Abstract
Living cells use a variety of regulatory network motifs for accurate gene expression in response to changes in their environment or during differentiation processes. In Rhodobacter sphaeroides, a complex regulatory network controls expression of photosynthesis genes to guarantee optimal energy supply on one hand and to avoid photooxidative stress on the other hand. Recently, we identified a mixed incoherent feed-forward loop comprising the transcription factor PrrA, the sRNA PcrZ and photosynthesis target genes as part of this regulatory network. This point-of-view provides a comparison to other described feed-forward loops and discusses the physiological relevance of PcrZ in more detail.
Keywords: photosynthesis, gene regulation, feed-forward loop, oxygen tension, redox regulation, small RNAs, Rhodobacter
Introduction
Bacterial small RNAs (sRNAs) are widely recognized as important regulators of gene expression and commonly operate at the post-transcriptional level by influencing mRNA stability and/or translation.1,2 The combination of transcriptional and post-transcriptional gene control by protein transcription factors and sRNAs, respectively, endues bacteria with elaborate switches for adaptive processes. Examples for regulatory networks consisting of transcription factors and sRNAs, which either concertedly control genes or regulate each other, are accumulating; sRNAs extend the regulatory scope of the networks and alter their dynamics.3 The combined networks are frequently employed by bacteria to adapt to sudden changes, such as upcoming stress factors or changes in nutrient availability. Especially bacteria living in quickly alternating environments have a need for sophisticated fine-tuning of gene regulation. In the case of purple bacteria like Rhodobacter, the amount of photosynthetic complexes is adjusted to the given oxygen tension and light intensities by a complex network of protein factors (Fig. 1).4,5 For example, the response regulator PrrA activates transcription of photosynthesis genes when oxygen tension drops. However, it was recently shown that the trans-encoded sRNA PcrZ (photosynthesis control RNA Z) of Rhodobacter sphaeroides is transcribed from a PrrA-dependent promoter and subsequently counteracts the induction of photosynthesis genes on the post-transcriptional level.6 This regulatory interplay constitutes a rare example of a mixed incoherent feed-forward loop (FFL) involving a protein regulator (PrrA) and an sRNA (PcrZ), which finally allows for balanced expression of photosynthetic complexes. In this point-of-view, we will compare expression kinetics of PcrZ with those of photosynthesis genes and draw a comparison to other described FFLs. The physiological relevance of PcrZ will be discussed in detail.
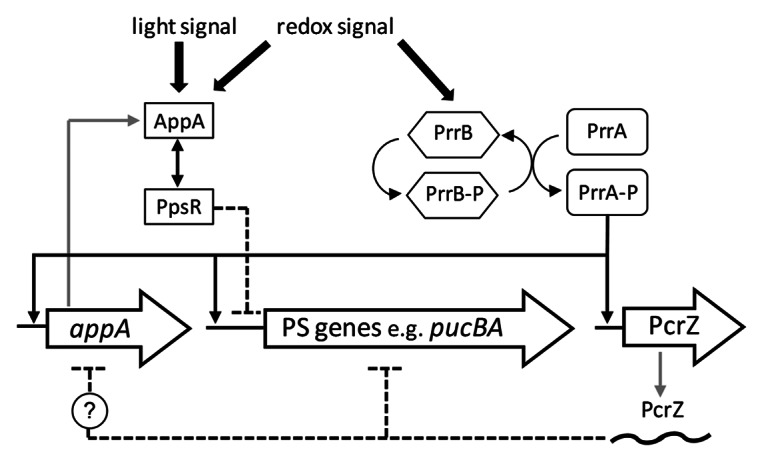
Figure 1. Schematic illustration of the regulatory network controlling photosynthesis (PS) gene expression, modified from Mank et al.6 PrrA activates the expression of PS genes and PcrZ at low oxygen tension, whereas PcrZ counteracts the activation of PS genes. PcrZ also reduces, directly or indirectly, the amount of AppA, leading to a stronger repression of PS genes by PpsR.
PcrZ: First trans-acting small RNA with a role in photosynthesis gene regulation
PcrZ is an sRNA with a size of 136 nt, which seems to be evolutionary restricted to R. sphaeroides strains. The pcrZ gene is located on chromosome 1 in the intergenic region between RSP_0819 (rhlE2 encoding DEAD/DEAH helicase) and RSP_6134 (hypothetical protein). Induction of PcrZ expression depends on the response regulator PrrA, which is activated by the sensor kinase PrrB when the oxygen tension drops and recognizes a fairly well-conserved PrrA-binding motif upstream of the pcrZ gene.6,7 PrrA also induces the expression of photosynthesis genes, like the puc and puf operon encoding structural elements of the photosynthetic apparatus.8 Constitutive overexpression of PcrZ resulted in a decrease of photosynthetic complexes, which was visible by a strongly reduced pigmentation of cells. Northern blot analysis revealed that processing fragments with sizes around 51–56 nt and different 3′-ends strongly accumulated in the PcrZ overexpression strain. However, overexpression of the 51 nt processed fragment was not sufficient to reduce photosynthetic complexes, which leads to the assumption that full-length PcrZ represents the functional unit. Microarray analysis suggested a negative effect of PcrZ on multiple photosynthesis genes, like RSP_0285 (bchN, encodes light-independent protochlorophyllide reductase subunit) and RSP_6158 (puc2A, encodes protein of the light-harvesting II complex). Direct PcrZ binding to bchN and puc2A mRNAs was supported by a lacZ-based reporter system combined with mutational analysis, which also validated the repressing effect of PcrZ. Given that PrrA induces expression of photosynthesis genes and, in parallel, PcrZ, which, in turn, represses photosynthesis genes, the regulatory architecture resembles a mixed incoherent FFL. The function of this regulatory loop is assumed to balance the synthesis of photosynthetic complexes. This aspect is discussed below in more detail.
Comparison to other feed-forward loops
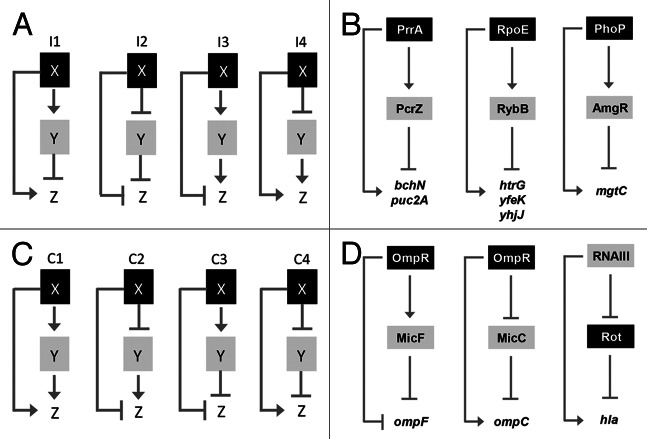
Regulatory networks control accurate gene expression and are therefore essential to fully adapt to various stimuli like stresses or nutrient changes. One of these networks is the so-called FFL, which consists of a transcription factor X regulating a second transcription factor Y. Both protein factors jointly regulate the transcription rate of a target gene Z (Fig. 2). Whenever one of the protein factors (X or Y) is replaced by an sRNA, a post-transcriptional regulation level is inserted into the FFL. In this case it is called a mixed FFL.9 In Escherichia coli and Saccharomyces cerevisiae, one of the most common network motifs is the incoherent FFL. There are four described types of this network motif, which have in common that direct (X to Z) and indirect (X over Y to Z) regulation paths are opposite (Fig. 2A). This type of gene regulation allows for speeding up inhibition of target gene activation, which is called pulse expression.9,10 This enables accurate adaptation to environmental changes. Incoherent FFLs that simply include protein regulators are common,10 whereas mixed FFLs are rarely known until now. Besides PrrA/PcrZ, the RpoE/RybB and PhoP/AmgR systems of E. coli and Salmonella enterica, respectively, exhibit features of incoherent FFLs. During envelope stress in E. coli RpoE is released from the membrane and induces the expression of the sRNA RybB and of about 100 other genes mainly involved in outer membrane modification and repair.11 Induction of three RpoE-dependent genes is counteracted by RybB, closing a potential incoherent FFL (Fig. 2B). In S. enterica, low magnesium concentrations lead to phosphorylation of the transcriptional regulator PhoP, which, in turn, activates the expression of mgtC and its cis-encoded sRNA AmgR simultaneously.12 AmgR counteracts mgtC expression resulting in reduced MgtC amounts (Fig. 2B).13 In all three cases (PrrA/PcrZ, RpoE/RybB and PhoP/AmgR), beneficial fine-tuning of gene expression is achieved. Another FFL is the coherent form, where the direct regulation path (X to Z) has the same outcome as the overall regulation (X over Y to Z). The four different types of coherent FFLs are illustrated in Figure 2C. Coherent FFLs can cause a sign-sensitive delay by using either AND- or OR-gates to regulate the transcription of target genes.9 In the case of an AND-gate, X and Y are required for entire Z transcription. If an OR-gate is present, X or Y is sufficient for Z transcription. Examples for mixed coherent FFLs are described in E. coli and Staphylococcus aureus. In E. coli, the sensor kinase EnvZ monitors changes in osmolarity and modulates the activity of the transcriptional regulator OmpR.14 Phosphorylated OmpR (OmpR-P) modulates the expression of its target genes ompC and ompF, encoding outer membrane proteins in a concentration-dependent manner.15,16 At high OmpR-P levels, ompC expression is activated, whereas ompF is repressed. Additionally, OmpR-P inhibits MicC and induces MicF expression, leading to an enhanced activation of ompC and an increased inhibition of ompF, respectively (Fig. 2D).16,17 The multifunctional RNAIII from S. aureus acts as both activator and repressor of mRNA translation. In stationary growth phase, the ArgA-ArgC two-component system activates transcription of RNAIII.18,19 On one hand, binding of the RNAIII 5′-end to a secondary structure, which blocks the ribosome-binding side (RBS) of hla (hemolysin α) mRNA, results in translation initiation.20 On the other hand RNAIII also binds to rot (repressor of toxins) mRNA and initiates RNase III cleavage of the sRNA/mRNA duplex.21 Since Rot represses hla, the inactivation of Rot by RNAIII further enhances transcription of hla (Fig. 2D).
Figure 2. (A and C) Structure and regulatory output of incoherent and coherent FFL motifs. Four different types (I1-I4 and C1-C4) are depicted, where X and Y refer to transcription factors (black boxes) and small RNAs (gray boxes), respectively. Z reflects target genes of X and Y, modified from Mangan and Alon.9 In the case of the RNAIII FFL, Rot stands on one hand for the rot mRNA, repressed by RNAIII, on the other hand for the Rot protein, repressing hla. (B) Schematic picture of the PrrA/PcrZ, RpoE/RybB and PhoP/AmgR mixed incoherent FFLs in R. sphaeroides, E. coli and S. enterica, respectively. (D) Schematic picture of mixed coherent FFLs. The OmpR-based and RNAIII-based FFLs from E. coli and S. aureus, respectively, are depicted.
A special property of mixed FFLs is that target genes are regulated simultaneously at the transcriptional and post-transcriptional level, leading to a tightly controllable inhibition or activation.22 A mixed FFL provides two benefits for the cells: (1) responding times in an sRNA-mediated manner are much faster compared with protein regulation because of the lack of translation; (2) recovery of a target gene, after external stimuli, is faster in an sRNA-mediated regulation, which clearly depends on the degradation rate of the sRNA and the ratio between sRNA and mRNA production.22 Future studies will reveal whether more of these sRNA-based networks exist, which is clearly expected, since the benefits of sRNA-mediated regulation are universal in all three kingdoms of life.
Oxygen-dependent expression kinetics of PcrZ and photosynthesis genes
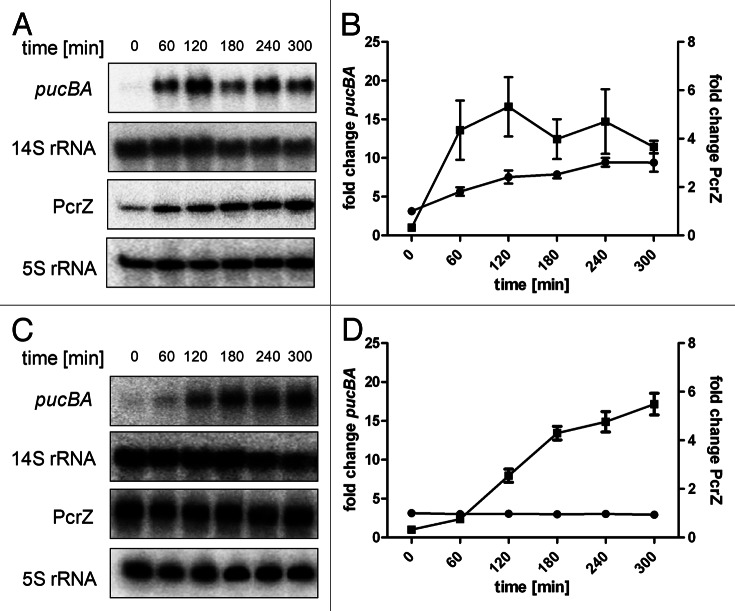
Oxygen tension and light are the most important stimuli controlling formation of photosynthetic complexes in R. sphaeroides. A drop of oxygen tension is an activating stimulus and corresponding expression kinetics of photosynthetic complexes were first monitored for R. capsulatus, which shares many factors involved in regulation of photosynthesis genes with R. sphaeroides.23 In R. capsulatus, mRNA levels of photosynthesis genes intensely increased after drop of oxygen tension, reached a maximum after 30–60 min and decreased again. A strong increase of photosynthesis gene expression (e.g., pucBA) is also observable in R. sphaeroides, but maximal expression is reached only 120 min following the drop of oxygen tension. After maximal expression levels are reached, we observed only a slow decrease of expression in R. sphaeroides (Fig. 3A and B). In the initial phase, the expression kinetic of PcrZ is similar to that of pucBA, but the factor of increase is smaller and a steady increase over the time of the experiment was observed (Fig. 3A and B). A constitutive overexpression of PcrZ results in elevated levels of PcrZ, irrespective of the oxygen tension (Fig. 3C and D).6 As a consequence, activation of photosynthesis gene expression is strongly reduced and maximal mRNA levels of pucBA are not reached within 240 min after drop of oxygen tension. It is likely that the steady increase of PcrZ in the R. sphaeroides wild-type helps to counteract excessive photosynthesis gene activation by PrrA. A slight pulse of pucBA expression with a maximum at 120 min is the characteristic feature of this particular incoherent FFL. Overexpression of PcrZ consequently eliminates this characteristic pulse expression. In R. capsulatus, photosynthesis gene expression exhibits an earlier and more precise pulse, although PcrZ is not present. Obviously, this type of FFL is not necessarily required for inhibition of photosynthesis gene activation. The protein-based regulatory system for controlling photosynthesis genes is however not identical in the two species and sRNAs in R. capsulatus have not been identified. Thus, a different factor may take over this inhibiting effect in R. capsulatus.
Figure 3. Representative northern blots for pucBA and PcrZ expression kinetics after drop of oxygen tension at time point 0. R. sphaeroides 2.4.1 wild-type (A) and R. sphaeroides PcrZ overexpression strain (pRKPcrZ) (C) pre-cultures were grown overnight under high oxygen conditions (8 mg/L soluble O2) to an OD660nm of 0.6–0.8 (t0). Samples were collected at indicated time points after a shift to low oxygen conditions (0.5 mg/L soluble O2). Fifteen µg of total RNA were separated on 1% agarose gels containing 2.2 M formaldehyde. For pucBA detection, a specific DNA fragment was radioactively labeled with (α-32P)-dCTP using the NEBlot kit (New England Biolabs). A 14S rRNA-specific oligonucleotide was end-labeled with (γ-32P)-ATP and served as loading control. In R. sphaeroides, 23S rRNA is processed to 16S, 14S and 5.8S rRNA fragments by RNase III.46,47 For PcrZ and 5S rRNA, 7.5 µg of total RNA was separated on 10% polyacrylamide gels containing 7 M urea. PcrZ and 5S rRNA-specific oligonucleotides were radioactively end-labeled with (γ-32P)-ATP and were used for detection. 5S rRNA served as loading control. (B) Quantification of northern blot signals for wild-type (A). (D) Quantification of northern blot signals for pRKPcrZ (C). The mean and the standard error are based on three independent biological experiments. The Y axis to the left shows the fold change of pucBA (black squares), the Y axis to the right displays the fold change of PcrZ (black circles).
Transcription activation of PcrZ and pucBA is achieved by the same two-component system (PrrB/PrrA). The different intensity of expression activation of PcrZ and pucBA (Fig. 3A and B) might be due to variable recognition elements in the PrrA-binding side.7 A different recognition of binding elements in the promoter region is also described for the protein regulator PhoP in S. enterica. PhoP binds to the mgtC promoter with a higher affinity than to the amgR promoter, since amgR exhibits only half of the PhoP-box motif in its promoter region.13 Besides PrrA, the puc operon of R. sphaeroides is controlled by the PpsR/AppA repressor anti-repressor system. Under high oxygen tension, PpsR represses transcription of puc genes.24 With decreasing oxygen tension, the anti-repressor AppA binds to PpsR, which will no longer bind to its target promoters.5 AppA is redox-responsive through a heme bound to its C-terminal SCHIC domain and also functions as a photoreceptor by sensing blue light through its N-terminal BLUF domain.25-28 Since the PpsR/AppA system does not act on the PcrZ promoter, it might contribute to the observed differences between PcrZ and pucBA expression kinetics. It should also be noted that in R. capsulatus and in R. sphaeroides, the half-life of the pucBA mRNA increase upon drop of oxygen tension (see ref. 29 and unpublished results), which also contributes to a stronger increase of the pucBA mRNA level.29
PcrZ obviously counteracts an unnecessarily high expression of photosynthesis genes when oxygen tension drops. Interestingly, PcrZ is already expressed in considerable amounts when oxygen tension is high (Fig. 3A). One characteristic feature of sRNAs is that they can establish thresholds for gene expression to avoid that a short, transient stimulus triggers an inappropriate response. Given that the transcription rate of the sRNA (αs) is higher than the target transcription (αm), while RNA stabilities are comparable, all target mRNAs pair with the sRNA and translation is abolished. After an increase of αm and constant αs, all sRNAs pair with its target, however unpaired mRNAs are accessible for translation.30,31 In the case of R. sphaeroides, PcrZ might tightly control photosynthesis genes under aerobic conditions and prevent their leaky expression (αs > αm). Slight and transient drops of oxygen tension may also be compensated (αs > αm) and only strong induction of photosynthesis genes allows to escape PcrZ repression (αs < αm). A similar tight control was reported for the iron stress-induced protein A (IsiA) in Synechocystis species. IsiA is produced in high amounts under iron-limiting conditions, high light and oxidative stress, to form a giant ring structure around photosystem I (PS-I) and to dissipate excess light energy.32-34 The isiA gene is under negative control of the cis-encoded sRNA IsrR when conditions are favorable, but escapes repression under stress, reflecting another example of RNA-based regulation of a photosynthetic component.35 Gogol et al. describe a mixed incoherent FFL for controlled gene expression of yfeK (arrests growth) and htrG (cell lysis) in E. coli.11 These genes are activated by RpoE and downregulated by the sRNA RybB. During low levels of stress, the repressing feature of RybB is sufficient to block yfeK and htrG expression. Increasing stress conditions, leading to highly damaged cells, result in accelerated RpoE expression. This overcomes the repressing effect of RybB, leading to expression of yfeK and htrG followed by cell death. If growth conditions get favorable again these cells are removed preventing them from competing for resources.11
Finally, tight control by an sRNA might cause retardation of the response to the external stimulus, which is indeed observed for R. sphaeroides; pucBA induction is clearly retarded when compared with R. capsulatus, which lacks PcrZ.23,36
PcrZ indirectly counteracts photooxidative stress
Why is regulation of photosynthesis genes so complex? The main reason might be found in the ATP yields of different energy producing pathways. Rhodobacter species grow best by performing aerobic respiration and photosynthesis is avoided as long as oxygen tension is elevated. Only when oxygen starts to deplete, formation of photosynthetic complexes is induced, which is mainly due to the action of the oxygen-dependent PrrB/PrrA two-component system.37 However, at intermediate oxygen tension, photosynthesis genes are still repressed by PpsR when blue light, even at very low fluence rates, is present.38 It is speculated that under these conditions, generation of highly toxic singlet oxygen is avoided by keeping bacteriochlorophyll (BChl) levels low. It was demonstrated that BChls can act as natural photosensitizers for the light-driven generation of singlet oxygen, a situation also known as photooxidative stress.39,40 In general it emerged that performing photosynthesis in the presence of oxygen confronts bacteria with photooxidative stress due to singlet oxygen production. Cyanobacteria like Synechocystis species, which perform oxygenic photosynthesis, permanently produce oxygen in the water-splitting complex of photosystem II (PS-II), which is consequently damaged by singlet oxygen. These damages, especially the D1 protein is targeted, accelerate selective protein turnover and subunit replacement.41 As mentioned above, the IsiA protein is produced in high amounts under iron-limiting conditions, high light and oxidative stress, to form a giant ring structure around PS-I and to dissipate excess light energy.32-34 Cyanobacteria obviously have to cope with singlet oxygen by constantly repairing damages and protecting their photosystems, which happens at the expense of resources. In contrast, facultative phototrophs like Rhodobacter species have the opportunity to avoid extensive singlet oxygen generation by reducing BChl levels and it is tempting to speculate that regulatory systems like AppA/PpsR and the newly identified PrrA/PcrZ system are in charge of counteracting photooxidative stress.5,6,24,27 This idea is supported by the observation that under anoxic conditions in the light, at the time when no singlet oxygen can be generated and photosynthesis is used for ATP generation, both systems seem to play negligible roles, as PpsR is completely inactivated by AppA and PcrZ is only expressed at basal levels.4,6
What comes next?
There are still several open questions, which have to be followed up in the future. As described above, the processed 51 nt 5′-fragment of PcrZ alone is not capable of producing the photosynthesis-related phenotype. We therefore propose that processing is linked to the regulatory mechanism. Processing of PcrZ could be coupled to degradation of its target mRNAs by RNase III as it is observed for, e.g., RyhB/sodB interaction in enterobacteria.42 However, the PcrZ processing pattern is normal in an RNase III deletion strain of R. sphaeroides (unpublished results). A major influence of RNase E, which performs initial cleavage, together with 3′ to 5′ exonucleases, which produce the different PcrZ 3′-ends, might represent a feasible scenario but needs further elucidation.
Several trans-acting sRNAs interact with Hfq for imperfect base-pairing with their targets.43 We have strong evidence that PcrZ is not Hfq-associated.6,44 However, it is likely that trans-regulation of multiple PcrZ targets relies, at least partially, on an RNA-binding protein, which facilitates structural rearrangements as well as influences RNA stabilities. The rhlE2 gene, which is located upstream of pcrZ, encodes a DEAD/DEAH box helicase. The RhlE2 helicase might interact with PcrZ in a fashion similar to Hfq and affect processing and interaction. An Argonaute-like protein with a function that resembles Hfq was recently identified in Sinorhizobium meliloti.45 We can therefore expect that new proteins with an impact on sRNA functions will be discovered in the future.
It was demonstrated that the appA gene is indirectly affected by PcrZ and it is likely that this accounts also for some of the other genes with altered expression in a PcrZ overexpression strain.6 Distinguishing direct from indirect targets as well as identification of further PcrZ targets are future tasks. Likewise, it will be an exciting topic to identify additional sRNAs that regulate photosynthesis genes in R. sphaeroides and other bacteria.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23769
References
- 1.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–82. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregor J, Klug G. Regulation of bacterial photosynthesis genes by oxygen and light. FEMS Microbiol Lett. 1999;179:1–9. doi: 10.1111/j.1574-6968.1999.tb08700.x. [DOI] [PubMed] [Google Scholar]
- 5.Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–23. doi: 10.1016/S0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 6.Mank NN, Berghoff BA, Hermanns YN, Klug G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc Natl Acad Sci USA. 2012;109:16306–11. doi: 10.1073/pnas.1207067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao L, Mackenzie C, Roh JH, Eraso JM, Kaplan S, Resat H. Combining microarray and genomic data to predict DNA binding motifs. Microbiology. 2005;151:3197–213. doi: 10.1099/mic.0.28167-0. [DOI] [PubMed] [Google Scholar]
- 8.Eraso JM, Kaplan S. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1996;178:7037–46. doi: 10.1128/jb.178.24.7037-7046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–5. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006;356:1073–81. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci USA. 2011;108:12875–80. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin D, Groisman EA. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem. 2005;280:4089–94. doi: 10.1074/jbc.M412741200. [DOI] [PubMed] [Google Scholar]
- 13.Lee EJ, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76:1020–33. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forst SA, Roberts DL. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res Microbiol. 1994;145:363–73. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 15.Egger LA, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells. 1997;2:167–84. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol. 2004;186:6689–97. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiba H, Matsuyama SI, Mizuno T, Mizushima S. Function of micF as an antisense RNA in osmoregulatory expression of the ompF gene in Escherichia coli. J Bacteriol. 1987;169:3007–12. doi: 10.1128/jb.169.7.3007-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–75. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–49. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 20.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–77. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–66. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, et al. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol. 2007;3:138. doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klug G, Kaufmann N, Drews G. Gene expression of pigment-binding proteins of the bacterial photosynthetic apparatus: Transcription and assembly in the membrane of Rhodopseudomonas capsulata. Proc Natl Acad Sci USA. 1985;82:6485–9. doi: 10.1073/pnas.82.19.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomelsky M, Kaplan S. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J Bacteriol. 1995;177:1634–7. doi: 10.1128/jb.177.6.1634-1637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Y, Meyer MH, Keusgen M, Klug G. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol Microbiol. 2007;64:1090–104. doi: 10.1111/j.1365-2958.2007.05724.x. [DOI] [PubMed] [Google Scholar]
- 26.Moskvin OV, Kaplan S, Gilles-Gonzalez MA, Gomelsky M. Novel heme-based oxygen sensor with a revealing evolutionary history. J Biol Chem. 2007;282:28740–8. doi: 10.1074/jbc.M703261200. [DOI] [PubMed] [Google Scholar]
- 27.Braatsch S, Gomelsky M, Kuphal S, Klug G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol Microbiol. 2002;45:827–36. doi: 10.1046/j.1365-2958.2002.03058.x. [DOI] [PubMed] [Google Scholar]
- 28.Gomelsky M, Klug G. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/S0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 29.Klug G. Endonucleolytic degradation of puf mRNA in Rhodobacter capsulatus is influenced by oxygen. Proc Natl Acad Sci USA. 1991;88:1765–9. doi: 10.1073/pnas.88.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine E, Hwa T. Small RNAs establish gene expression thresholds. Curr Opin Microbiol. 2008;11:574–9. doi: 10.1016/j.mib.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cadoret JC, Demoulière R, Lavaud J, van Gorkom HJ, Houmard J, Etienne AL. Dissipation of excess energy triggered by blue light in cyanobacteria with CP43′ (isiA) Biochim Biophys Acta. 2004;1659:100–4. doi: 10.1016/j.bbabio.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Yeremenko N, Kouril R, Ihalainen JA, D’Haene S, van Oosterwijk N, Andrizhiyevskaya EG, et al. Supramolecular organization and dual function of the IsiA chlorophyll-binding protein in cyanobacteria. Biochemistry. 2004;43:10308–13. doi: 10.1021/bi048772l. [DOI] [PubMed] [Google Scholar]
- 34.Ihalainen JA, D’Haene S, Yeremenko N, van Roon H, Arteni AA, Boekema EJ, et al. Aggregates of the chlorophyll-binding protein IsiA (CP43′) dissipate energy in cyanobacteria. Biochemistry. 2005;44:10846–53. doi: 10.1021/bi0510680. [DOI] [PubMed] [Google Scholar]
- 35.Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci USA. 2006;103:7054–8. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legewie S, Dienst D, Wilde A, Herzel H, Axmann IM. Small RNAs establish delays and temporal thresholds in gene expression. Biophys J. 2008;95:3232–8. doi: 10.1529/biophysj.108.133819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eraso JM, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metz S, Jäger A, Klug G. In vivo sensitivity of blue-light-dependent signaling mediated by AppA/PpsR or PrrB/PrrA in Rhodobacter sphaeroides. J Bacteriol. 2009;191:4473–7. doi: 10.1128/JB.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchoa AF, Knox PP, Turchielle R, Seifullina NKh, Baptista MS. Singlet oxygen generation in the reaction centers of Rhodobacter sphaeroides. Eur Biophys J. 2008;37:843–50. doi: 10.1007/s00249-008-0287-y. [DOI] [PubMed] [Google Scholar]
- 40.Glaeser J, Nuss AM, Berghoff BA, Klug G. Singlet oxygen stress in microorganisms. Adv Microb Physiol. 2011;58:141–73. doi: 10.1016/B978-0-12-381043-4.00004-0. [DOI] [PubMed] [Google Scholar]
- 41.Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J. Recent advances in understanding the assembly and repair of photosystem II. Ann Bot. 2010;106:1–16. doi: 10.1093/aob/mcq059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonyushkin T, Vecerek B, Moll I, Bläsi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–89. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F, Vogel J, et al. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol Microbiol. 2011;80:1479–95. doi: 10.1111/j.1365-2958.2011.07658.x. [DOI] [PubMed] [Google Scholar]
- 45.Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39:4691–708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kordes E, Jock S, Fritsch J, Bosch F, Klug G. Cloning of a gene involved in rRNA precursor processing and 23S rRNA cleavage in Rhodobacter capsulatus. J Bacteriol. 1994;176:1121–7. doi: 10.1128/jb.176.4.1121-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marrs BL, Kaplan S. 23 s precursor ribosomal RNA of Rhodopseudomonas spheroides. J Mol Biol. 1970;49:297–317. doi: 10.1016/0022-2836(70)90247-0. [DOI] [PubMed] [Google Scholar]