Abstract
Graphene is a novel material whose application in the biomedical sciences has only begun to be realized. In the present study, we have employed three-dimensional graphene foams as culture substrates for human mesenchymal stem cells and provide evidence that these materials can maintain stem cell viability and promote osteogenic differentiation.
Graphene, a single atomic monolayer of graphite, has received unprecedented attention over the last decade due to its unique electronic and nanoscale properties.1, 2 Recently, three-dimensional graphene foams (3D GFs) were developed3 and since have been employed for various applications, including battery technology4 and electrochemical sensing.5 However, the applicability of 3D GFs as cell culture platforms or the ability of 3D GFs to direct stem cell fate are yet to be determined. 3D GFs are cheap to produce and highly-scalable for industrial applications, thus exhibiting great potential as a new class of cell-instructive materials for tissue engieering scaffolds and implantable medical devices.
Despite the biological applicability of graphene,6 studies aiming to elucidate the response of living cells to graphene culture substrates are limited, and only a few reports have investigated stem cell behavior on two-dimensional graphene sheets.7–9 Human bone marrow-derived mesenchymal stem cells (hMSCs) are a clinically-safe adult stem cell source whose allogenic/autologous availability, immunomodulatory capabilities, and multilineage differentiation potential have stimulated substantial interest for their applications in tissue engineering and regenerative medicine.10 When cultured on two-dimensional graphene sheets in the presence of osteogenic induction medium, hMSCs have been shown to exhibit accelerated osteogenic differentiation.7 Furthermore, Wang et al. recently reported the ability of fluorinated graphene sheets to promote the neuronal differentiation of hMSCs.8 In a 2009 study by Oh et al., titanium nanotubes (TiNTs) of specific dimensions were shown to be sufficient for spontaneous osteogenic induction of hMSCs in the absence of biochemical stimulation,11 suggesting an intrinsic material property driving this event similar to that which was observed on graphene and fluorinated graphene sheets. These reports demonstrate that material-derived cues can guide hMSC fate decisions, but the ability of 3D GFs in regulating cell behavior remains unknown. Therefore, in the present study, GFs have been investigated as novel cell culture substrates for hMSCs and the resulting cellular behavior and phenotype are reported. GFs are shown to maintain hMSC viability, and stimulate changes in morphology and protein expression patterns that indicate spontaneous osteogenic differentiation without extrinsic biochemical inputs. This is the first study to utilize 3D GFs as cell culture substrates and represents the first step for incorporating 3D GFs into biomedical studies. The findings reported in this study will allow us to develop a novel platform of cheap, graphene-based osteoconductive grafts, thereby opening a new avenue of bone tissue engineering research.
Multilayer GFs were fabricated by growing graphene on 3D Ni scaffolds. Nickel was subsequently removed by FeCl3 etching. Ni/graphene foams and pristine GFs were characterized by scanning electron microscopy (SEM). SEM images reveal a robust, three-dimensional structure of the foam before nickel etching (Fig. 1A) and retention of the 3D structure following etching, but composed of a much thinner outer layer (Fig. 1B). After growth, Raman spectroscopy was used to verify the formation of multilayer graphene on the Ni foams (Fig. 1C–D). Raman spectroscopyis currently considered as the standard to compare the quality of graphene because it is non-destructive and provides detailed structural and electronic information about a material by measuring energy shift that results from inelastic scattering with phonon vibrations in a graphene lattice.3 The Raman spectra of multilayer graphene films are shown in in Fig. 1C (as grown on Ni) and Fig. 1D (after etching away Ni), respectively. Two characteristic peaks are observed at ~1580cm−1(G) and ~2700cm−1(2D).12 There is also a D peak at ~1350cm−1. The D peak arises due to the carbon ring breathing mode and is seen due to defects.12, 13 Finally, energy dispersive x-ray spectroscopy (EDX) was used to verify the dissolution of Ni from the 3D scaffold. Our results (Fig. 1E) show that no Ni is detected and the scaffold is made completely out of interconnected graphene layers.
Figure 1. Fabrication and characterization of 3D GFs from nickel foam templates.
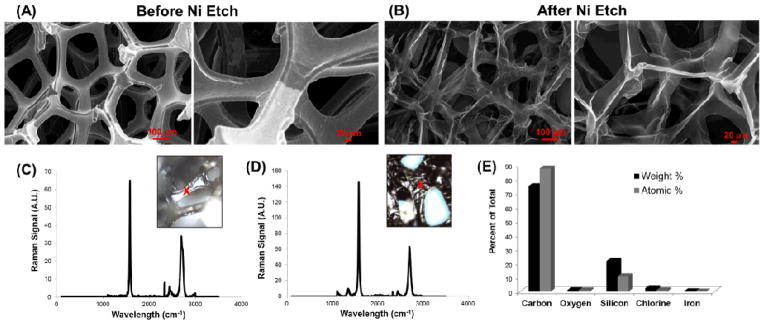
SEM images of three-dimensional foams (A) before and (B) after the nickel template was removed by etching. All SEM images were taken at an accelerating voltage of 10kV. Raman spectra (C) before and (D) after nickel etching, along with (E) EDX data after etching, confirm the removal of nickel and the purity of the graphene foams (GFs). Red X’s in the insets of (C–D) represent the area on the material at which the Raman was taken. Weight and atomic percentages in the EDX chart from silicon are derived from the underlying glass (SiO2) substrates upon which the GFs were immobilized.
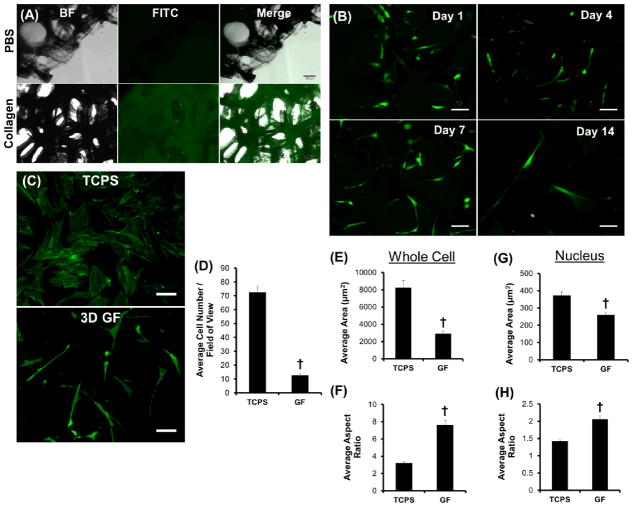
GFs were next investigated for promoting hMSC attachment, maintaince of cell viability, and spontaneous morphological changes (Fig. 2). Protein adsorption at a material GFs were incubated with fluorescein isothiocyanate (FITC)-labeled collagen, a common extracellular matrix protein to which hMSCs are known to bind,14 in phosphate buffered saline (PBS) to evaluate protein adsorption before cellular interactions were studied. As seen in Fig. 2A, labeled collagen adsorbed to the GF surface and co-localized with the 3D GF architecture, but no fluorescence was observed for GFs incubated with PBS (vehicle control) alone. hMSCs were then seeded on 3D GFs and cell viability was observed over a 14 day period (Fig. 2B). GFs were shown to maintain viable hMSCs (green) with virtually no deadsurface is critical for cell attachment and spreading.15 Therefore, cells (red) for up to two weeks, and morphological changes immediately became apparent. When compared to tissue culture polystyrene (TCPS), hMSCs cultured upon GFs for 7 days were shown to be much fewer in number (Fig. 2C–D) and exhibit drastic differences in whole cell and nuclear morphologies (Fig. 2C& E–H). Specfically, when compared to hMSCs cultured on TCPS, whole cells and individual nuclei became significantly more elongated as shown in Fig. 2C, which was quantitavely demonstrated with decreased area (Fig. 2E & G) and increased aspect ratio (Fig. 2F & H) for both nuclei and cytoplam.
Figure 2. Protein adsorption, hMSC attachment, and morphological changes on 3D GFs.
(A) Brightfield (BF), fluorescent (FITC), and merged images reveal that FITC-labeled collagen homogeneously adsorbed to the surface of graphene foams (GFs). In the absence of collagen (PBS control), virtually no background FITC signal was observed. (B) GFs support the attachment and viability of hMSCs over 14 days. The majority of attached hMSCs remained viable (green) and few dead cells (red) were observed. (C) hMSC number and morphology is significantly different on GFs as compared to tissue culture polystyrene (TCPS) control after seven days in culture. (D) When quantified, ~6x more cells were present on TCPS compared to GFs. (E–F) Cellular and (G–H) nuclear morphology revealed a significant increase in aspect ratio with a concurrent decrease in total area for hMSCs cultured on GFs as compared to TCPS control. †p < 0.01 between groups; all scale bars = 100 μm.
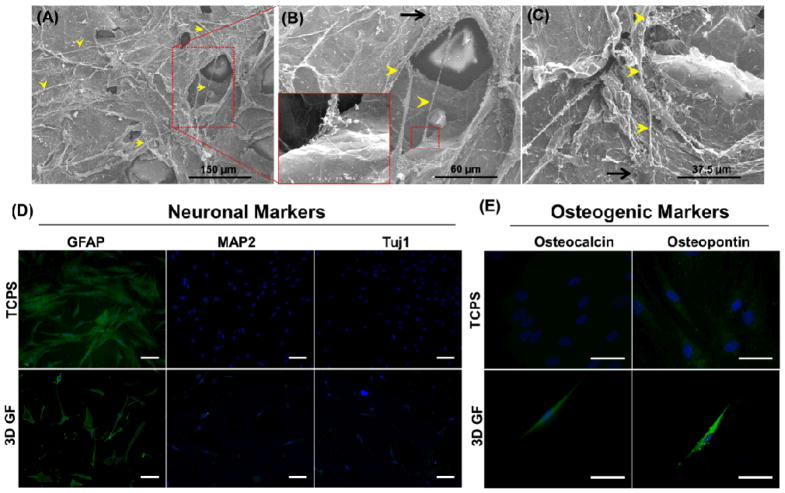
Because substantial morphological changes were observed for hMSCs on GFs, as compared to TCPS (Fig. 2C–H), cell morphology and interaction with 3D GFs were further observed in higher resolution by SEM. As seen in Fig. 3A–C, hMSCs on GFs exhibited long protrusions up to 100 μm in length (yellow arrowheads) that extended from defined cell bodies (black arrows). Interestingly, in some instances, the protrusions extended over porous regions of the GF (Fig. 3B) and interacted directly with the material (Fig. 3B inset). We then sought to identify the phenotypic changes in hMSCs cultured on 3D GFs (Fig. 3D–E). Since pristine 2D graphene sheets have been shown to accelerate osteogenic differentiation7 and fluorinated graphene sheets promote neuronal differentiation,8 immunostaining for relevant neuronal and osteogenic markers was next performed (Fig. 3D–E). After seven days in culture on GFs, hMSCs were stained for neuronal markers, such as glial fibrillary acidic protein (GFAP), microtubule-associated protein 2 (MAP2), and neuronal class III β-tubulin (Tuj1), or osteogenic markers, such as osteocalcin (OC) and osteopontin (OP). When compared to TCPS control, hMSCs on GFs did not alter the expression of neuronal markers (positive for GFAP, negative for both MAP2 and Tuj1) (Fig. 3D). hMSCs on TCPS did not express detectable levels of OC, and OP expression was faint and punctate (Fig. 3E); however, hMSCs cultured on GFs strongly expressed both OC and OP, and exhibited a spindle-shaped, elongated morphology with thin, aligned nuclei, representative of osteoprogenitor cells.16
Figure 3. Morphologic and phenotypic changes in hMSCs cultured on graphene foams (GFs) confirm spontaneous osteogenic differentiation.
(AC) hMSCs cultured on GFs for four days formed protrusions up to 100 μm in length (yellow arrowheads) that extended from small cell bodies (black arrows). Interestingly, several of these protrusions spanned large pores in the GF and interacted directly with the material surface. All SEM images were taken at an accelerating voltage of 5kV. Immunostaining for (D) neuronal and (E) osteogenic markers for hMSCs cultured on GFs or tissue culture polystyrene (TCPS) control for seven days. Culture of hMSCs on GFs did not affect the expression of neuronal markers, but stimulated de novo expression of the osteogenic marker osteocalcin and upregulated expression of osteopontin. These data confirm that 3D GFs promote the spontaneous osteogenic differentiation of hMSCs. (D) Scale bars = 100 μm, (E) Scale bars = 50 μm
The results presented in this study indicate that 3D GFs induce spontaneous osteogenic differentiation of hMSCs without the need for extrinsic biochemical manipulation. Previous work demonstrating accelerated osteogenic differentiation of hMSCs on 2D graphene sheets required defined medium,7 demonstrating potential for graphene in clinically-relevant applications but limiting its use relative to other osteo-inductive materials. It is well-established that hMSC differentiation into osteoprogenitors/osteoblasts occurs on stiff culture substrates,14 and the elongated morphology that was observed on GFs is identical to that of hMSCs undergoing osteogenic differentiation on TiNTs.11 The mechanism driving spontaneous differentiation remains unclear, but is suspected to be an intrinsic cellular response to the stress derived from the interaction with a high stiffness material.11 Indeed, cytoskeletal tension and cell shape have been shown to regulate hMSC fate, specifically with the Rho/ROCK cascade controlling osteo-/adipogenic lineage commitments in which extreme intracellular tension was shown to bias the cell towards osteogenesis.17 However, cell spreading and a total increase in area were not observed for hMSCs on GFs, suggesting that material properties alone were responsible for increased cytoskeletal tension, presumably through forced hMSC elongation along the bulk of the GF structure (i.e. around/across pores). Although these studies provide crucial evidence for guided cell behavior, further elucidation of the fundamental biological mechanisms regulating material-mediated hMSC differentiation is crucial for the safe and effective clinical translation of hMSC/biomaterial technologies.
In this study, hMSC attachment on GFs was seen to be much lower than on TCPS (Fig. 2C), due in part to the high porosity (i.e. void space) of the material. Because homogeneous cell attachment, spreading, and proliferation over the scaffold are critical parameters for osteoconductive applications, this is an important issue that must be addressed for translation of GFs as large-scale tissue engineering culture substrates. The cell seeding efficiency can be improved through two potential avenues: bioreactor application and pore architecture/size optimization. For example, application of established bioreactor designs with 3D GFs could enhance cell attachment and spreading, although fluid shear in rotating bioreactors could impact cell fate by either helping to improve differentiation or inhibiting specific lineage commitments. The size of individual pores likely plays an integral role in modulating the cellular response in terms of spreading and differentiation, and the overall porosity of the 3D GF will dictate the surface area that is available for cell attachment. The 3D GFs used in this study were highly porous with individual pore sizes ≥100 μm, and cells that sensed and spread across the pores were seen in certain areas (i.e. Fig. 3A–C); however, pores of much smaller size (~1–10 μm) might provide a local microenvironmental cue that can be used as an additional parameter to manipulate cell behavior. Furthermore, surface functionalization with specific chemical groups (such as fluorine8) could improve cell attachment and also influence lineage specification.8 By altering each of these parameters and observing the resulting cellular response, the ideal properties of 3D GFs for osteoconductive applications, as well as other avenues for biomedical therapies, can be determined.
Conclusions
3D GFs support the attachment and viability of hMSCs, and induce spontaneous osteogenic differentiation. The findings from this study will be used to develop novel grapheme-based strategies for osteoconductive tissue engineered constructs. The fabrication of 3D GFs presented here represents a low cost and highly-scalable approach for developing tissue engineered constructs that, along with the autologous availability and multilineage differentiation potential of hMSCs, holds great promise for advanced strategies in regenerative medicine.
Materials and Methods
Fabrication and Characterization of Three-Dimensional, Porous Graphene Foams
Graphene was grown on a 3D nickel scaffold, allowing for the growth of an interconnected scaffold of 3D graphene according to a previous protocol.3 Ni foams with a thickness of 1.2mm and ~320g/m2 areal density were purchased from Alantum Advanced Technology Materials (Shanghai, China). The nickel foams were cut into small pieces and placed inside a 25mm quartz tube for growth. The foam was initially annealed at 1000°C under 500 sccm of Ar and 100 sccm of H2 to remove any contaminants. After annealing, 7 sccm of CH4 was introduced for 5 minutes which decomposes the CH4 and deposits carbon atoms on the Ni foam. After growth, Raman microscopy was used to verify the growth of multilayer graphene on the Ni foams. Ni was removed by FeCl3 etching: a drop of A7 poly(methyl methacrylate) (PMMA) was placed on top of the freshly grown graphene/Ni hybrid structure and heated on a hotplate at 90°C for 5 minutes. The PMMA provided a physical support and stops the graphene layers from collapsing onto itself upon Ni etching. After the Ni was etched, the PMMA was removed in an acetone bath. GFs were characterized by SEM (S4200 SEM Hitachi Ltd., Tokyo, Japan), EDX (Oxford Instruments INCA Energy 250, Abingdon, Oxfordshire, UK), and Raman spectroscopy (DXR Confocal Raman Microscope, Thermo Scientific, Waltham, MA).
Cell Culture
hMSCs were purchased from Lonza (Walkersville, MD) and used between passages 4 and 7. Cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco, Carlsbad, CA) with 10% heat-inactivated fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco).
To visualize protein adsorption on the GF surface, GFs were incubated with FITC-conjugated collagen (100 μg mL−1, Elastin Products Company Inc., Owensville, MO) overnight at 37°C, washed three times with sterile PBS (Gibco), and imaged under a Nikon Ti inverted microscope (Nikon Instruments Inc., Melville, NY).
For cell culture experiments, GFs were autoclaved and then incubated with sterile-filtered porcine gelatin (0.2% weight/volume, Sigma Aldrich, St, Louis, MO) in deionized water for 2 hours at 37°C. GFs on glass coverslips were placed into a 24-well plate and held in place with silicon O-rings (McMaster Carr, Atlanta, GA), washed twice with sterile PBS to remove unattached gelatin, and cells were seeded at a density of 7,500 cells cm−2. Culture media was replaced every 3–4 days during culture on the GF foams.
Cell Viability and Immunofluorescence
At 1, 4, 7, or 14 days after seeding, cells in culture were treated with Calcein AM (4μM, Invitrogen, Carlsbad, CA) and ethidium homodimer-1 (2 μM, Invitrogen) in sterile PBS for 15 min at 37°C to stain live and dead cells, respectively. Cells were washed three times with sterile PBS and imaged under a Nikon Ti inverted microscope.
For immunofluorescence staining, hMSCs were fixed with 4% paraformaldehyde (Sigma) in deinonized water for 15 minutes at room temperature, permeabilized with 0.2% Triton-X in PBS and blocked with 10% goat serum (Sigma) with 0.05% Triton-X in PBS. For staining the actin cytoskeleton, cells were incubated with fluorescein-labelled phalloidin (Invitrogen) according to the manufacturer’s instructions. For all other experiments, cells were incubated with primary antibodies overnight at 4°C, incubated with secondary antibodies for 2 hours at room temperature in the dark, and counterstained with Hoechst (5μg mL−1, Sigma). Primary antibodies included the neuronal markers tubulin β3 (Millipore, raised in mouse, 1:250 dilution), microtubule-associated protein 2 (MAP2, raised in rabbit, 1:200 dilution), and glial fibrillary acidic protein (GFAP, raised in rabbit, 1:1000 dilution), and the osteogenic markers osteocalcin (OC, raised in mouse, 1:250 dilution, Abcam, Cambridge, MA) and osteopontin (OP, raised in rabbit, 1:500, Abcam). Secondary antibodies included DyLight550-conjugated goat anti-mouse (1:500 dilution, Abcam) and DyLight488-conjugated goat anti-rabbit (1:500 dilution, Abcam). All cells were imaged under a Nikon Ti inverted microscope and images were post-processed in ImageJ software. Whole cell and nuclear shape descriptors were measured with ImageJ software and include cells from n ≥ 5 images.
hMSC Fixation for SEM Imaging
hMSCs were fixed in 2.5% gultaraldehyde buffered with a 0.1M sodium cacodylate (NaC) solutions for 1 hour at room temperature and then overnight at 4°C. Samples were then rinsed with 0.1M NaC, underwent secondary fixation with 1% osmium tetroxide in 0.1M NaC for 1 hour, dehydrated with graded ethanol washes in water for 15 min each (30%, 50%, 75%, 85%, 95%, 100% three times), dried in a critical point dryer (EMS 850, Electron Microscopy Sciences, Hatfield, PA), and mounted onto an aluminun SEM peg with colloidal silver paint. Samples were imaged on a Hitachi S-4200 SEM with an accelerating voltage of 5kV.
Statistical Analysis
In all experiments, results are presented as mean ± standard error of the mean (SEM). Results from each experiment were analyzed using an unpaired Student’s t-test. p < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by NIH UH2TR000491, NSF CBET 1056046, and NSF CAREER DMR-1056859. The authors acknowledge the Vanderbilt Institute of Nanoscale Science and Engineering for SEM, Raman spectroscopy (NSF EPS-1004083), and EDX techniques. Fixation of hMSC samples for SEM was performed in part through the use of the VUMC Cell Imaging Shared Resource.
Notes and references
- 1.Geim AK, Novoselov KS. Nat Mater. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 2.Novoselov KS, Geim AK, Morozov SV, Jiang D, Zhang Y, Dubonos SV, Grigorieva IV, Firsov AA. Science. 2004;306:666–669. doi: 10.1126/science.1102896. [DOI] [PubMed] [Google Scholar]
- 3.Chen ZP, Ren WC, Gao LB, Liu BL, Pei SF, Cheng HM. Nature Materials. 2011;10:424–428. doi: 10.1038/nmat3001. [DOI] [PubMed] [Google Scholar]
- 4.Li N, Chen ZP, Ren WC, Li F, Cheng HM. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17360–17365. doi: 10.1073/pnas.1210072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong XC, Wang XW, Wang LH, Song H, Zhang H, Huang W, Chen P. Acs Applied Materials & Interfaces. 2012;4:3129–3133. doi: 10.1021/am300459m. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen P, Berry V. Journal of Physical Chemistry Letters. 2012;3:1024–1029. doi: 10.1021/jz300033g. [DOI] [PubMed] [Google Scholar]
- 7.Nayak TR, Andersen H, Makam VS, Khaw C, Bae S, Xu XF, Ee PLR, Ahn JH, Hong BH, Pastorin G, Ozyilmaz B. Acs Nano. 2011;5:4670–4678. doi: 10.1021/nn200500h. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Lee WC, Manga KK, Ang PK, Lu J, Liu YP, Lim CT, Loh KP. Advanced Materials. 2012;24:4285-+. doi: 10.1002/adma.201200846. [DOI] [PubMed] [Google Scholar]
- 9.Chen GY, Pang DW, Hwang SM, Tuan HY, Hu YC. Biomaterials. 33:418–427. doi: 10.1016/j.biomaterials.2011.09.071. [DOI] [PubMed] [Google Scholar]
- 10.Chamberlain G, Fox J, Ashton B, Middleton J. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 11.Oh S, Brammer KS, Li YSJ, Teng D, Engler AJ, Chien S, Jin S. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari AC, Meyer JC, Scardaci V, Casiraghi C, Lazzeri M, Mauri F, Piscanec S, Jiang D, Novoselov KS, Roth S, Geim AK. Physical Review Letters. 2006;97:187401. doi: 10.1103/PhysRevLett.97.187401. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari AC. Solid State Communications. 2007;143:47–57. [Google Scholar]
- 14.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DIC, Whitesides GM, Ingber DE. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 16.Clarke B. Clin J Am Soc Nephrol. 2008;33(Suppl):S131–139. doi: 10.2215/CJN.04151206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]