Abstract
We have developed an amplified chemiluminescence turn-on sensing platform that relies on single-walled carbon nanotube for ultrasensitive DNA detection. This new type of assay exhibits high detection sensitivity over traditional biosensors three orders of magnitude and high specificity for target molecules.
Various types of DNA sensors, including electrochemical and fluorescent have been developed.1 Although intensive studies on DNA sensors based on fluorescence measurements have been carried out,2 reports on chemiluminescent DNA sensors are few.3 Chemiluminescence (CL) differs from fluorescence in that the electronic excited state is a product of a chemical reaction rather than derived from photon absorption. A significant advantage of chemiluminescent sensors is, therefore, without needing an external light source. This is very significant in imaging and sensing operations because 1) background signals caused by an external light excitation is minimized; and 2) simple and miniaturized instruments can be used. CL resonance energy transfer (CRET) occurs by the oxidation of a chemiluminescent donor in a CL reaction.4 Thanks to its various advantages, CRET becomes an attractive light measuring scheme in bioassays as demonstrated in recent years.5
Carbon nanotubes (CNTs) have emerged as a fascinating nanomaterial because of their unique chemical, electrical, and mechanical properties, as well as the possibility of versatile applications.6 In particular, the high specific surface area and biocompatibility of CNTs make them a suitable and promising platform for numerous biological applications, such as interaction studies, drug delivery, biomolecule detection, and biosensor design.7 Recent photophysical studies have shown that CNTs are a class of excellent energy acceptors with a long-range energy transfer property.8 The rate of energy transfer between dye molecules and CNTs was suggested to have an R−5 dependence on distance,9 in contrast to the R−6 dependence for traditional donor-acceptor FRET.10 Tan and co-workers applied a FRET system involving fluorescence quenching of excited fluorescent dye molecules by the π system of CNTs to biomolecule detection.11 As with general fluorescence techniques, however, FRET based bioassays suffer from drawbacks associated with the requirement of an external light excitation source.12
Herein, we report on the first amplified single-walled carbon nanotube (SWNT)-mediated CL turn-on sensing platform for ultrasensitive DNA assay. The sensing platform was based on the modulation in CRET efficiency between SWNTs acceptor and the chemiluminescent donor, N-(4-aminobutyl)-N-ethylisoluminol (ABEI)-tagged DNA recognition probe, by the target DNA that blocks the donor-acceptor interaction. The CL of the sensor is turned off by CRET on the SWNTs surface and switched on by the exonuclease-recycled DNA cleavage, and results in the amplification of the readout signal. Furthermore, use of appropriate DNA hairpins to increase the versatility of the sensing platform was explored. Scheme 1 shows a schematic presentation of the amplified SWNT-mediated CL turn-on sensing platform for DNA detection. DNA-1 probe is labeled at the 3′-terminus with ABEI, the chemiluminescent donor, and SWNTs are used as the acceptor. As strong noncovalent binding occurs between SWNTs and single-stranded DNA molecules,11 the ABEI-labeled DNA-1 molecules are adsorbed onto the SWNTs surface, resulting in quenching of CL from ABEI-labeled DNA-1 molecules due to CRET. However, in the presence of the target DNA-2, it hybridizes with ABEI-labeled DNA-1 absorbed on SWNTs, forming a stable DNA/DNA duplex, and thus releasing the duplex from SWNTs. This diminishes CRET efficiency between SWNTs and ABEI-DNA-1 molecules, resulting in a CL restoration from ABEI. In the presence of the Exo III as a biocatalyst, Exo III selectively catalyzes the stepwise removal of mononucleotides from 3′-hydroxyl termini of duplex DNAs, resulting in the removal of ABEI and the release of the target DNA-2. The released target DNA-2 then can hybridize with another ABEI-labeled DNA-1 adsorbed on the SWNTs surface, and the cycle starts anew, resulting in the continuous cleavage of DNA/DNA duplex. Consequently, each target DNA can initiate the cleavage of many ABEI-labeled DNA-1 probes, generating a concomitant increase in the CL due to the block of the interaction between ABEI and SWNTs. This provides an amplification path for the CL detection of target DNA.
Scheme 1.
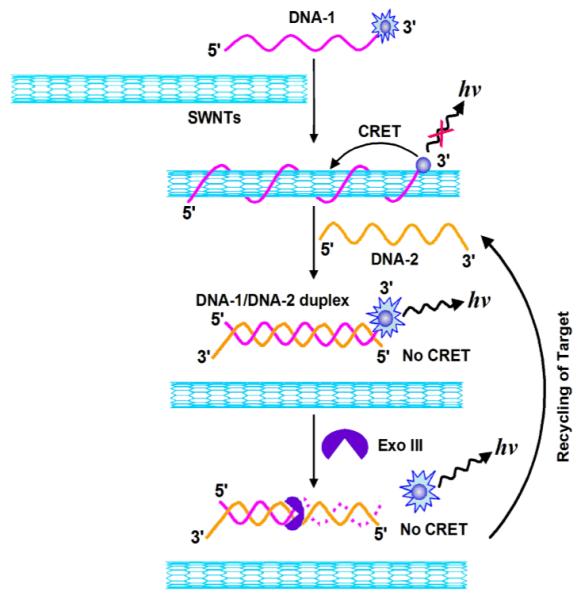
Schematic illustration of the sensing principle with the proposed amplified SWNT-mediated CRET platform for DNA detection.
To evaluate the probe designed, we first confirmed the effectiveness of SWNTs as a CRET acceptor. In this work, the ABEI-hydrogen peroxide (H2O2) CL reaction catalyzed by horseradish peroxidase (HRP) was used. This reaction is one of the most sensitive CL reactions. More importantly, the CL spectrum of ABEI was largely overlapped with the absorption of SWNTs (See Figure S1), which satisfied the essential condition for CRET. Figure 1 depicts the CL titration profile of ABEI-DNA-1 by adding different amount of SWNTs. With the increasing amount of SWNTs, CL intensity of ABEI was gradually quenched. The degree of CL quenching at 450 nm reached approximately 93% with 45 μg/mL SWNTs. The CL quenching was well correlated to the relative concentration of ABEI-DNA-1 immobilized on SWNTs. Binding between the acceptor (i.e. SWNTs) and the donor (i.e. ABEI-DNA-1 excited by HRP-catalyzed oxidation) reduced their distance and thus enabled CRET at a higher energy transfer efficiency from the donor to the acceptor. We carried out a control experiment by mixing a free ABEI solution with SWNTs. It was found that CL intensity remained almost unchanged with increasing the amount of SWNTs (See Figure S2). This indicated that the proximity of ABEI to SWNTs was critical for an efficient energy transfer, which was in consistent with fluorescence quenching by SWNTs.9, 11
Figure 1.
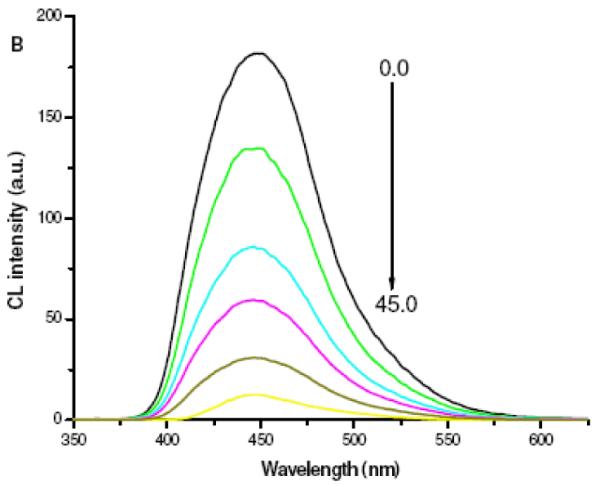
CL titration profile of 50 nM ABEI-DNA-1 by SWMTs with concentrations ranging from 0 to 45 μg/mL at an interval of 9 μg/mL, confirming ABEI-DNA-1/SWNTs hybridization. Response time was 3 s. Reaction buffer for CRET assay is 20 mM Tris-HCl solution (pH 7.4) containing 50 mM NaCl, 10 mM MgCl2 and 10 μM HRP. The oxidizer solution was 40 mM Na2CO3 buffer (pH 11.0) containing 30 mM H2O2.
Importantly, the sensitivity of the proposed assay can be dramatically increased by signal amplification using Exo III as a biocatalyst. Figure S3 depicts the time-dependent CL changes of the ABEI-DNA-1/SWNTs hybrid subjected to the target DNA-2 in the presence of Exo III. The CL of ABEI was intensified as the reaction time increased. Control experiments showed that only slight CL changes occurred in the absence of Exo III (Figure S4). This suggested that the observed CL changes were mainly due to the cleavage of ABEI-DNA-1 in the resulting DNA-1/DNA-2 duplex by Exo III. A plot of the CL signal versus the concentration of DNA-2 using Exo III amplification for 2 h showed a linear response from 100 fM to 1 nM. The amplified assay had a detection limit of 45 fM, representing a 1000-times improvement compared with the assay without amplification. As far as we know, the present assay was far more sensitive than most of the assays reported so far whose detection limits were at the pM level. The sequence specificity of the amplified assay was also investigated by testing the response to other DNA molecules, including a single-base mismatch target DNA-3 and a random target DNA-4. The perfectly-matched complementary target DNA-2, single-base mismatch target DNA-3 and a random target DNA-4 were distinctly discriminated in the same detection condition (Figure 2). These results clearly indicated the high specificity of the proposed SWNT-mediated CRET amplified assay.
Figure 2.
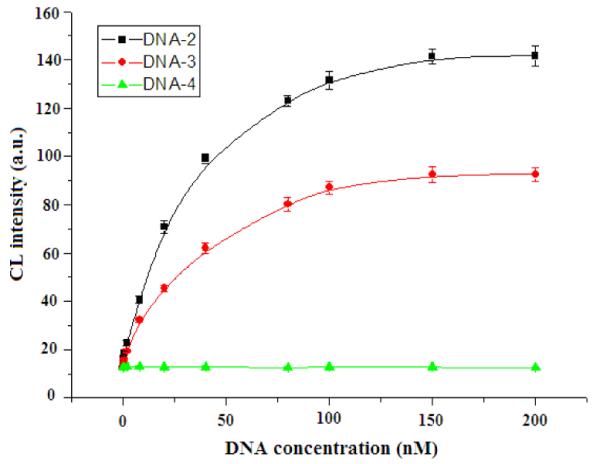
CL response curves from assaying a perfectly matched complementary target DNA-2, a single mismatched complementary target DNA-3 and a random target DNA-4. Target concentrations ranged from 100 fM to 200 pM. Experimental conditions were as in Figure 1.
The versatility of the amplified sensing platform can be greatly enhanced by introducing an appropriate DNA hairpin into the system. The principle is illustrated by Scheme 2. An ABEI-DNA-5/SWNTs hybrid serves as the transducer for sensing. A DNA hairpin structure (DNA-6) that recognizes the target DNA-7 is introduced into the system. The hairpin structure include the encoded base sequence enabling the versatile sensing of any DNA through Exo III-recycled target DNA. The domain I consists of the recognition sequence of the target DNA-7, whereas the region II is a conserved sequence complementary to DNA-5 linked to the SWNTs. The conserved sequence, II, is caged in the stem region of the hairpin. In the presence of the target DNA-7, the hairpin opens to form a DNA-6/DNA-7 duplex with overhangs at the 3′-ends of both DNA-6 and DNA-7, which is resistant to the cleavage by Exo III. The open hairpin duplex then hybridizes with DNA-5 associated with the SWNTs, forming a DNA-5/DNA-6/DNA-7 duplex structure. Which triggers the selective cleavage of the 3′-end of DNA-5 by Exo III, resulting in the removal of the chemiluminescent units and ultimately release of the open hairpin duplex. The released open hairpin duplex can further hybridize with another ABEI-DNA-5 probe linked to the SWNTs, and the cycle starts anew. This results in the continuous digestion of ABEI-DNA-5 linked to the SWNTs, generating a concomitant increase in CL intensity, and thus achieving target assisted Exo III-catalyzed signal amplification. By monitoring the increase in CL intensity, the target DNA can be detected with a very high sensitivity. Using ABEI-DNA-5/SWNT hybrid as the signal transducer, CL signal was monitored at different times from a DNA-7 solution in the presence of DNA-6 and Exo III. Response linearity of the assay was evaluated by analyzing a series of DNA-7 solutions at concentrations varying from 200 fM to 2 nM. From the calibration curve obtained, the detection limit was estimated to be 88 fM DNA-7.
Scheme 2.
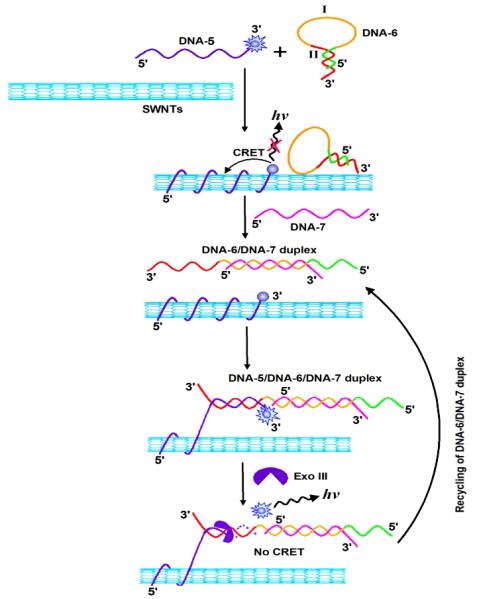
Schematic illustration of a versatile SWNT-mediate CRET platform for amplified detection of DNA using a hairpin structure and Exo III-catalyzed recycling of the analyte /hairpin complex.
In summary, we have developed an amplified SWNT-mediated CL turn-on sensing platform for ultrasensitive DNA assay. We first confirmed that ABEI-labeled DNA molecules were effectively adsorbed onto SWNTs surface, resulting in the quenching of CL from ABEI due to CRET. However, in the presence of the target DNA, it hybridizes with ABEI-labeled DNA absorbed on SWNTs, forming a stable DNA/DNA duplex, and thus releasing the duplex from SWNTs. The sensitivity could be increased by using Exonuclease III-catalyzed target recycling for 2 h thanks to the extremely low background signal from the “turned off” chemiluminescent probes. The Exo III amplification assay had a limit of detection of 45 fM that was by far more sensitive than most of the DNA assays reported so far. Further, by introducing appropriate DNA hairpins into the system, the proposed chemiluminescent probe became versatile for DNA quantification. It’s also worth mentioning that since hairpin structures may be opened by other analytes, e.g. aptamer-substrate complexes, the proposed sensing platform may be applicable to sensitive quantification of analytes other than DNAs.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundations of China (No. 21175030), the Natural Science Foundations of Guangxi Province (No. 2010GXNSFF013001) as well as BAGUI Scholar Program and US National Institutes of Health (GM089557 to YML).
Footnotes
† Electronic Supplementary Information (ESI) available: Experimental section and supporting figures and tables. See DOI: 0.1039/b000000x/
Notes and references
- 1(a).Martínez M, Tseng YC, Ormategui N, Loinaz I, Eritja R, Bokor J. Nano Lett. 2009;9:530. doi: 10.1021/nl8025604. [DOI] [PubMed] [Google Scholar]; (b) Yoo SM, Yoon I, Lee SY, Kim B. Nano Lett. 2010;10:T. Kang, 1189. doi: 10.1021/nl1000086. [DOI] [PubMed] [Google Scholar]; (c) Wang F, Elbaz J, Willner I. J. Am. Chem. Soc. 2012;134:5504. doi: 10.1021/ja300616w. [DOI] [PubMed] [Google Scholar]; (d) Kim HN, Lee E-H, Xu Z, Kim H-E, Lee H-S, Lee J-H, Yoon J. Biomaterials. 2012;33:2282. doi: 10.1016/j.biomaterials.2011.11.073. [DOI] [PubMed] [Google Scholar]
- 2.Schäferling M. Angew Chem Int Ed Engl. 2012;51:3532. doi: 10.1002/anie.201105459. [DOI] [PubMed] [Google Scholar]
- 3(a).Freeman R, Liu X, Willner I. J Am Chem Soc. 2011;133:11597. doi: 10.1021/ja202639m. [DOI] [PubMed] [Google Scholar]; (b) Bi S, Zhou H, Zhang S. Chem. Sci. 2010;1:681. [Google Scholar]
- 4(a).Huang X, Li L, Qian H, Dong C, Ren J. Angew. Chem. Int. Ed. 2006;45:5140. doi: 10.1002/anie.200601196. [DOI] [PubMed] [Google Scholar]; (b) Zhao S, Huang Y, Liu R, Shi M, Liu YM. Chem. Eur. J. 2010;16:6142. doi: 10.1002/chem.201000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5(a).Liu X, Freeman R, Golub E, Willner I. ACS Nano. 2011;5:7648. doi: 10.1021/nn202799d. [DOI] [PubMed] [Google Scholar]; (b) Lee JS, Joung HA, Kim MG, Park CB. ACS Nano. 2012;6:2978. doi: 10.1021/nn300684d. [DOI] [PubMed] [Google Scholar]; (c) Qin G, Zhao S, Huang Y, Jiang J, Ye F. Anal. Chem. 2012;84:2708. doi: 10.1021/ac202959d. [DOI] [PubMed] [Google Scholar]
- 6(a).Tasis D, Tagmatarchis N, Bianco A, Prato M. Chem. Rev. 2006;106:1105. doi: 10.1021/cr050569o. [DOI] [PubMed] [Google Scholar]; (b) Britz DA, Khlobystov AN. Chem. Soc. Rev. 2006;35:637. doi: 10.1039/b507451g. [DOI] [PubMed] [Google Scholar]; (c) Avouris P, Chen Z, Perebeinos V. Nat. Nanotechnol. 2007;2:605. doi: 10.1038/nnano.2007.300. [DOI] [PubMed] [Google Scholar]; (d) Kanungo M, Lu H, Malliaras GG, Blanchet GB. Science. 2009;323:234. doi: 10.1126/science.1166087. [DOI] [PubMed] [Google Scholar]; (e) Zelada-Guillén GA, Riu J, Düzgün A, Rius FX. Angew. Chem. Int. Ed. 2009;48:7334. doi: 10.1002/anie.200902090. [DOI] [PubMed] [Google Scholar]; (f) Yang W, Ratinac KR, Ringer SP, Thordarson P, Gooding JJ, Braet F. Angew. Chem. Int. Ed. 2010;49:2114. doi: 10.1002/anie.200903463. [DOI] [PubMed] [Google Scholar]; (g) Clavé G, Campidelli S. Chem. Sci. 2011;2:1887. [Google Scholar]
- 7(a).Zhang M, Jagota A, Semke ED, Bruce A, Diner BA, Mclean RS, Lustig SR, Richardson RE, Tassi NG. Nat. Mater. 2003;2:338. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]; (b) Wang S, Humphreys ES, Chung SY, Delduco DF, Lustig SR, Wang H, Parker K, Rizzo NW, Subramoney S, Chiang YM, Jagota A. Nat. Mater. 2003;2:196. doi: 10.1038/nmat833. [DOI] [PubMed] [Google Scholar]
- 8(a).Jeng ES, Moll AE, Roy AC, Gastala JB, Strano MS. Nano Lett. 2006;6:371. doi: 10.1021/nl051829k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kim NWS, O’Connell M, Wisdom JA, Dai HJ. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11600. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhao C, Qu K, Song Y, Xu C, Ren J, Qu X. Chem. Eur. J. 2010;16:8147. doi: 10.1002/chem.201000306. [DOI] [PubMed] [Google Scholar]
- 9.Zhen SJ, Chen LQ, Xiao SJ, Li YF, Hu PP, Zhan L, Peng L, Song EQ, Huang CZ. Anal. Chem. 2010;82:8432. doi: 10.1021/ac100709s. [DOI] [PubMed] [Google Scholar]
- 10(a).Swathi RS, Sebastian KL. J. Chem. Phys. 2008;129:054703. doi: 10.1063/1.2956498. [DOI] [PubMed] [Google Scholar]; (b) Swathi RS, Sebastian KL. J. Chem. Sci. 2009;121:777. [Google Scholar]
- 11.Yang R, Jin J, Chen Y, Shao N, Kang H, Xiao Z, Tang Z, Wu Y, Zhu Z, Tan W. J. Am. Chem. Soc. 2008;130:8351. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 12(a).Ciruela F. Curr. Opin. Biotechnol. 2008;19:338. doi: 10.1016/j.copbio.2008.06.003. [DOI] [PubMed] [Google Scholar]; (b) Tu D, Liu L, Ju Q, Liu Y, Zhu H, Li R, Chen X. Angew. Chem. Int. Ed. 2011;50:6306. doi: 10.1002/anie.201100303. [DOI] [PubMed] [Google Scholar]; (c) Huang Y, Zhao S, Liang H, Chen ZF, Liu YM. Chem. Eur. J. 2011;17:7313. doi: 10.1002/chem.201003765. [DOI] [PubMed] [Google Scholar]
- 13(a).You M, Chen Y, Peng L, Han D, Yin B, Ye B, Tan W. Chem. Sci. 2011;2:1003. [Google Scholar]; (b) Lu CH, Wang F, Willner I. Chem. Sci. 2012;3:2616. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


