.
Our MR approach was based on the use of aggregate results for both the gene–iron and gene–PD associations: for each polymorphism, we performed a meta-analysis of studies investigating its effect on iron levels and a meta-analysis of studies investigating its effect on PD risk, with no studies contributing to both meta-analyses (see next sections). Three separate MR estimates of the effect of iron on PD were obtained for the three polymorphisms, and they were subsequently pooled by meta-analysis to provide a single MR estimate. Heterogeneity between the three MR estimates was investigated to detect the possible presence of pleiotropy.
Data on Gene Associations with Iron
Estimates of the effect sizes of the three polymorphisms in HFE and TMPRSS6 on total serum iron levels was based on the findings of a recent GWA meta-analysis on iron parameters performed by the Genetics of Iron Status (GIS) Consortium (Table 1) (unpublished data). The GIS meta-analysis includes ten cohorts from eight participating research groups. The individual datasets included in the meta-analysis are described in Table S1.
Table 1. Characteristics of the studies included for the gene–iron and gene–PD associations.
| Data Source | n studies | Type of Study | Maximum Sample Size |
| Gene–iron association | |||
| GIS Consortiuma | 10 | GWA | 21,567 |
| Gene–PD association | |||
| PDGene database [13]–[21] | 9 | Candidate gene studies (HFE rs1800562 and HFE rs1799945) | 2,384 cases; 6,908 controls |
| PD GWAS Consortium [22] | 5 | GWA | 4,238 cases; 4,239 controls |
| 23andMe [23] b | 1 | GWA | 4,127 cases; 62,037 controls |
| IPDGC [24],[25] c | 4 | GWA | 4,258 cases; 10,152 controls |
| IPDGC [24],[25] | 5 | Immunochip genotyping | 5,802 cases; 5,556 controls |
Unpublished data. The original sample size was 22,444, but genotype and phenotype data were available only for 21,567.
23andMe: slightly expanded version of the cohort used in [23].
IPDGC: USA-NIA and USA-dbGAP studies were not included in our analysis due to overlap with PD GWAS Consortium; the Icelandic dataset was not available for analysis.
Data on Gene Associations with PD Risk
To estimate the association of the three polymorphisms with PD risk, we performed a meta-analysis of both candidate gene and GWA studies (Table 1).
Candidate gene studies were identified using PDGene (http://www.pdgene.org), a database providing a regularly updated synopsis of genetic association studies performed in PD [12]. These studies provided data for the two polymorphisms in HFE, rs1800562 and rs1799945. A total of nine studies were included in our analysis for both rs1800562 [13]–[20] and rs1799945 [13]–[17],[19]–[21] (Tables 1 and S2).
Three large international GWA studies recently published, the PD GWAS Consortium [22], the 23andMe study [23], and the International Parkinson's Disease Genomics Consortium (IPDGC) [24],[25], provided data for all three polymorphisms (Table 1). The PD GWAS Consortium includes data from five studies: PROGENI/GenePD [26], NIA Phase I [27], NIA Phase II [28], HIHG [29], and NGRC [30]. The 23andMe data come from a slightly expanded version of the cohort used in [23], including more than 4,000 PD cases and 60,000 controls. From the IPDGC, four GWA studies were included in our analysis, together with five studies genotyped with a custom genotyping array (Immunochip Illumina iSelect array); the USA-NIA and the USA-dbGAP studies were not included because of overlap with the PD GWAS dataset, and the Icelandic study was not available for analysis.
A detailed description of the individual datasets is reported in Text S1 and in Table S2.
Statistical Analyses
GIS meta-analysis results for the gene–iron association were expressed in terms of Z-score, that is the number of standard deviations (SDs) above the mean iron level associated with each copy of the allele.
Study results for the candidate gene studies investigating the gene–PD risk association were obtained either from the PDGene website or directly from the original papers [18]–[20]. For two studies, estimates of the associations of interest were not provided, but they could be calculated from the data reported, by performing a per-genotype analysis based on an additive genetic model [19], or a per-allele analysis when genotype data were not available [20]. For the gene–PD meta-analysis, estimates of the (log) odds ratio (OR) were combined across studies using an inverse-variance-weighted fixed-effect model and assuming an additive genetic model, consistently with the gene–iron meta-analysis.
As for the instrumental variable analysis, an MR estimate of the effect of iron on PD risk was obtained for each of the three instruments separately, and the three estimates were combined using an inverse-variance-weighted fixed-effect meta-analysis. We evaluated the presence and magnitude of heterogeneity across the three instruments with the I2 statistics, a measure defined as the percentage of total variation in study estimates explained by heterogeneity rather than sampling error [31]. MR estimates were derived using the Wald-type estimator [32]:
where log ORPD/iron is the (log) increase of PD risk by SD unit increase in iron (MR estimate), log ORPD/allele is the (log) increase in PD risk per allele (gene–PD association), and betairon/allele is the number of SDs above the mean iron level per allele (gene–iron association). The standard error of the MR estimate was derived using the Delta method [33],[34]. The MR estimate is presented in terms of OR, by exponentiating the log ORPD/iron.
We evaluated the strength of each instrument using the F statistics, which is a function of the magnitude and precision of the genetic effect on the biomarker (iron):
where R2 is the variance of iron blood levels explained by the genetic variant and n is the sample size for the gene–iron association. We also evaluated the overall F statistics for the three combined instruments assuming that their effects were independent, as are expected to be given that the three gene variants are not in linkage disequilibrium.
A sensitivity analysis was performed to investigate the possible impact on our findings of population stratification in any of the studies included in the gene–iron or gene–PD analyses, by excluding studies which had not adjusted for population stratification.
All analyses were performed using Stata 10 (StataCorp LP).
Results
Gene Association with Iron
The GIS meta-analysis for iron levels included 21,567 individuals from Europe and Australia (Table S1). The effect on iron levels, expressed as number of SDs from the mean, was 0.37 (95% CI 0.33–0.41; p = 4.0×10−77) for each copy of the A allele of HFE rs1800562, 0.19 (95% CI 0.17–0.21; p = 1.7×10−42) for the G allele of HFE rs1799945, and 0.19 (95% CI 0.17–0.21; p = 4.3×10−77) for the G allele of TMPRSS6 rs855791 (Figure 1; Table S3). With a SD for serum iron levels of 37.6 µg/dl, these figures correspond to an increase in iron per allele of approximately 13.9, 7.1. and 7.1 µg/dl, respectively. HFE rs1800562, HFE rs1799945, and TMPRSS6 rs855791 explained 1.7%, 0.9%, and 1.7% of iron total variance, respectively (Table S3).
The F statistics was very high for all genetic variants, as can be expected given the sample size of more than 21,000 individuals [35]: 382, 199, and 379 for HFE rs1800562, HFE rs1799945, and TMPRSS6 rs855791, respectively. The F statistics for all combined instruments was 987.
Gene Association with PD Risk
All datasets available for the analysis of the effects of the three genetic polymorphisms on PD risk (Table S2) were checked for the presence of overlapping studies, and duplicates were removed. The meta-analysis, which included a total of 20,809 PD cases and 88,892 controls from Europe and North America (Table S2), revealed a significant association for TMPRSS6 rs855791 with PD risk, with an OR of 0.97 (95% CI 0.94–0.99; p = 0.034) per copy of the G allele. As shown in the Forest plot of the meta-analysis for this polymorphism (Figure S3), there was no statistical evidence of heterogeneity across studies, with a heterogeneity test p-value of 0.86 and an I2 of 0% (95% CI 0%–85%). In particular, although the 23andMe study was based on self-reported disease status and therefore differed from the rest, its results were consistent with those of the other PD studies. The association with PD risk for the two polymorphisms in HFE was not statistically significant, with an OR of 0.97 (95% CI 0.92–1.02; p = 0.281) for the A allele of rs1800562 and 0.99 (95% CI 0.96–1.03; p = 0.715) for the G allele of rs1799945 (Figures 1, S1, and S2; Table S4). This might be explained by the much lower statistical power for the two HFE variants compared with the TMPRSS6 variant due to their lower minor allele frequency (1,000 Genomes project: 0.02 and 0.08 versus 0.40), as suggested by their wide confidence intervals.
Mendelian Randomization Estimate of Iron Association with PD Risk
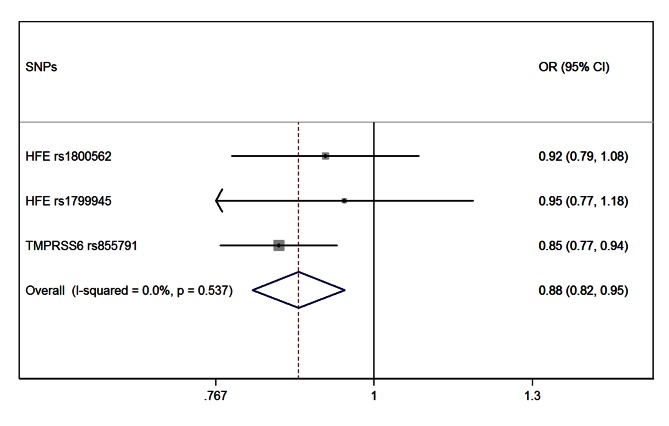
The meta-analysis of the three MR estimates resulted in a statistically significant combined estimate of 0.88 (95% CI 0.82–0.95; p = 0.001), representing the OR for PD per SD unit increase in iron (Figure 1). Again, with a SD for iron levels of 37.6 µg/dl, this corresponds approximately to an OR of 0.997 (95% CI 0.994–0.999) per 1 µg/dl increase in iron, that is a 0.3% (95% CI 0.1%–0.6%) relative risk reduction. The Forest plot in Figure 2 shows how the meta-analysis result was driven by the TMPRSS6 rs855791 variant, and that there was no statistical evidence of heterogeneity across instruments (p = 0.54; I2: 0%, 95% CI 0%–90%), suggesting that the assumption of no pleiotropy might hold.
Figure 2. Forest plot of the MR estimates from the three instruments.
The size of the squares is proportional to the precision of the MR estimates for each polymorphism, with the horizontal lines indicating their 95% confidence intervals. The combined MR estimate is represented by the centre of the diamond, with the lateral tips indicating its 95% confidence interval. The solid vertical line is the line of no effect.
The sensitivity analysis investigating the impact of population stratification excluded the nine studies from PDGene, which had not reported any adjustment for population stratification, while there were no exclusions from the GIS consortium on iron since all studies had adjusted for population stratification (Table S2). The result of the sensitivity analysis was similar to that of the main analysis, with a combined MR estimate of 0.91 (95% CI 0.83–0.99; p = 0.032) (Figure S4).
Discussion
Our study shows a protective effect of serum iron levels on PD, with a 3% (95% CI 1%–6%; p = 0.001) relative reduction in PD risk per 10 µg/dl increase in iron. If we hypothesise increasing serum iron levels of one SD unit (38 µg/dl in our study) in a population of Caucasians older than 60, where PD risk is around 1% [36], a corresponding relative risk reduction of 12% would translate to a decrease in PD cases from 100/10,000 to 88/10,000. Since genotype influences on serum iron levels represent differences that generally persist throughout adult life, the estimate of our MR study reflects an effect of iron over the course of a lifetime. These findings are important since evidence on the association between serum iron levels and PD risk collected so far has been controversial. Although iron is generally thought of as a risk factor for PD, in line with the well-known phenomenon of iron accumulation in the brain of PD patients [2],[3], epidemiological studies have shown effects of iron in opposite directions. A recent meta-analysis of epidemiological studies suggests a possible protective role of serum iron levels on PD risk, but its findings are difficult to interpret owing to the very large degree of heterogeneity across studies [6]. Epidemiological studies suffer from confounding and reverse causation, which are intrinsic to their observational nature, so that they can hardly provide conclusive evidence on the causality of an observed association. Tobacco smoking and coffee drinking, which have been suggested as protective factors for PD [37],[38], represent two potential confounders for the association between iron and PD, since both might have an effect on iron levels. Nicotine might decrease the availability of free reactive iron [39], and coffee is known to inhibit the intestinal absorption of iron [40],[41]. Reverse causation could also produce spurious associations in epidemiological studies if the phenotype level can be influenced by the presence of the disease. An example is that of monoamine oxidase (MAO) inhibitors used to treat PD. MAO inhibitors may have iron-chelating effects and thus reduce iron blood levels, which could lead to spurious epidemiological evidence of a difference in iron levels between PD cases and controls [42]. Although causality is usually assessed by use of randomized clinical trials, the MR approach represents a valuable alternative whenever these are not feasible [7]. It is based on the concept that genetic variation modifying the concentration of a biomarker should also affect the disease risk if (and only if) the biomarker is directly and causally involved in the disease pathogenesis. Being genes randomly allocated at conception, their effects on biomarkers are unaffected by classical confounding factors and reverse causation [8].
The protective effect of higher serum iron levels on PD risk found in our study may seem somewhat counterintuitive at first sight. However, there are several reports in the literature in line with our findings. A recent study showed a negative correlation between SN echogenicity, a marker for increased SN iron content [43], and serum iron levels in PD patients [44]. A case-control study suggested an increased risk of PD in men who reported multiple recent blood donations and thus experienced depleted systemic iron stores [45], and another study showed an association of anemia experienced early in life with increased PD risk, with the authors hypothesizing that anemia could be a surrogate marker for iron deficiency [46]. Finally, in dietary iron-restricted mice impaired motor behavior and a marked decrease of striatal dopamine levels was observed, which was explained with the fact that iron is essential for the activity of tyrosine hydroxylase, the rate-limiting enzyme in the dopamine synthesis [47]. Consistent with these findings, a recent study performed in Japan found an association between higher iron intake and reduced PD risk [48].
The underlying mechanisms of the protective effect of iron on PD risk observed in our study remains unclear, as does the mechanism that regulates the relationship between serum and brain iron levels. Low peripheral iron levels may reduce the functioning of neuronal enzymes or receptors, since iron is a crucial cofactor of tyrosine hydroxylase [49], plays a role in the synthesis of monoamine neurotransmitters, and is involved in dopaminergic neurodevelopment [50]. Furthermore, low iron levels may decrease neuronal iron storage in the form of ferritin [51], which was found to be inappropriately low in SN neurons in PD [1]. A reduction in ferritin could decrease neuronal iron utilization by decreasing the pool of iron available for neuronal enzymes [47], thus leading to the accumulation of free iron in SN [1]. Similar large-scale MR studies investigating other markers of iron metabolism, such as ferritin and transferrin, could contribute to our understanding of the role of peripheral iron homeostasis in the pathophysiology of PD.
To our knowledge, this is the first MR study aimed at estimating the magnitude of the effect of serum iron levels on PD risk. Previous case-control studies have tried to assess causality and direction of the association by investigating the effect on PD risk of genes involved in iron metabolism and homeostasis, although their findings are somewhat inconsistent with only some supporting the hypothesis of a causal association. Among the many genes evaluated, which include FTL, FTH1, TF, TFRC, IREB2, LTF, CP, FXN, HFE [52], HPX, HAMP, HFE2 [53], and FTMT [54], only the G258S polymorphism in the TF gene showed a statistically significant association with PD [17], although the finding was not replicated in a subsequent study [55], and a haplotype in the SLC11A2 gene was found to occur more frequently in PD [56]. However, all these previous studies were relatively small and therefore underpowered to detect modest genetic effects on PD risk. Our MR study used three polymorphisms in the HFE and TMPRSS6 genes as instruments. Evidence on their association with PD risk was obtained through meta-analysis of several candidate gene studies and three large GWA studies, including a total of more than 20,000 patients and 88,000 controls, which represents the largest PD case-control sample with genetic data meta-analyzed to date. Similarly, estimates of the effect of the three polymorphisms on serum iron levels were based on results from a recent GWA meta-analysis including more than 21,000 individuals. Unlike similar MR investigations that have combined multiple instruments into a single allele score using individual data analyses from all contributing studies, our analyses required only aggregate results for the effect of each genetic variant on both biomarker and disease. This may have practical importance, since it allows inclusion of results from ongoing genetic consortia without requiring further analyses, as well as inclusion of previous findings from published reports. However, methodological work will be needed to assess the relative benefits of the two approaches under different scenarios.
The crucial aspect of a MR study, and more generally of any study based on an instrumental variable approach, is the choice of the gene (instrument) that needs to have a strong effect on the intermediate phenotype of interest. We used three polymorphisms as instrumental variables, since the use of multiple instruments influencing the intermediate phenotype of interest can increase the statistical power of the MR analysis [10]. The instrument strength was high for all of them, as shown by their very large F-statistic values. Two of them, rs1800562 (C282Y) and rs1799945 (H63D), are non-synonymous polymorphisms in HFE, a gene with well known effects in the modulation of iron blood levels [57]. The third non-synonymous polymorphism, rs855791 (V736A), is located in TMPRSS6, a gene whose role in iron regulation was demonstrated more recently [58]. The two variants in the HFE gene are responsible for most cases of hereditary hemochromatosis [59],[60], and they are associated with iron overload when present in the homozygous (C282Y/C282Y) or compound heterozygous (C282Y/H63D) state. The C282Y variant prevents the altered HFE protein from reaching the cell surface and interacting with the transferrin receptor (TfR) [61],[62]. As a result, iron regulation is disrupted. The exact functional effect of the H63D variant is as yet unclear, but some evidence suggests that it may alter an intramolecular salt bridge, possibly affecting the interaction of the HFE protein with the TfR [63]. The TMPRSS6 V736A variant was found associated with iron-deficiency anemia [64]. Furthermore, the A allele has been shown to inhibit hepcidin more efficiently than the V allele in in vitro experiments, and to affect hepcidin levels in healthy individuals [65]. Interestingly, TMPRSS6 rs855791 was by far the most influential and was the one driving the result of the meta-analysis of MR estimates from the three instruments. The wide confidence intervals of the MR estimates for HFE rs1800562 and rs1799945 suggest that the power of their MR analysis was very limited due to their low allele frequency. This illustrates the importance of balancing the strength of the effect on the intermediate phenotype with allele frequency and statistical power when choosing the instruments for a MR study.
A potential source of bias specific to MR studies is pleiotropy, whereby the HFE or TMPRSS6 genotypes could influence PD risk through another mechanism that is independent of their effect on serum iron levels. Although we cannot completely exclude pleiotropic effects of the three polymorphisms used in our study because of incomplete knowledge of the underlying biology, we can indirectly investigate the presence of such effects through the simultaneous use of the three polymorphisms as multiple instruments. In a MR study, if all instruments are valid, their MR estimates should differ only as a result of sampling error [10], so that there should be no heterogeneity in the meta-analysis of MR estimates. In our meta-analysis of MR estimates there was no evidence of heterogeneity, although the statistical power to detect heterogeneity is limited when only three estimates are included in the meta-analysis [66]. As more evidence on genes influencing iron blood levels becomes available, MR studies investigating the effects of iron on the risk of PD and other diseases will be able to include many more genetic variants as instruments. This will ensure that pleiotropy can be ruled out with greater confidence. Selection of genes to be used as instruments requires careful consideration, since inclusion of variants with small genetic effects on the biomarker may introduce a “weak instrumental variable” bias [35]. Another potential issue in MR investigations is developmental canalization, the ability to produce the same phenotype regardless of genetic (or environmental) variation. If a genetic polymorphism is expressed during fetal development, compensatory processes may influence development in a way that can protect against the effect of the polymorphism [8]. Although canalization of genetic effects needs to be considered when interpreting MR findings, this problem is very difficult to investigate. Finally, one could speculate that the observed association of the subject's iron-related genotype with PD risk might actually reflect an intrauterine effect of iron due to a similar iron-related maternal genotype. Some evidence suggests that maternal iron deficiency could result in an altered iron status of the newborn, with possible negative effects on the neurophysiologic development [67].
Despite all the possible limitations discussed above, MR offers a valuable approach to derive causal effect estimates whenever randomized trials are very difficult to perform, as in the case of iron and PD. A trial investigating the long-term effect of changes in a subject's iron status, obtained by some means, on the risk of developing PD would require not only a very long follow-up but also a huge sample size, given the low frequency of the disease and the magnitude of the effect that might realistically be expected.
In our study, the MR analysis to combine the OR of the gene–PD association with the effect of the gene–iron association was based on a Wald-type estimator, which works under a “rare disease assumption” that is appropriate in the case of PD. However, the use of a Wald-type estimator for the MR analysis of binary outcomes represents only an approximate method and may produce biased MR estimates [32]. Although such bias has been recently shown to be small, typically within 10% of the MR estimate [68], methods in this area are still under active development.
In summary, our MR study suggests a causal association between increased serum iron levels and decreased risk of developing PD, suggesting that disrupted iron metabolism may be an important factor in the pathogenesis of PD. However, further research is needed to elucidate the pathophysiological mechanism of action underlying our findings. The effect of dietary iron or drugs capable of altering the balance between serum iron and iron storage compartments, might prove to be suitable to test in experimental models. The development of such disease models is therefore necessary before any public health or clinical recommendation can be made for primary prevention in subjects at high risk of developing PD.
Supporting Information
Forest plot of the meta-analysis of the studies included for the effect of HFE rs1800562 on PD risk. The boxes indicate the genetic (additive) effects of individual studies, with the size of the box being inversely proportional to the variance and horizontal lines indicating 95% confidence intervals. The diamond indicates the pooled effect estimate, obtained using inverse-variance weighted fixed-effect meta-analysis, and its 95% confidence interval. The full vertical line shows the value for no effect, as opposed to the dashed line indicating the estimated pooled effect.
(TIF)
Forest plot of the meta-analysis of the studies included for the effect of HFE rs1799945 on PD risk. The boxes indicate the genetic (additive) effects of individual studies, with the size of the box being inversely proportional to the variance and horizontal lines indicating 95% confidence intervals. The diamond indicates the pooled effect estimate, obtained using inverse-variance weighted fixed-effect meta-analysis, and its 95% confidence interval. The full vertical line shows the value for no effect, as opposed to the dashed line indicating the estimated pooled effect.
(TIF)
Forest plot of the meta-analysis of the studies included for the effect of TMPRSS6 rs855791 on PD risk. The boxes indicate the genetic (additive) effects of individual studies, with the size of the box being inversely proportional to the variance and horizontal lines indicating 95% confidence intervals. The diamond indicates the pooled effect estimate, obtained using inverse-variance weighted fixed-effect meta-analysis, and its 95% confidence interval. The full vertical line shows the value for no effect, as opposed to the dashed line indicating the estimated pooled effect.
(TIF)
Sensitivity analysis: Forest plot of the mendelian randomization estimates after exclusion of nine studies from the PDGene dataset that had not adjusted for population stratification (see Table S2).
(TIF)
Characteristics and sample size of the individual studies included for the gene–iron association. In all studies, the analyses were adjusted for age and sex, as well as for the first five MDS (multidimensional scaling) or principal components to control for population stratification.
(DOC)
Characteristics and sample size of the individual studies included for the gene–PD association.
(DOC)
Gene–iron association: GIS-consortium meta-analysis. The effect size for the genetic effects on iron levels is expressed as number of SDs from the mean (Z-scores).
(DOC)
Gene–PD association: meta-analysis of all available candidate gene and GWA studies.
(DOC)
Detailed description of the studies included in the three GWA investigations of PD risk.
(DOC)
Acknowledgments
The authors are grateful to the study participants. We are grateful to Esther Meissner and Maria Liebsch (Max Planck Institute forMolecular Genetics, Berlin, Germany) for assembling data of the PDGene database. We thank the GIS, the PD GWAS and the IPDGC consortia, and the 23andMe study for providing data for the gene–iron and gene–PD associations. The IPDGC acknowledges the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD, USA, and DNA panels, samples, and clinical data from the National Institute of Neurological Disorders and Stroke Human Genetics Resource Center DNA and Cell Line Repository were used. People who contributed samples are acknowledged in descriptions of every panel on the repository website. It thanks the French Parkinson's Disease Genetics Study Group: Y Agid, M Anheim, A-M Bonnet, M Borg, A Brice, E Broussolle, J-C Corvol, P Damier, A Destée, A Dürr, F Durif, S Klebe, E Lohmann, M Martinez, P Pollak, O Rascol, F Tison, C Tranchant, M Vérin, F Viallet, and M Vidailhet. It also thanks the members of the French 3C Consortium: A Alpérovitch, C Berr, C Tzourio, and P Amouyel for allowing us to use part of the 3C cohort, and D Zelenika for support in generating the genome-wide molecular data. IPDGC thanks P Tienari (Molecular Neurology Programme, Biomedicum, University of Helsinki), T Peuralinna (Department of Neurology, Helsinki University Central Hospital), L Myllykangas (Folkhalsan Institute of Genetics and Department of Pathology, University of Helsinki), and R Sulkava (Department of Public Health and General Practice Division of Geriatrics, University of Eastern Finland) for the Finnish controls (Vantaa85+ GWAS data). It also thanks Jeffrey Barrett for assistance with the design of the ImmunoChip. We thank the Nijmegen Biomedical Study (Principal investigators L.A.L.M. Kiemeney, M. den Heijer, A.L.M. Verbeek, D.W. Swinkels, and B. Franke).
Additional PD GWAS consortium members providing data to this study: GenePD Investigators and Coordinators: M. Lew, University Southern California School of Medicine; O. Suchowersky, University of Calgary; C. Klein, University of Lübeck, Germany; L. Golbe, M.H. Mark, UMDNJ-Robert Wood Johnson Medical School; J. Growdon, N. Huggins, Massachusetts General Hospital, Harvard Medical School; G.F. Wooten, University of Virginia Health System; R. Watts, University of Alabama at Birmingham; M. Guttman, University of Toronto; B. Racette, J. Perlmutter, Washington University School of Medicine; L. Marlor, Barrow Neurological Institute; H. Shill, Sun Health Research Institute; C. Singer, University of Miami; S. Goldwurm, G. Pezzoli, Parkinson Institute, Istituti Clinici di Perfezionamento, Milano, Italy; M.H. Saint-Hilaire, T. Massood, Boston University School of Medicine; K. Baker, I. Itin, Cleveland Clinic Foundation; I. Litvan, University of Louisville School of Medicine; G. Nicholson, A. Corbett, University of Sydney ANZAC Research Institute, Concord Hospital, Sydney, Australia; M. Nance, Struthers Parkinson's Center, Minneapolis; E. Drasby, Port City Neurology, Scarborough, ME; S. Isaacson, Parkinson's Disease and Movement Disorder Center of Boca Raton; D. Burn, P. Chinnery, Newcastle University, Newcastle upon Tyne, UK; S. Sherman, University of Arizona; R. Roxburgh, B. Snow, Auckland City Hospital, Auckland, New Zealand; J. Slevin, F. Cambi, University of Kentucky College of Medicine. GenePD Molecular Genetics Laboratories: J.F. Gusella, M.E. McDonald, M. Sun, L. Mysore, M.A, Anderson, D. Lucente, Center for Human Genetic Research, Massachusetts General Hospital, Harvard Medical School; S. Williamson, M.W. Nagle, Neurogenetics Laboratory, Boston University School of Medicine).
PSG-PROGENI Investigators and Coordinators: S Factor, D Higgins, S Evans, Albany Medical College; H Shill, M Stacy, J Danielson, L Marlor, K Williamson, Barrow Neurological Institute; J Jankovic, C Hunter, Baylor College of Medicine; D Simon, P Ryan, L Scollins, Beth Israel Deaconess Medical Center; R Saunders-Pullman, K Boyar, C Costan-Toth, E Ohmann, Beth Israel Medical Center; L Sudarsky, C Joubert, Brigham & Women's Hospital; J Friedman, K Chou, H Fernandez, M Lannon, Brown University (Memorial Hospital of RI); N Galvez-Jimenez, A Podichetty, K Thompson, Cleveland Clinic Florida-Weston; P Lewitt, M DeAngelis, Clinical Neuroscience Center; C O'Brien, L Seeberger, C Dingmann, D Judd, Colorado Neurological Institute; K Marder, J Fraser, J Harris, Columbia University Medical Center; J Bertoni, C Peterson, Creighton University; M Rezak, G Medalle, Evanston Northwestern Healthcare; S Chouinard, M Panisset, J Hall, H Poiffaut, Hotel Dieu Hospital-Chum; V Calabrese, P Roberge, Hunter Homes McGuire Veterans Medical Center; J Wojcieszek, J Belden, Indiana University School of Medicine; D Jennings, K Marek, S Mendick, Institute For Neurodegenerative Disorders; S Reich, B Dunlop, Johns Hopkins University; M Jog, C Horn, London Health Sciences Centre; R Uitti, M Turk, Mayo Clinic Jacksonville; T Ajax, J Mannetter, McFarland Neurosciences; K Sethi, J Carpenter, B Dill, L Hatch, K Ligon, S Narayan, Medical College of Georgia; K Blindauer, K Abou-Samra, J Petit, Medical College of Wisconsin; L Elmer, E Aiken, K Davis, C Schell, S Wilson, Medical University of Ohio; M Velickovic, W Koller (deceased), S Phipps, Mount Sinai School of Medicine; A Feigin, M Gordon, J Hamann, E Licari, M Marotta-Kollarus, B Shannon, R Winnick, North Shore-LIJ Health System; T Simuni, A Videnovic, A Kaczmarek, K Williams, M Wolff, Northwestern University; J Rao, M Cook, Ochsner Clinic Foundation; M Fernandez, S Kostyk, J Hubble, A Campbell, C Reider, A Seward, Ohio State University; R Camicioli, J Carter, J Nutt, P Andrews, S Morehouse, C Stone, Oregon Health & Science University; T Mendis, D Grimes, C Alcorn-Costa, P Gray, K Haas, J Vendette, Ottawa Hospital Civic Site; J Sutton, B Hutchinson, J Young, Pacific Neuroscience Medical Group; A Rajput, A Rajput, L Klassen, T Shirley, Saskatoon Dist Health Board Royal Univ Hosp; B Manyam, P Simpson, J Whetteckey, B Wulbrecht, Scott & White Hospital/Texas A&M University; D Truong, M Pathak, K Frei, N Luong, T Tra, A Tran, J Vo, The Parkinson's & Movement Disorder Institute; A Lang, G Kleiner-Fisman, A Nieves, L Johnston, J So, Toronto Western Hospital, University Health; L Giffin, UMDNJ-School of Osteopathic Medicine: G Podskalny; P Atchison, C Allen, University of Alabama at Birmingham; W Martin, M Wieler, University of Alberta; O Suchowersky, S Furtado, M Klimek, University of Calgary; N Hermanowicz, S Niswonger, University of California Irvine; C Shults (deceased), D Fontaine, University of California San Diego; M Aminoff, C Christine, M Diminno, J Hevezi, University of California San Francisco; A Dalvi, U Kang, J Richman, S Uy, J Young, University of Chicago; A Dalvi, A Sahay, M Gartner, D Schwieterman, University of Cincinnati; D Hall, M Leehey, S Culver, T Derian, University of Colorado Health Sciences Center; T Demarcaida, S Thurlow, University of Connecticut; R Rodnitzky, J Dobson, University of Iowa; K Lyons, R Pahwa, T Gales, S Thomas, University of Kansas Medical Center; L Shulman, S Reich, W Weiner, K Dustin, University of Maryland School of Medicine; K Lyons, C Singer, W Koller (deceased), W Weiner, L Zelaya, University of Miami; P Tuite, V Hagen, S Rolandelli, R Schacherer, J Kosowicz, University of Minnesota; P Gordon, J Werner, University of New Mexico; C Serrano, S Roque, University of Puerto Rico School of Medicine; R Kurlan, D Berry, I Gardiner, University of Rochester; R Hauser, J Sanchez-Ramos, T Zesiewicz, H Delgado, K Price, P Rodriguez, S Wolfrath, University of South Florida; R Pfeiffer, L Davis, B Pfeiffer, University of Tennessee Health Science Center; R Dewey, B Hayward, A Johnson, M Meacham, B Estes, University of Texas Southwestern Medical Center; F Walker, V Hunt, C O'Neill, Wake Forest University School of Medicine; B Racette, L Good, M Rundle, Washington University.
NGRC Investigators and Coordinators: D Kay, J Montimurro, V Kusel, New York State Department of Health Wadsworth Center; A Samii, E Martinez, D Yearout, VA Puget Sound Health Care System and University of Washington; J Nutt, Oregon Health and Sciences University; P Agarwal, A Griffith, Evergreen Hospital Medical Center; JW Roberts, Virginia Mason Medical Center; DS Higgins, Samuel Stratton VA Medical Center and Albany Medical Center; Eric Molho, Albany Medical Center; Ami Rosen, Emory University.
HIHG Investigators and Coordinators: CA Jauregui, MA Nance, RL Watts, JP Hubble, WC Koller, K Lyons, R Pahwa, MB Stern, A Colcher, BC Hiner, J Jankovic, WG Ondo, FH Allen, Jr., CG Goetz, GW Small, D Masterman, F Mastaglia, BL Scott, C Singer, F Nahab, MA Pericak-Vance, and JL Haines.
Additional IPDGC consortium members providing data to this study: Vincent Plagnol, UCL Genetics Institute, London, UK; Dena G Hernandez, Laboratory of Neurogenetics, National Institute on Aging; and Department of Molecular Neuroscience, UCL Institute of Neurology, London, UK; Manu Sharma, Department for Neurodegenerative Diseases, Hertie Institute for Clinical Brain Research, University of Tübingen, and DZNE, German Center for Neurodegenerative Diseases, Tübingen, Germany; Una-Marie Sheerin, Department of Molecular Neuroscience, UCL Institute of Neurology; Mohamad Saad, INSERM U563, CPTP, Toulouse, France; and Paul Sabatier University, Toulouse, France; Javier Simón-Sánchez, Department of Clinical Genetics, Section of Medical Genomics, VU University Medical Centre, Amsterdam, Netherlands; Claudia Schulte, Department for Neurodegenerative Diseases, Hertie Institute for Clinical Brain Research; Suzanne Lesage, INSERM, UMR_S975 [formerly UMR_S679], Paris, France; Université Pierre et Marie Curie-Paris, Centre de Recherche de l'Institut du Cerveau et de la Moelle épinière, Paris, France; and CNRS, Paris, France; Sampath Arepalli, Laboratory of Neurogenetics, National Institute on Aging; Roger Barker, Department of Neurology, Addenbrooke's Hospital, University of Cambridge, Cambridge, UK; Yoav Ben-Shlomo, School of Social and Community Medicine, University of Bristol; Henk W Berendse, Department of Neurology and Alzheimer Center, VU University Medical Center; Daniela Berg, Department for Neurodegenerative Diseases, Hertie Institute for Clinical Brain Research; Kailash Bhatia, Department of Motor Neuroscience, UCL Institute of Neurology; Rob M A de Bie, Department of Neurology, Academic Medical Center, University of Amsterdam, Amsterdam, Netherlands; Alessandro Biffi, Center for Human Genetic Research and Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; and Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA; Bas Bloem, Department of Neurology, Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands; Zoltan Bochdanovits, Department of Clinical Genetics, Section of Medical Genomics, VU University Medical Centre; Michael Bonin, Department of Medical Genetics, Institute of Human Genetics, University of Tübingen, Tübingen, Germany; Jose M Bras, Department of Molecular Neuroscience, UCL Institute of Neurology; Kathrin Brockmann, Department for Neurodegenerative Diseases, Hertie Institute for Clinical Brain Research; Janet Brooks, Laboratory of Neurogenetics, National Institute on Aging; David J Burn, Newcastle University Clinical Ageing Research Unit, Campus for Ageing and Vitality, Newcastle upon Tyne, UK; Gavin Charlesworth, Department of Molecular Neuroscience, UCL Institute of Neurology; Honglei Chen, Epidemiology Branch, National Institute of Environmental Health Sciences, National Institutes of Health, NC, USA; Patrick F Chinnery, Neurology M4104, The Medical School, Framlington Place, Newcastle upon Tyne, UK; Sean Chong, Laboratory of Neurogenetics, National Institute on Aging; Carl E Clarke, School of Clinical and Experimental Medicine, University of Birmingham, Birmingham, UK; and Department of Neurology, City Hospital, Sandwell and West Birmingham Hospitals NHS Trust, Birmingham, UK; Mark R Cookson, Laboratory of Neurogenetics, National Institute on Aging; J Mark Cooper, Department of Clinical Neurosciences, UCL Institute of Neurology; Jean Christophe Corvol, INSERM, UMR_S975; Université Pierre et Marie Curie-Paris; CNRS; and INSERM CIC-9503, Hôpital Pitié-Salpêtrière, Paris, France; Carl Counsell, University of Aberdeen, Division of Applied Health Sciences, Population Health Section, Aberdeen, UK; Philippe Damier, CHU Nantes, CIC0004, Service de Neurologie, Nantes, France; Jean-François Dartigues, INSERM U897, Université Victor Segalen, Bordeaux, France; Panos Deloukas, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, UK; Günther Deuschl, Klinik für Neurologie, Universitätsklinikum Schleswig-Holstein, Campus Kiel, Christian-Albrechts-Universität Kiel, Kiel, Germany; David T Dexter, Parkinson's Disease Research Group, Faculty of Medicine, Imperial College London, London, UK; Karin D van Dijk, Department of Neurology and Alzheimer Center, VU University Medical Center; Allissa Dillman, Laboratory of Neurogenetics, National Institute on Aging; Frank Durif, Service de Neurologie, Hôpital Gabriel Montpied, Clermont-Ferrand, France; Alexandra Dürr (INSERM, UMR_S975; Université Pierre et Marie Curie-Paris; CNRS; and AP-HP, Pitié-Salpêtrière Hospital; Sarah Edkins, Wellcome Trust Sanger Institute; Jonathan R Evans, Cambridge Centre for Brain Repair, Cambridge, UK; Thomas Foltynie, UCL Institute of Neurology; Jianjun Gao, Epidemiology Branch, National Institute of Environmental Health Sciences; Michelle Gardner, Department of Molecular Neuroscience, UCL Institute of Neurology; J Raphael Gibbs, Laboratory of Neurogenetics, National Institute on Aging; and Department of Molecular Neuroscience, UCL Institute of Neurology; Alison Goate, Department of Psychiatry, Department of Neurology, Washington University School of Medicine, MI, USA; Emma Gray, Wellcome Trust Sanger Institute;, Rita Guerreiro, Department of Molecular Neuroscience, UCL Institute of Neurology; Clare Harris, University of Aberdeen; Jacobus J van Hilten, Department of Neurology, Leiden University Medical Center, Leiden, Netherlands; Albert Hofman, Department of Epidemiology, Erasmus University Medical Center, Rotterdam, Netherlands; Albert Hollenbeck, AARP, Washington DC, USA; Janice Holton, Queen Square Brain Bank for Neurological Disorders, UCL Institute of Neurology; Michele Hu, Department of Clinical Neurology, John Radcliffe Hospital, Oxford, UK; Xuemei Huang, Departments of Neurology, Radiology, Neurosurgery, Pharmacology, Kinesiology, and Bioengineering, Pennsylvania State University–Milton S Hershey Medical Center, Hershey, PA, USA; Heiko Huber, Department for Neurodegenerative Diseases, Hertie Institute for Clinical Brain Research; Gavin Hudson, Neurology M4104, The Medical School, Newcastle upon Tyne, UK; Sarah E Hunt, Wellcome Trust Sanger Institute; Thomas Illig, Institute of Epidemiology, Helmholtz Zentrum München, German Research Centre for Environmental Health, Neuherberg, Germany; Jean-Charles Lambert, INSERM U744, Lille, France; and Institut Pasteur de Lille, Université de Lille Nord, Lille, France; Cordelia Langford, Cambridge Centre for Brain Repair; Andrew Lees, Queen Square Brain Bank for Neurological Disorders; Peter Lichtner, Institute of Human Genetics, Helmholtz Zentrum München, German Research Centre for Environmental Health, Neuherberg, Germany; Patricia Limousin, Institute of Neurology, Sobell Department, Unit of Functional Neurosurgery, London, UK; Grisel Lopez, Section on Molecular Neurogenetics, Medical Genetics Branch, NHGRI, National Institutes of Health; Delia Lorenz, Klinik für Neurologie, Universitätsklinikum Schleswig-Holstein; Alisdair McNeill, Department of Clinical Neurosciences, UCL Institute of Neurology; Catriona Moorby, School of Clinical and Experimental Medicine, University of Birmingham; Matthew Moore, Laboratory of Neurogenetics, National Institute on Aging), Huw R Morris, MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University School of Medicine, Cardiff, UK; Karen E Morrison, School of Clinical and Experimental Medicine, University of Birmingham; and Neurosciences Department, Queen Elizabeth Hospital, University Hospitals Birmingham NHS Foundation Trust, Birmingham, UK; Ese Mudanohwo, Neurogenetics Unit, UCL Institute of Neurology and National Hospital for Neurology and Neurosurgery; Sean S O'Sullivan, Queen Square Brain Bank for Neurological Disorders; Justin Pearson, MRC Centre for Neuropsychiatric Genetics and Genomics; Joel S Perlmutter, Department of Neurology, Radiology, and Neurobiology at Washington University, St Louis; Pierre Pollak, Service de Neurologie, CHU de Grenoble, Grenoble, France; Bart Post, Department of Neurology, Radboud University Nijmegen Medical Centre; Simon Potter, Wellcome Trust Sanger Institute; Bernard Ravina, Translational Neurology, Biogen Idec, MA, USA; Tamas Revesz, Queen Square Brain Bank for Neurological Disorders; Olaf Riess, Department of Medical Genetics, Institute of Human Genetics, University of Tübingen; Fernando Rivadeneira, Departments of Epidemiology and Internal Medicine, Erasmus University Medical Center; Patrizia Rizzu, Department of Clinical Genetics, Section of Medical Genomics, VU University Medical Centre; Mina Ryten, Department of Molecular Neuroscience, UCL Institute of Neurology; Stephen Sawcer, University of Cambridge, Department of Clinical Neurosciences, Addenbrooke's hospital, Cambridge, UK; Anthony Schapira, Department of Clinical Neurosciences, UCL Institute of Neurology; Hans Scheffer, Department of Human Genetics, Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands; Karen Shaw, Queen SquareBrain Bank for Neurological Disorders; Ira Shoulson, Department of Neurology, University of Rochester, Rochester, NY, USA; Ellen Sidransky, Section on Molecular Neurogenetics, Medical Genetics Branch, NHGRI; Colin Smith, Department of Pathology, University of Edinburgh, Edinburgh, UK; Chris C A Spencer, Wellcome Trust Centre for Human Genetics, Oxford, UK; Joanna D Stockton, School of Clinical and Experimental Medicine; Amy Strange, Wellcome Trust Centre for Human Genetics; Kevin Talbot, University of Oxford, Department of Clinical Neurology, John Radcliffe Hospital, Oxford, UK; Carlie M Tanner, Clinical Research Department, The Parkinson's Institute and Clinical Center, Sunnyvale, CA, USA; Avazeh Tashakkori-Ghanbaria (Wellcome Trust Sanger Institute; François Tison, Service de Neurologie, Hôpital Haut-Lévêque, Pessac, France; Daniah Trabzuni, Department of Molecular Neuroscience, UCL Institute of Neurology; Bryan J Traynor, Laboratory of Neurogenetics, National Institute on Aging; André G Uitterlinden, Departments of Epidemiology and Internal Medicine, Erasmus University Medical Center; Daan Velseboer, Department of Neurology, Academic Medical Center; Marie Vidailhet, INSERM, UMR_S975, Université Pierre et Marie Curie-Paris, CNRS, UMR 7225; Robert Walker, Department of Pathology, University of Edinburgh; Bart van de Warrenburg, Department of Neurology, Radboud University Nijmegen Medical Centre; Mirdhu Wickremaratchi, Department of Neurology, Cardiff University, Cardiff, UK; Nigel Williams, MRC Centre for Neuropsychiatric Genetics and Genomics; Caroline H Williams-Gray, Department of Neurology, Addenbrooke's Hospital; Sophie Winder-Rhodes, Department of Psychiatry and Medical Research Council and Wellcome Trust Behavioural and Clinical Neurosciences Institute, University of Cambridge; Maria Martinez, INSERM U563; and Paul Sabatier University; John Hardy, Department of Molecular Neuroscience, UCL Institute of Neurology; Peter Heutink, Department of Clinical Genetics, Section of Medical Genomics, VU University Medical Centre; Alexis Brice, INSERM, UMR_S975, Université Pierre et Marie Curie-Paris, CNRS, UMR 7225, AP-HP, Pitié-Salpêtrière Hospital; Thomas Gasser, Department for Neurodegenerative Diseases, Hertie Institute for Clinical Brain Research, and DZNE, German Center for Neurodegenerative Diseases; Andrew B Singleton, Laboratory of Neurogenetics, National Institute on Aging; Nicholas W Wood, UCL Genetics Institute; and Department of Molecular Neuroscience, UCL Institute of Neurology. Wellcome Trust Case-Control Consortium 2 members: Peter Donnelly (Chair), Wellcome Trust Centre for Human Genetics, Oxford, UK; Dept Statistics, University of Oxford, Oxford, UK; Ines Barroso (Deputy Chair), Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK; Jenefer M Blackwell, Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia, Subiaco, Western Australia; Cambridge Institute for Medical Research, University of Cambridge School of Clinical Medicine, Cambridge, UK; Elvira Bramon, Department of Psychosis Studies, NIHR Biomedical Research Centre for Mental Health at the Institute of Psychiatry, King's College London and The South London and Maudsley NHS Foundation Trust, Denmark Hill, London, UK; Matthew A Brown, Diamantina Institute of Cancer, Immunology and Metabolic Medicine, Princess Alexandra Hospital, University of Queensland, Brisbane, Queensland, Australia; Juan P Casas, Dept Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London and Dept Epidemiology and Public Health, University College London, UK; Aiden Corvin, Neuropsychiatric Genetics Research Group, Institute of Molecular Medicine, Trinity College Dublin, Dublin, Eire; Panos Deloukas, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK; Audrey Duncanson, Molecular and Physiological Sciences, The Wellcome Trust, London; Janusz Jankowski, Centre for Digestive Diseases, Queen Mary University of London, London, UK and Digestive Diseases Centre, Leicester Royal Infirmary, Leicester, UK and Department of Clinical Pharmacology, Old Road Campus, University of Oxford, Oxford, UK; Hugh S Markus, Clinical Neurosciences, St George's University of London, London; Christopher G Mathew, King's College London Dept Medical and Molecular Genetics, School of Medicine, Guy's Hospital, London, UK; Colin NA Palmer, Biomedical Research Centre, Ninewells Hospital and Medical School, Dundee, UK; Robert Plomin, King's College London Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Denmark Hill, London, UK; Anna Rautanen, Wellcome Trust Centre for Human Genetics, Oxford, UK; Stephen J Sawcer, University of Cambridge Dept Clinical Neurosciences, Addenbrooke's Hospital, Cambridge, UK; Richard C Trembath, King's College London Dept Medical and Molecular Genetics, School of Medicine, Guy's Hospital, London, UK; Ananth C Viswanathan, NIHR Biomedical Research Centre for Ophthalmology, Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London, UK; Nicholas W Wood, Dept Molecular Neuroscience, Institute of Neurology, Queen Square, London, UK (Management Committee). Chris C A Spencer, Gavin Band, Céline Bellenguez, Colin Freeman, Garrett Hellenthal, Eleni Giannoulatou, Matti Pirinen, Richard Pearson, Amy Strange, Zhan Su, Damjan Vukcevic, Wellcome Trust Centre for Human Genetics, Oxford, UK; Peter Donnelly, Wellcome Trust Centre for Human Genetics, Oxford, UK; Dept Statistics, University of Oxford, Oxford, UK (Data and Analysis Group). Cordelia Langford, Sarah E Hunt, Sarah Edkins, Rhian Gwilliam, Hannah Blackburn, Suzannah J Bumpstead, Serge Dronov, Matthew Gillman, Emma Gray, Naomi Hammond, Alagurevathi Jayakumar, Owen T McCann, Jennifer Liddle, Simon C Potter, Radhi Ravindrarajah, Michelle Ricketts, Matthew Waller, Paul Weston, Sara Widaa, Pamela Whittaker, Ines Barroso, Panos Deloukas, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK (DNA, Genotyping, Data QC and Informatics Group). Christopher G Mathew (Chair), King's College London Dept Medical and Molecular Genetics, School of Medicine, Guy's Hospital, London, UK; Jenefer M Blackwell, Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia, Subiaco, Western Australia; 5 Cambridge Institute for Medical Research, University of Cambridge School of Clinical Medicine, Cambridge, UK; Matthew A Brown, Diamantina Institute of Cancer, Immunology and Metabolic Medicine, Princess Alexandra Hospital, University of Queensland, Brisbane, Queensland, Australia; Aiden Corvin, Neuropsychiatric Genetics Research Group, Institute of Molecular Medicine, Trinity College Dublin, Dublin, Eire; Mark I McCarthy, Oxford Centre for Diabetes, Endocrinology and Metabolism (ICDEM), Churchill Hospital, Oxford, UK; Chris C A Spencer, Wellcome Trust Centre for Human Genetics, Oxford, UK (Publications Committee).
Additional GIS consortium members providing data to this study: Willem H Ouwehand, Department of Haematology, University of Cambridge, UK; Aparna Radhakrishnan, Department of Haematology, University of Cambridge, UK; Jennifer Sambrook, Department of Haematology, University of Cambridge, UK; Daniela Toniolo, Division of Genetics and Cell Biology, San Raffaele Research Institute, Milano Italy; Michela Traglia, Division of Genetics and Cell Biology, San Raffaele Research Institute, Milano Italy; Clara Camaschella, Vita and Salute University, Milano, Italy; Andres Metspalu, Estonian Biobank, Estonian Genome Centre, University of Tartu; Tonu Esko, Estonian Biobank, Estonian Genome Centre, University of Tartu; Christian Gieger, Institute of Genetic Epidemiology, Helmholtz Center Munich, German Research Center for Environmental Health, Neuherberg, Germany; Janina Ried, Institute of Genetic Epidemiology, Helmholtz Center Munich, German Research Center for Environmental Health, Neuherberg, Germany; Thomas Meitinger, Institute of Human Genetics, Helmholtz Center Munich, German Research Center for Environmental Health, Neuherberg, Germany; Konrad Oexle, Institute of Human Genetics, Klinikum Rechts der Isar, Technische Universität München, Munich, Germany; Juliane Winkelmann, Institute of Human Genetics, Klinikum Rechts der Isar, Technische Universität München, Munich, Germany, Institute of Human Genetics, Helmholtz Center Munich, Neuherberg, Germany; Dorine Swinkels, Laboratory of Genetic, Endocrine and Metabolic Diseases, Department of Laboratory Medicine, Radboud University Medical Centre, Nijmegen, The Netherlands; Sita Vermeulen, Department for Health Evidence, Radboud University Medical Centre, Nijmegen, The Netherlands; Cornelia van Duijn, Erasmus University Medical Center, the Netherlands; Linda Broer, Erasmus University Medical Center, the Netherlands; John Beilby, Pathwest, Queen Elizabeth II Medical Centre, Perth, Australia; Jennie Hui, Pathwest, Queen Elizabeth II Medical Centre, Perth, Australia; Denise Anderson, Pathwest, Queen Elizabeth II Medical Centre, Perth, Australia; Peter Visscher, Queensland Institute of Medical Research, Brisbane, Queensland, Australia; Nick Martin, Queensland Institute of Medical Research, Brisbane, Queensland, Australia.
Abbreviations
- GIS
Genetics of Iron Status Consortium
- GWA
genome-wide association
- IPDGC
International Parkinson's Disease Genomics Consortium
- MR
mendelian randomization
- PD
Parkinson disease
- SD
standard deviation
- SN
substantia nigra
Funding Statement
This study was supported by the Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano and the South Tyrolean Sparkasse Foundation. The PDGene database has been made possible by the kind support of the Michael J. Fox Foundation (MJFF) for Parkinson's Research, with additional support from Cure Alzheimer's Fund (CAF), the Fidelity Biosciences Research Initiative, Prize4Life, the National Alliance for Research on Schizophrenia and Depression (NARSAD), and EMD Serono. RHM received grants from the Robert P. & Judith N. Goldberg Foundation, the Cogan Family Foundation and R01-NS076843, Characterization of the Role of cyclin G-Associated Kinase in Parkinson disease and an unrestricted grant from the Karyopharm Therapuetics Inc. TF has received research grants from NIH, MJFF for Parkinson's Research, and the Susan G. Komen Foundation. IPDGC was supported by the Intramural Research Programs of the National Institute on Aging, National Institute of Neurological Disorders and Stroke, National Institute of Environmental Health Sciences, and National Human Genome Research Institute of the US National Institutes of Health (NIH), Department of Health and Human Services (project numbers Z01-AG000949-02 and Z01-ES101986), human subjects protocol 2003-077. IPDGC was also supported by the US Department of Defense (award number W81XWH-09-2-0128); NIH (grants NS057105 and RR024992); American Parkinson Disease Association (APDA); Barnes Jewish Hospital Foundation; Greater St. Louis Chapter of the APDA; Hersenstichting Nederland; Neuroscience Campus Amsterdam; and the section of medical genomics, the Prinses Beatrix Fonds. The KORA (Cooperative Research in the Region of Augsburg) research platform was started and financed by the Forschungszentrum für Umwelt und Gesundheit, which is funded by the German Federal Ministry of Education, Science, Research, and Technology and by the State of Bavaria. It was also funded by the German National Genome Network (NGFNplus number 01GS08134, German Ministry for Education and Research); by the German Federal Ministry of Education and Research (NGFN 01GR0468, PopGen); and 01EW0908 in the frame of ERA-NET NEURON and Helmholtz Alliance Mental Health in an Ageing Society (HA-215), which was funded by the Initiative and Networking Fund of the Helmholtz Association. The French GWAS work was supported by the French National Agency of Research (ANR-08-MNP-012). It was also sponsored by the European Community Framework Programme 7, People Programme, and IAPP on novel genetic and phenotypic markers of Parkinson's disease and Essential Tremor (MarkMD), contract number PIAP-GA-2008-230596 MarkMD (to HP and JHu). The WTCCC2 project was funded by the Wellcome Trust (083948/Z/07/Z). WTCCC1 study was supported by the Medical Research Council and Wellcome Trust disease centre (grant WT089698/Z/09/Z to NW, JHa, and ASc). This study was also supported by Parkinson's UK (grants 8047 and J-0804) and the Medical Research Council (G0700943). DNA extraction work that was done in the UK at University College London Hospitals, University College London, which received a proportion of funding from the Department of Health's National Institute for Health Research Biomedical Research Centres funding. This study was supported in part by the Wellcome Trust/Medical Research Council Joint Call in Neurodegeneration award (WT089698) to the Parkinson's Disease Consortium (UKPDC), whose members are from the UCL Institute of Neurology, University of Sheffield, and the Medical Research Council Protein Phosphorylation Unit at the University of Dundee. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Crichton RR, Dexter DT, Ward RJ (2011) Brain iron metabolism and its perturbation in neurological diseases. J Neural Transm 118: 301–314. [DOI] [PubMed] [Google Scholar]
- 2. Zecca L, Youdim MB, Riederer P, Connor JR, Crichton RR (2004) Iron, brain ageing and neurodegenerative disorders. Nat Rev Neurosci 5: 863–873. [DOI] [PubMed] [Google Scholar]
- 3. Dusek P, Jankovic J, Le W (2012) Iron dysregulation in movement disorders. Neurobiol Dis 46: 1–18. [DOI] [PubMed] [Google Scholar]
- 4. Forno LS (1996) Neuropathology of Parkinson's disease. J Neuropathol Exp Neurol 55: 259–272. [DOI] [PubMed] [Google Scholar]
- 5. Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, et al. (1997) Alpha-synuclein in Lewy bodies. Nature 388: 839–840. [DOI] [PubMed] [Google Scholar]
- 6. Mariani S, Ventriglia M, Simonelli I, Donno S, Bucossi S, et al. (2013) Fe and Cu do not differ in Parkinson's disease: a replication study plus meta-analysis. Neurobiol Aging 34: 632–633. [DOI] [PubMed] [Google Scholar]
- 7. Davey Smith G, Ebrahim S (2005) What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ 330: 1076–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davey Smith G, Ebrahim S (2003) ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32: 1–22. [DOI] [PubMed] [Google Scholar]
- 9. Pierce BL, Ahsan H, Vanderweele TJ (2011) Power and instrument strength requirements for mendelian randomization studies using multiple genetic variants. Int J Epidemiol 40: 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, et al. (2012) Using multiple genetic variants as instrumental variables for modifiable risk factors. Stats Methods Med Res 21: 223–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benyamin B, Ferreira MA, Willemsen G, Gordon S, Middelberg RP, et al. (2009) Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet 41: 1173–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, et al. (2012) Comprehensive research synopsis and systematic meta-analyses in parkinson's disease genetics: The PDGene database. PLoS Genet 8: e1002548 doi:10.1371/journal.pgen.1002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greco V, De Marco EV, Rocca FE, Annesi F, Civitelli D, et al. (2011) Association study between four polymorphisms in the HFE, TF and TFR genes and Parkinson's disease in southern Italy. Neurol Sci 32: 525–527. [DOI] [PubMed] [Google Scholar]
- 14. Halling J, Petersen MS, Grandjean P, Weihe P, Brosen K (2008) Genetic predisposition to parkinson's disease: CYP2D6 and HFE in the Faroe Islands. Pharmacogenet Genomics 18: 209–212. [DOI] [PubMed] [Google Scholar]
- 15. Guerreiro RJ, Bras JM, Santana I, Januario C, Santiago B, et al. (2006) Association of HFE common mutations with Parkinson's disease, Alzheimer's disease and mild cognitive impairment in a portuguese cohort. BMC Neurol 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dekker MC, Giesbergen PC, Njajou OT, van Swieten JC, Hofman A, et al. (2003) Mutations in the hemochromatosis gene (HFE), Parkinson's disease and parkinsonism. Neurosci Lett 348: 117–119. [DOI] [PubMed] [Google Scholar]
- 17. Borie C, Gasparini F, Verpillat P, Bonnet AM, Agid Y, et al. (2002) Association study between iron-related genes polymorphisms and Parkinson's disease. J Neurol 249: 801–804. [DOI] [PubMed] [Google Scholar]
- 18. Buchanan DD, Silburn PA, Chalk JB, Le Couteur DG, Mellick GD (2002) The Cys282Tyr polymorphism in the HFE gene in Australian Parkinson's disease patients. Neurosci Lett 327: 91–94. [DOI] [PubMed] [Google Scholar]
- 19. Aamodt AH, Stovner LJ, Thorstensen K, Lydersen S, White LR, et al. (2007) Prevalence of haemochromatosis gene mutations in Parkinson's disease. J Neurol, Neurosurg Psychiatry 78: 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biasiotto G, Goldwurm S, Finazzi D, Tunesi S, Zecchinelli A, et al. (2008) HFE gene mutations in a population of Italian Parkinson's disease patients. Parkinsonism Relat Disord 14: 426–430. [DOI] [PubMed] [Google Scholar]
- 21. Akbas N, Hochstrasser H, Deplazes J, Tomiuk J, Bauer P, et al. (2006) Screening for mutations of the HFE gene in Parkinson's disease patients with hyperechogenicity of the substantia nigra. Neurosci Lett 407: 16–19. [DOI] [PubMed] [Google Scholar]
- 22. Pankratz N, Beecham GW, DeStefano AL, Dawson TM, Doheny KF, et al. (2012) Meta-analysis of Parkinson's disease: Identification of a novel locus, RIT2. Ann Neurol 71: 370–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, et al. (2011) Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet 7: e1002141 doi:10.1371/journal.pgen.1002141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. International Parkinson Disease Genomics Consortium (2011) Nalls MA, Plagnol V, Hernandez DG, Sharma M, et al. (2011) Imputation of sequence variants for identification of genetic risks for Parkinson's disease: A meta-analysis of genome-wide association studies. Lancet 377: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. International Parkinson's Disease Genomics Consortium (IPDGC), Wellcome Trust Case Control Consortium 2 (WTCCC2) (2011) A two-stage meta-analysis identifies several new loci for Parkinson's disease. PLoS Genet 7: e1002142 doi:10.1371/journal.pgen.1002142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pankratz N, Wilk JB, Latourelle JC, DeStefano AL, Halter C, et al. (2009) Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet 124: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fung HC, Scholz S, Matarin M, Simon-Sanchez J, Hernandez D, et al. (2006) Genome-wide genotyping in Parkinson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol 5: 911–916. [DOI] [PubMed] [Google Scholar]
- 28. Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, et al. (2009) Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet 41: 1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, et al. (2010) Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet 74: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, et al. (2010) Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet 42: 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D Didelez V, Meng S (2010) Assumptions of IV methods for observational epidemiology. Stat Sci 25: 22–40. [Google Scholar]
- 33. Bautista LE, Smeeth L, Hingorani AD, Casas JP (2006) Estimation of bias in nongenetic observational studies using “mendelian triangulation”. Ann Epidemiol 16: 675–680. [DOI] [PubMed] [Google Scholar]
- 34. Thomas DC, Lawlor DA, Thompson JR (2007) Re: Estimation of bias in nongenetic observational studies using “mendelian triangulation” by Bautista et al. Ann Epidemiol 17: 511–513. [DOI] [PubMed] [Google Scholar]
- 35. Pierce BL, Ahsan H, Vanderweele TJ (2011) Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 40: 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tanner CM, Goldman SM (1996) Epidemiology of Parkinson's disease. Neurol Clin 14: 317–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ritz B, Ascherio A, Checkoway H, Marder KS, Nelson LM, et al. (2007) Pooled analysis of tobacco use and risk of Parkinson disease. Arch Neurol 64: 990–997. [DOI] [PubMed] [Google Scholar]
- 38. Hernan MA, Takkouche B, Caamano-Isorna F, Gestal-Otero JJ (2002) A meta-analysis of coffee drinking, cigarette smoking, and the risk of Parkinson's disease. Ann Neurol 52: 276–284. [DOI] [PubMed] [Google Scholar]
- 39. Linert W, Bridge MH, Huber M, Bjugstad KB, Grossman S, et al. (1999) In vitro and in vivo studies investigating possible antioxidant actions of nicotine: Relevance to Parkinson's and Alzheimer's diseases. Biochim Biophy Acta 1454: 143–152. [DOI] [PubMed] [Google Scholar]
- 40. Morck TA, Lynch SR, Cook JD (1983) Inhibition of food iron absorption by coffee. Am J Clin Nutr 37: 416–420. [DOI] [PubMed] [Google Scholar]
- 41. Zijp IM, Korver O, Tijburg LB (2000) Effect of tea and other dietary factors on iron absorption. Crit Rev Food Sci Nutr 40: 371–398. [DOI] [PubMed] [Google Scholar]
- 42. Kupershmidt L, Amit T, Bar-Am O, Youdim MB, Weinreb O (2012) Neuroprotection by the multitarget iron chelator M30 on age-related alterations in mice. Mech Ageing Dev 133: 267–274. [DOI] [PubMed] [Google Scholar]
- 43. Berg D, Roggendorf W, Schroder U, Klein R, Tatschner T, et al. (2002) Echogenicity of the substantia nigra: Association with increased iron content and marker for susceptibility to nigrostriatal injury. Arch Neurol 59: 999–1005. [DOI] [PubMed] [Google Scholar]
- 44. Walter U, Witt R, Wolters A, Wittstock M, Benecke R (2012) Substantia nigra echogenicity in Parkinson's disease: Relation to serum iron and C-reactive protein. J Neural Transm 119: 53–57. [DOI] [PubMed] [Google Scholar]
- 45. Logroscino G, Chen H, Wing A, Ascherio A (2006) Blood donations, iron stores, and risk of Parkinson's disease. Mov Disord 21: 835–838. [DOI] [PubMed] [Google Scholar]
- 46. Savica R, Grossardt BR, Carlin JM, Icen M, Bower JH, et al. (2009) Anemia or low hemoglobin levels preceding Parkinson disease: a case-control study. Neurology 73: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Levenson CW, Cutler RG, Ladenheim B, Cadet JL, Hare J, et al. (2004) Role of dietary iron restriction in a mouse model of Parkinson's disease. Exp Neurol 190: 506–514. [DOI] [PubMed] [Google Scholar]
- 48. Miyake Y, Tanaka K, Fukushima W, Sasaki S, Kiyohara C, et al. (2011) Dietary intake of metals and risk of Parkinson's disease: a case-control study in Japan. J Neurol Sci 306: 98–102. [DOI] [PubMed] [Google Scholar]
- 49. Ramsey AJ, Hillas PJ, Fitzpatrick PF (1996) Characterization of the active site iron in tyrosine hydroxylase. Redox states of the iron. J Biol Chem 271: 24395–24400. [DOI] [PubMed] [Google Scholar]
- 50. Beard J, Erikson KM, Jones BC (2003) Neonatal iron deficiency results in irreversible changes in dopamine function in rats. J Nutr 133: 1174–1179. [DOI] [PubMed] [Google Scholar]
- 51. Levenson CW, Tassabehji NM (2004) Iron and ageing: an introduction to iron regulatory mechanisms. Ageing Res Rev 3: 251–263. [DOI] [PubMed] [Google Scholar]
- 52. Rhodes SL, Ritz B (2008) Genetics of iron regulation and the possible role of iron in Parkinson's disease. Neurobiol Dis 32: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Castiglioni E, Finazzi D, Goldwurm S, Pezzoli G, Forni G, et al. (2010) Analysis of nucleotide variations in genes of iron management in patients of Parkinson's disease and other movement disorders. Parkinsons Dis 2011: 827693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Castiglioni E, Finazzi D, Goldwurm S, Levi S, Pezzoli G, et al. (2010) Sequence variations in mitochondrial ferritin: Distribution in healthy controls and different types of patients. Genet Test Mol Biomarkers 14: 793–796. [DOI] [PubMed] [Google Scholar]
- 55. Ezquerra M, Campdelacreu J, Munoz E, Tolosa E (2005) Association study of the G258S transferrin gene polymorphism and Parkinson's disease in the Spanish population. J Neurol 252: 1269–1270. [DOI] [PubMed] [Google Scholar]
- 56. He Q, Du T, Yu X, Xie A, Song N, et al. (2011) DMT1 polymorphism and risk of Parkinson's disease. Neurosci Lett 501: 128–131. [DOI] [PubMed] [Google Scholar]
- 57. Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, et al. (2003) The discovery of the new haemochromatosis gene. 1996. J Hepatol 38: 704–709. [DOI] [PubMed] [Google Scholar]
- 58. Du X, She E, Gelbart T, Truksa J, Lee P, et al. (2008) The serine protease TMPRSS6 is required to sense iron deficiency. Science 320: 1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, et al. (1996) A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 13: 399–408. [DOI] [PubMed] [Google Scholar]
- 60. Bradley LA, Johnson DD, Palomaki GE, Haddow JE, Robertson NH, et a (1998) Hereditary haemochromatosis mutation frequencies in the general population. J Med Screen 5: 34–36. [DOI] [PubMed] [Google Scholar]
- 61. Waheed A, Parkkila S, Zhou XY, Tomatsu S, Tsuchihashi Z, et al. (1997) Hereditary hemochromatosis: effects of C282Y and H63D mutations on association with beta2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proc Natl Acad Sci U S A 94: 12384–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Feder JN, Tsuchihashi Z, Irrinki A, Lee VK, Mapa FA, et al. (1997) The hemochromatosis founder mutation in HLA-H disrupts beta2-microglobulin interaction and cell surface expression. J Biol Chem 272: 14025–14028. [DOI] [PubMed] [Google Scholar]
- 63. Lebrón JA, Bennett MJ, Vaughn DE, Chirino AJ, Snow PM, et al. (1998) Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell 93: 111–123. [DOI] [PubMed] [Google Scholar]
- 64. An P, Wu Q, Wang H, Guan Y, Mu M, et al. (2012) TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum Mol Genet 21: 2124–2131. [DOI] [PubMed] [Google Scholar]
- 65. Nai A, Pagani A, Silvestri L, Campostrini N, Corbella M, et al. (2011) TMPRSS6 rs855791 modulates hepcidin transcription in vitro and serum hepcidin levels in normal individuals. Blood 118: 4459–4462. [DOI] [PubMed] [Google Scholar]
- 66. Fleiss JL (1993) The statistical basis of meta-analysis. Stat Methods Med Res 2: 121–145. [DOI] [PubMed] [Google Scholar]
- 67. Tamura T, Goldenberg RL, Hou J, Johnston KE, Cliver SP, Ramey SL, et al. (2002) Cord serum ferritin concentrations and mental and psychomotor development of children at five years of age. J Ped 186: 458–463. [DOI] [PubMed] [Google Scholar]
- 68. Harbord RM, Didelez V, Palmer TM, Meng S, Sterne JA, et al. (2013) Severity of bias of a simple estimator of the causal odds ratio in mendelian randomization studies. Stat Med 32: 1246-1258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plot of the meta-analysis of the studies included for the effect of HFE rs1800562 on PD risk. The boxes indicate the genetic (additive) effects of individual studies, with the size of the box being inversely proportional to the variance and horizontal lines indicating 95% confidence intervals. The diamond indicates the pooled effect estimate, obtained using inverse-variance weighted fixed-effect meta-analysis, and its 95% confidence interval. The full vertical line shows the value for no effect, as opposed to the dashed line indicating the estimated pooled effect.
(TIF)
Forest plot of the meta-analysis of the studies included for the effect of HFE rs1799945 on PD risk. The boxes indicate the genetic (additive) effects of individual studies, with the size of the box being inversely proportional to the variance and horizontal lines indicating 95% confidence intervals. The diamond indicates the pooled effect estimate, obtained using inverse-variance weighted fixed-effect meta-analysis, and its 95% confidence interval. The full vertical line shows the value for no effect, as opposed to the dashed line indicating the estimated pooled effect.
(TIF)
Forest plot of the meta-analysis of the studies included for the effect of TMPRSS6 rs855791 on PD risk. The boxes indicate the genetic (additive) effects of individual studies, with the size of the box being inversely proportional to the variance and horizontal lines indicating 95% confidence intervals. The diamond indicates the pooled effect estimate, obtained using inverse-variance weighted fixed-effect meta-analysis, and its 95% confidence interval. The full vertical line shows the value for no effect, as opposed to the dashed line indicating the estimated pooled effect.
(TIF)
Sensitivity analysis: Forest plot of the mendelian randomization estimates after exclusion of nine studies from the PDGene dataset that had not adjusted for population stratification (see Table S2).
(TIF)
Characteristics and sample size of the individual studies included for the gene–iron association. In all studies, the analyses were adjusted for age and sex, as well as for the first five MDS (multidimensional scaling) or principal components to control for population stratification.
(DOC)
Characteristics and sample size of the individual studies included for the gene–PD association.
(DOC)
Gene–iron association: GIS-consortium meta-analysis. The effect size for the genetic effects on iron levels is expressed as number of SDs from the mean (Z-scores).
(DOC)
Gene–PD association: meta-analysis of all available candidate gene and GWA studies.
(DOC)
Detailed description of the studies included in the three GWA investigations of PD risk.
(DOC)