Abstract
miRNAs, a subclass of small regulatory RNAs, are present from ancient unicellular protozoans to parasitic helminths and parasitic arthropods. The miRNA-silencing mechanism appears, however, to be absent in a number of protozoan parasites. Protozoan miRNAs and components of their silencing machinery possess features different from other eukaryotes, providing some clues on the evolution of the RNA-induced silencing machinery. miRNA functions possibly associate with neoblast biology, development, physiology, infection and immunity of parasites. Parasite infection can alter host miRNA expression that can favor both parasite clearance and infection. miRNA pathways are, thus, a potential target for the therapeutic control of parasitic diseases.
Keywords: miRNA, protozoan, helminth, snoRNA, neoblast
Introduction
microRNAs (miRNAs) are one subtype of small endogenous single-stranded RNAs and have been so far reported in many viruses, animals and plants such as Mareks diseased virus, fruit flies, humans, zebra fish and Arabidopsis. But the miRNA-induced silencing mechanism may be lost in yeast and some unicellular organisms.1 miRNA-mediated regulation of gene expression may be pervasively distributed across the parasite kingdom, although miRNAs are absent in some unicellular parasites.2,3 Individual miRNAs can directly or indirectly repress hundreds or even thousands of genes, affecting protein production mostly on a fine scale via translational repression or/and mRNA degradation.4,5 A systematic survey showed that more than 20% human genes were regulated by miRNAs.6 Although a study in which 95 miRNA genes were mutated in Caenorhabiditis elegans, no abnormal phenotypes were observed, indicating major miRNAs are not vital for the development or viability.7 Increasing evidence supports the idea that miRNAs participate in complex regulatory networks and the abnormality of some miRNAs is related to occurrence of many human diseases, including cancer.8-12
Silencing of the main RNA-induced silencing complex (RISC) component was shown to promote the susceptibility of Anopheles gambiae to Plasmodium infection.13 Similarly, dysfunction in Dicer and RNA-dependent RNA polymerase resulted in a loss of the regulation control of specific surface proteins in Giardia lamblia.14 These observations indicate that regulatory small RNAs are essential in parasite infection. We herein outline our current understanding of miRNAs in unicellular and multicellular parasites and provide examples of some unique properties of protozoan miRNAs. The relationship between snoRNAs and miRNA precursors, miRNA markers for neoblasts and the potential roles of miRNAs in parasites are also discussed.
Biogenesis of miRNAs
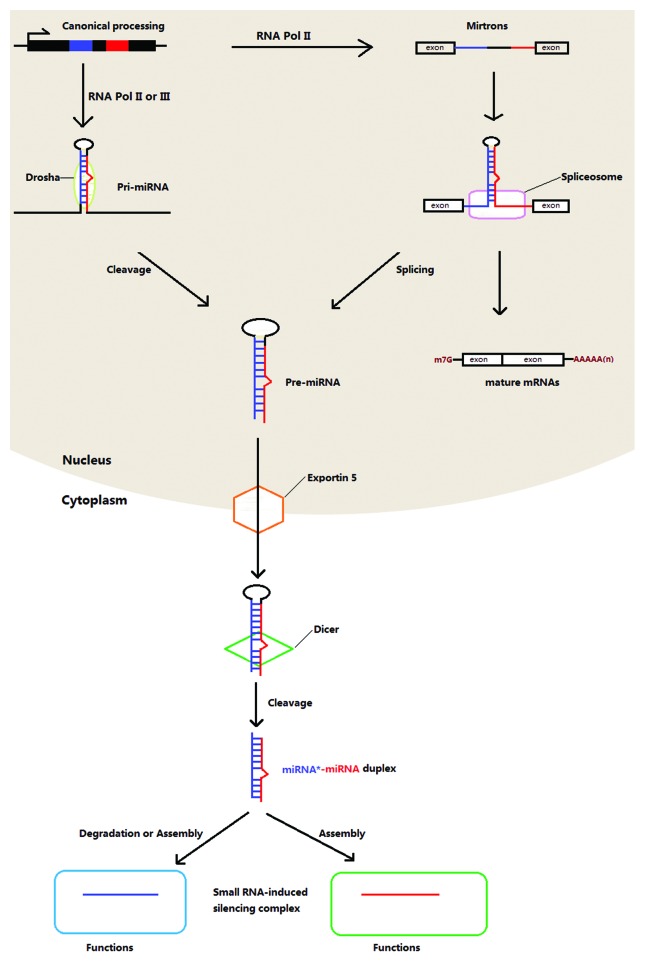
In the canonical approach, miRNA genes are first transcribed into long pri-miRNA transcripts by RNA polymerase (Pol), usually Pol II. These pri-miRNAs are specifically spliced by Drosha, a nuclear RNase III, giving rise to the precursors of miRNAs with a size of approximately 70 nucleotides (pre-miRNAs). With the help of a nuclear transport receptor (exportin-5) that acts with the molecules with a stem and a short overhang at the 3′ end, the pre-miRNAs in the nucleus are transported to the cytoplasm. Afterwards, the molecules exported are cleaved into smaller duplexes, which are mediated by RNase III Dicer. To be functional, double-stranded shortened miRNAs, which are swiftly changed into single strand during assembly, are loaded onto an Argonaut-containing RNA-induced silencing complex (RISC).15,16 In most cases, the partner of the mature miRNA incorporated into the complex, named as miRNA*, is degraded. Recently, it has been shown that miRNAs* are also present at a relatively high level and have capacity of repressing targets (Fig. 1).17 This finding adds the complexity of regulatory networks where miRNAs or miRNAs* dominate.
Figure 1. Biogenesis of canonical and mirtron miRNAs in animals.
By contrast, the biogenesis of mirtron miRNAs, which are found in C. elegans, Drosophila melanogster,15 chicken18 and several mammals,19 is different. The short intron-derived pri-miRNAs are spliced into intron-removed mRNAs and intron lariats. Without the cleavage by Drosha, the introns are folded directly to form pre-miRNAs with the hairpin structure15 and further processed as described above. Murine herpesvirus miRNAs are produced in yet another way.20 RNA Pol III, instead of Pol II, is functional to produce pri-miRNAs that contain a tRNA-like structure at the 5′ end and two stem loops 1 in the middle and 2 at the 3′ end. The pri-miRNAs are cleaved by tRNase Z but not Drosha to yield the pre-miRNAs that are then further edited in a typical way to liberate mature viral miRNAs.21 In Giardia lamblia, an ancient parasitic protozoan, miRNAs are derived from small nucleolar RNAs (snoRNAs) that participate in modification of other types of RNA.22 Together with other facts that Drosha and Exportin 5 homologs are absent but two vital RISC factors, Argonaute and Dicer, do exist in Giardia raise the possibility of another Drosha-independent pathway is involved in miRNA biogenesis.
The biogenesis of miRNAs from transcription to mature miRNAs is tightly controlled via modifications of miRNA-producing RNA and factors that form functional RISC or other complexes and these modifications can be offered by internal or even external molecules.23 Together with other proteins, such as TRBP and PACT, Dicer constitutes a machinery to process pre-miRNAs and its elevating level can result from the stabilized TRBP through phosphorylation.24 In infected T cells, the abundance of mature miR-27 was alternatively downregulated by binding to one of non-coding RNAs of Herpesvirus saimiri, HSUR1, although the consequent effects of induced decay have not been determined.25
miRNAs in parasite kingdom
At present, miRNAs have been computationally or experimentally investigated in seven protozoans, three trematodes, two nematodes, one cestode and three arthropods (Table 1). The lack of coding genes for Dicer and Argonaute proteins, key factors of RISC, in the genome of a number of protozoan parasites, implies the absence of miRNA-induced regulation of gene expression.3 Alignment analysis of Argonaute showed that the expression modulation by small RNA is extensively present across parasitic cestodes and nematodes (Table 1). Interestingly, apart from active Argonaute-coding genes, there are Argonaute pseudogenes present in three Leishmania species (Table 1), suggesting the shrinking of RNA-inducing silencing functions during evolution. Conversely, the expansion of miRNA pathways has been demonstrated in the plant parasite, Acyrthosiphon pisum, although the biological significance of the miRNA machinery gene duplication events is elusive.26
Table 1. miRNAs and/or RNA-induced silencing machinery in parasites.
| Species | Approacha | Num of miRNAs | Main RISC componentsb | Ref. |
|---|---|---|---|---|
|
Protozoanc |
|
|
|
|
| Plasmodium falciparum |
Ex |
No |
No |
2
|
| Trypanosoma brucei |
Com |
1,162 |
Yes |
3
,
69
|
| Trypanosoma congolense |
- |
ND |
Yes |
3
|
| Trypanosoma cruzi |
- |
ND |
Yes |
3
,
27
|
| Trypanosoma gambiense |
- |
ND |
Yes (sanger) |
|
| Trypanosoma vivax |
- |
ND |
Yes (sanger) |
|
| Leishmania major |
- |
ND |
Yes/Pseudogene |
|
| Leishmania infantum |
- |
ND |
Yes/Pseudogene |
|
| Leishmania braziliensis |
- |
ND |
Yes (sanger) |
|
| Leishmania mexicana |
- |
ND |
Yes/Pseudogene |
|
| Leishmania tarentolae |
- |
ND |
Yes (TriTrypDB) |
|
| Leishmania braziliensis |
- |
ND |
Yes |
3
|
| Cryptosporidium spp |
- |
ND |
No |
3
|
| Theileria spp |
- |
ND |
No |
3
|
| Babesia bovis |
- |
ND |
No |
3
|
| Eimeria tenella |
- |
ND |
No |
3
|
| Giardia lamblia |
Ex |
4 |
Yes |
22
|
| Neospora caninum |
- |
ND |
Yes |
|
| Giardia intestinalis |
Ex |
10 |
Yes |
3
,
49
|
| Trichomonas vaginalis |
Ex |
11 |
Yes |
3
,
49
|
| Entamoeba histolytica |
Com |
17 |
Yes |
3
,
78
|
| Toxoplasma gondii |
Ex |
35 |
Yes |
3
,
30
|
|
Trematode |
|
|
|
|
| Clonorchis sinensis |
Ex/Com |
62,518 |
- |
79
|
| Schistosoma mansoni |
Ex |
5 |
Yes |
58
,
59
|
| Schistosoma japonicum |
Ex |
55 |
Yes |
80
|
|
Nematode |
|
|
|
|
| Bursaphelenchus xylophilus |
Ex |
810 |
- |
61
|
| Brugia malayi |
Ex/Com |
32 |
Yes |
81
|
| Hemonchus contortus |
- |
ND |
Yes (sanger) |
|
| Globodera pallida |
- |
ND |
Yes (sanger) |
|
| Trichuris trichiura |
- |
ND |
Yes (sanger) |
|
| Onchocerca volvulus |
- |
ND |
Yes (sanger) |
|
| Strongyloides ratti |
- |
ND |
Yes (sanger) |
|
| Trichuris muris |
- |
ND |
Yes (sanger) |
|
| Nippostrongylus brasiliensis |
- |
ND |
Yes (sanger) |
|
| Heligmosomoides polygyrus |
- |
ND |
Yes (GenePool) |
|
|
Cestode |
|
|
|
|
| Echinococcus multilocularis |
Ex |
22 |
Yes (sanger) |
82
|
| Echinococcus granulosus |
- |
23 |
Yes (sanger) |
82
|
| Hymenolepis microstoma |
- |
ND |
Yes (sanger) |
|
|
Arthropod |
|
|
|
|
| Pediculus humanus humanus |
Com |
57 |
Yes (VectorBase) |
83
|
| Acyrthosiphon pisum |
Com |
163 |
Yes |
84
|
| Ixodes scapularis | Ex | 34 | Yes (VectorBase) | 85 |
a miRNAs are identified experimentally (Ex) or computationally (Com) or both (Ex/Com);
b Argonaute or Dicer or both are present in species indicated. Argonaute homolog(s) in parasitic nematodes, cestodes and arthropods were searched using C. elegans Argonaute (ABA18180) in the databases shown in brackets;
c Drosha homolog(s) in protozoans were searched using C. elegans Drosha (NP_001122460) and there were no hits with an e value > 10−10. Due to the fact that protozoan Drosha homologs may be highly heterogeneous in an amino acid level, it can’t be ruled out the possibility of factor(s) that are functionally similar to Drosha;
d Our unpublished data;
ND, not determined; -, not applicable; ?, unclear.
The loss of this mechanism should be evaluated cautiously; however, in that these coding genes may have undergone great variations in domain(s) during evolution, leading to low similarity and difficulties in identification of putative protein homologs. For instance, Trypanosoma cruzi, the causative pathogen of Chagas disease, was thought to be unable to utilize siRNAs to alter gene expression;3 more recently, however, the presence of a ubiquitously expressed Argonaute/Piwi protein-coding gene has been described.27 Although no potential miRNAs have been found in a small RNA subpopulation,28 it is still hard to rule out the possibility of that this protozoan parasite has the ability to produce miRNAs because of limitations of the methods used.
Characteristics of protozoan miRNA-induced silencing network
Growing studies have demonstrated that miRNA-related pathways in protozoan parasites have features different to other metazoan organisms, possibly shedding light on the evolution of miRNA-induced silencing networks. The RISC components are more heterogeneous in sequence, compared with other animals. Protozoan Argonaute- and Piwi-like genes form an individual lineage, distinct from other Argonaute-like subfamily and Piwi-like subfamily of other multicellular organisms.27 Similarly, phylogenetic analysis of Dicer and RNA-dependent RNA polymerase homologs of Toxoplasma gondii that play a role in magnifying RNA silencing have shown common origins with Chlamydomonas reinhardtii, a unicellular alga that has a capacity of orchestrating gene expression using miRNAs,29 and the fungus Neurospora crassa, respectively.30 It can be partly explained by the concept that the ancestor of apicomplexan animals is proposed to be an endosymbiont of red alga.30 These evidences reinforce the idea that the appearance of the RNA-induced silencing machinery is a primitive event.
Extensive alignment analysis reveals that parasitic protozoans lack Drosha homologs (Table 1), indicating that most miRNA biogenesis in these organisms is independent of Drosha. Alternatively, other factor(s) that are significantly divergent in the sequence execute the catalytic activity to liberate pre-miRNAs from primary transcripts. Without the functions of Drosha and Exportin 5, G. lamblia bypasses the canonical Drosha-dependent way to generate miRNAs using snoRNAs as precursors.22
In the target recognition, animal miRNAs are partly complementary to sites of mRNA, majority of which are localized at 3′ UTRs, while plant miRNAs show near-full or full pairing. It has been shown that the extensive base pairing in animal miRNAs induces the turnover of bound small RNA by tailing or trimming.31 In plants, the modification of 2’-O-methyl at the 3′ last nucleotide of miRNAs and siRNAs by Hen1, a methyltransferase, allows these silencing molecules to be immune to uridylation-elicited degradation. The 2’-O-methyl group addition is also required to form the active siRNA-argonaute complex in flies.31 Of particular interest, it was predicted that Toxoplasma miRNAs bind to target regions via nearly perfect or perfect matching.30 In addition to this, more than half of the target sites (20/34) are distributed at 5′ UTRs or in coding regions. Nevertheless, miRNAs are not methylated in T. gondii and the lack of methylation raises the question of how these miRNA are protected. Although the resultant effects are controversial, adenylation at 3′ ends of miRNAs may be a feasible approach adopted to enhance miRNA stabilization.32 Consistent with this, the addition of untemplated adenine found in some Toxoplasma miRNAs render the possibility that miRNAs that are perfectly paired with targets function normally although 3′ adenylation may have an adverse effect on loading miRNA into RISC.33 This may be reflected by the components of the RNA-silencing machinery in T. gondii, of which Dicer does not possess typical DSRM and PAZ domains that associate with RNA binding, are remarkably distinct in structures.
snoRNAs: Original precursors of miRNAs?
snoRNAs are comprised of two types, box C/D and box H/ACA snoRNAs, and combine with other specific factors to be able to modify other types of RNA or function in splicing. In human, both classes of snoRNAs can be a source of small regulatory RNAs (sd-RNAs) that are functionally similar to miRNAs.34,35 Nonetheless, C/D sd-RNAs are distinguished from these out of H/ACA snoRNAs in terms of length and location.36 Conservation of most snoRNAs that can be processed to generate miRNA-like molecules is observed across evolutionarily unrelated species.34,36 sd-RNAs are extensively present in many organisms, including plants, virus and unicellular eukaryotes.36,37 The production of miRNA-like sd-RNAs is Drosha-independent but does require Dicer. Moreover, it has been demonstrated that the capacity of small RNA production is a common characteristic of a portion of snoRNAs.34,38 The extensive discovery that sd-RNAs modulate gene expression in a miRNA-like pattern adds the complexity of RNA-induced silencing mechanism reservoir.
Though the classification of sd-RNAs as miRNAs is unresolved, sd-RNAs and miRNAs are evolutionarily related. snoRNAs and precursors of miRNAs have similar genomic locations. A large number of human snoRNAs reside in introns of genes as well as transposable elements, from which many miRNAs are derived.39,40 Moreover, both types of small silencing molecules exhibit conservation throughout evolution.34,36,37 Furthermore, like miRNAs, upon loading into Argonaute-containing RISC, sd-RNAs induce gene silencing via Watson-Crick base pairing to target sites of mRNA they regulate. Not surprisingly, some registered human miRNAs (miRBase) are really derived from snoRNAs.38,41 Of note, some sd-RNAs are expressed in a specific cell-type manner, likely due to the post-transcriptional process of snoRNAs as do some miRNA precursors.38,42 However, the second structures of snoRNAs contain two hairpins, obviously different from classical miRNA precursors. Taken together suggests the evolutionary relationship between snoRNAs and miRNA pathways.
Functional similarity of these silencing RNAs is supported by the interactions between Argonaute proteins and some core components of snoRNPs, such as NOP56.30,43 Although the real functions are unclear, the presence of NOP56 in RISC may reveal that sd-RNAs are expressed in T. gondii. The fact that miRNAs are exclusively derived from snoRNAs in unicellular animal, G. lamblia, results in the hypothesis that snoRNAs are the original source of miRNAs.22 In agreement with this, a portion of miRNA precursors did have box H/ACA snoRNA properties and all five snoRNA-like miRNA precursors investigated showed capacity of binding with dyskerin,41 a pseudouridine synthase in snoRNA-containing nucleolar ribonucleoparticles (snoRNPs),44 suggesting the preservation of snoRNA functions in these miRNA precursors. snoRNAs being derived from transposable elements as miRNA precursors may be likely during evolution because the rapid generation of miRNAs through transposable elements could be a driving force.39 It is no doubt that the clarification of how snoRNAs are processed into sd-RNAs allows us to profoundly understand the relationship between sd-RNAs and miRNAs.
snoRNAs are also reported in Plasmodium falciparum,45,46 Trypanosoma spp,47,48 Trichomonas vaginalis49 and S. mansoni and S. japonicum50 but their ability to encode miRNA-like sd-RNAs remains unclear. It is noticed that P. falciparum and Saccharomyces cervisiae express snoRNAs but no miRNAs, in agreement with the idea that primitive snoRNAs may have produced certain types of miRNA-like RNA.41
Potential miRNA biomarkers for flatworm neoblasts
In the phylum Platyhelminthes, planarians are characterized by the ability to regenerate51 and neoblasts are thought to contribute to this regeneration. The neoblasts of E. multilocularis are able to generate mature metacestodes in vitro under specific conditions,52 suggesting the preservation of regenerative capacity in the neoblasts.
Fifteen miRNAs, including let-7a and 7b, have been implicated in the functions of the planaria neoblasts,51,53-55 although there is some disparity probably due to methodological differences. Aligned with planarian miRNAs, there are six potential miRNA neoblast markers commonly found in S. japonicum and E. multilocularis, in all of which the seed regions are intact (Table 2). Interestingly, miR-71 with other three miRNAs miR-2d, miR-752 and miR-13 are clustered in planarians but miR-752 is completely lost in S. japonicum and E. multilocularis. Moreover, two copies of the miR71 clusters are observed in S. japonicum56 but not in E. multilocularis (our unpublished data), suggesting duplication of miR71 cluster after speciation. What the loss of miR-752 influences the regenerative ability of neoblasts is unknown and remains to be identified experimentally.
Table 2. Potential neoblast-specific miRNAs.
| miRNAa | Homologb | Homologc | Alignment (5′-3”)d | Ref |
|---|---|---|---|---|
| sme-miR-36b |
miR-36 |
miR-36-3p |
TCACCGGGTAGACATTAATCATG ----------------CC-TGC? C---------------C--TCGC |
54
,
55
|
| sme-miR-2a |
miR-2a |
miR-2b-3p |
TATCACAGCCCCGCTTGGAACGCT A----------T---------C—? -----------T------G--A-A |
54
|
| sme-miR-2d |
miR-2b |
miR-2a-3p |
TCACAGCCAAATTTGATGTCC? ----------TA------AA-G ---------GTA------AA-G |
54
,
55
|
| sme-miR-13 |
miR-2c |
miR-2c-3p |
TATCACAGTCATGCTAAAGAGC? --------C-C----TGG—CACA --------C-G----T---G--? |
53
,
54
|
| sme-miR-71b |
miR-71 |
miR-71 |
TGAAAGACACAGGTAGTGGGAC --------GAT-------A--? --------GAT-------A--? |
53
-
55
|
| sme-miR-124a |
miR-124 | miR-124-3p | TAAGGCACGCGGTGAATGCTT -----------------A-CA ------------------TCA |
55 |
| sme-miR-124b |
aS. mediterranea miRNAs closely related to neoblast biology. Apart from those listed in this table, another eight sem-miR-7b, 7c, 752, 92, let-7a, let-7b, 2160 and 756 are considered to participate in neoblast functions;
b Cognate miRNAs in E. multilocularis;
c Cognate miRNAs in S. japonicum;
d Sequences of miRNAs of both species are aligned. Consensus nucleotides are masked by dash and gaps, and are filled by question markers. Variants in homologs of E. multilocularis (second sequence) and S. japonicum (third sequence) are shown.
Possible roles of parasite miRNAs
An exactly spatial and temporal control of gene expression is crucial for animals whose life cycles are fulfilled through several different developmental stages. microRNAs, such as the let-7 family and lin-4, were first revealed to be a master switch of development in C. elegans,11 targeting hb1-7 and lin-14 mRNAs, respectively, to ensure proper larval developmental transition. The transcription of let-7 family is controlled by the interaction between the nuclear hormone receptor DAF-12 and its ligands, dafachronic acids that are naturally produced in favorable environments.57 The striking discrepancy of expression of Dicer and Argonaute at different stages in S. mansoni is an indication that small RNA-induced silencing represents a mechanism of modulation of developmental transitions.58 The idea that S. japonicum let-7 may associate with transition from miracidium to sporocyst is indirectly supported by higher expression of let-7 in miracidium. Along with let-7, the miRNA bantam that is extraordinarily highly expressed in cercaria is supposed to be active in developmental processes through regulation of a cellular population size.59 However, a recent study preferably supports the notion that let-7 has other functions rather than controlling developmental timing.60 It is clear that these presumptions need to be further experimentally tested.
Some miRNAs are anticipated to take part in other processes of parasites, such as reproduction and tissue development (Table 3). Of interest is the very high expression of miR-71(a/b) in stages in trematodes56,59,60 and a cestode (our unpublished data) but not in a nematode.61 Apart from functional connections with neoblast biology,53-55 miR-71 is expected to have an additional role probably related to development. Further experiments will be worthwhile specifying the connections between its expression and functions.
Table 3. Potential functions of parasite miRNAs.
| miRNA | Species | Possible functionsa | Expressionb | Ref |
|---|---|---|---|---|
| let-7 |
S. japonicum |
Transition from miracidium to sporocyst? |
? |
59
,
60
|
| Bantamc |
Regulation of cell proliferation and apoptosis/sex development or reproduction |
Cercaria, female |
59
,
60
|
|
| miR-7 |
Tissue development |
Cercaria |
60
|
|
| miR-36 |
Developmental transition |
Cercaria |
||
| miR-71 |
Sexual development |
Male |
||
| miR-nov-70d |
Sexual development/reproduction |
Female |
||
| let-7 |
B. xylophilus |
Regulation of worm activity |
Cold-stressed worm |
61
|
| miR-1 |
|
|||
| miR-nov-10d miR-29 miR-40 miR-72 |
|
Response to cold stress |
Downregulated in cold-stressed worm |
|
| miR-4 miR-49 miR-60 |
T. gondii |
Related to virulence? |
Lethal strain |
30
|
| miR-40 miR-56 |
|
Hypo-virulent strain |
||
| /e |
T. brucei |
Antigenic variation | ND |
69
|
| miR-2 | G. lamblia | 22 |
a None of miRNA functions was experimental validated;
b miRNA(s) are highly or exclusively expressed in tissues or strains listed, if not clearly stated;
c miR-nov-110 are highly homogenous to bantam and therefore the latter is listed here;
d Novel miRNAs;
e Many miRNAs are predicted to target VSG mRNA and therefore not listed here;
?, controversial or not clear; ND, not determined.
miRNAs are modulators in immune systems, affecting differentiation, development, homeostasis and functions of immune cells.62,63 It is well-studied that miRNAs play important roles in viral infection and immunology.25,64-67 The temporal regulation of variant surface proteins (VSGs), one of which covers the entire surface of a parasite at any time, is essential to survive host immune attacks in free living protozoans such as Trypanosomes and G. lamblia.68,69 Various VSGs expressed on the surface were observed in individual G. lamblia with silenced Dicer and RNA-dependent RNA polymerase, indicating the key role of silencing machinery in VSG expression regulation.14 miRNAs may be partly responsible for this antigenic variation by targeting 3′ VSG-coding gene UTRs that contain highly conserved fragments.22,68 The mechanism of how miRNA(s) modulate several hundreds of VSG genes to ensure that only one is retained is unknown.
Unfortunately, little has been done to experimentally investigate the functions of parasite miRNAs but a wealth of the genome, transcriptome and proteome data will accelerate to characterize their roles in parasites and parasitic infection.
Host miRNAs in response to parasite infection
The alterations of host miRNA expression reflect the roles of host-derived miRNAs in parasite infection. Cryptosporidium parvum, a protozoan parasite which lacks the RNA-induced silencing mechanism (Table 1), can induce a decrease of let-7 expression in infected cells, leading to upregulation of Toll-like receptor 4 (TLR4) expression, which contains a let-7 binding site in the 3′ UTR.70 In vitro suppression of let-7 gives rise to a significantly lower parasite burden, suggesting that it contributes to immune responses against the infection. The parasite-induced downregulation of let-7 expression is executed by a repressor binding to the Let-7 promoter, which is comprised of transcription factors NFκB p50 and CCAAT/enhancer-binding protein β.71 By contrast, a number of miRNAs was upregulated in a NFκB p65-dependent or -independent manner in response to C. parvum invasion (Fig. 2).72 Functional silencing of selected miRNAs resulted in an increase of C. parvum burden in vitro without effects on cell attachment and invasion, suggesting a role of host miRNAs in epithelial defense against parasite infection.
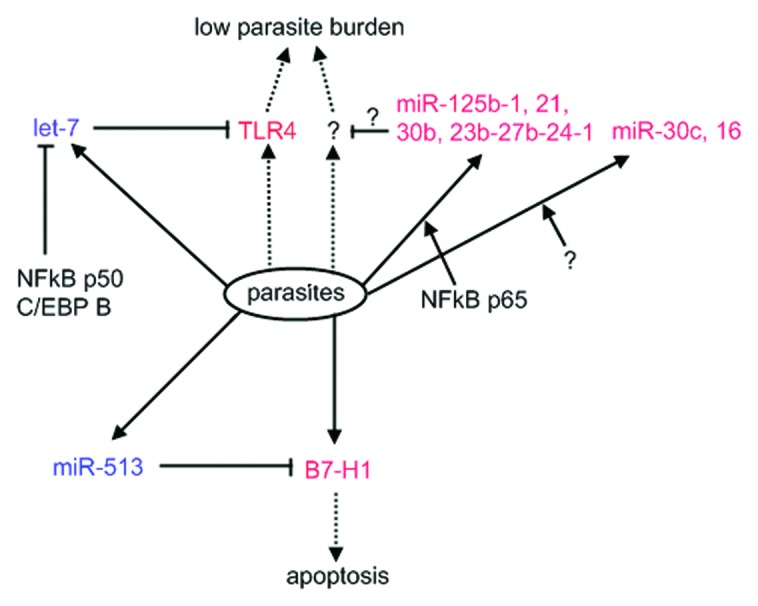
Figure 2. Host miRNAs response to Cryptosporidium infection. During the period of infection, Cryptosporidum can elicit alterations of host-origin miRNA expression. Some miRNAs are repressed (in blue), whereas others upregulated (in red) in NFκB p65-dependent or -independent (?) manner. Protein levels of some key players involved in infection and immunology will be subsequently increased (in red), facilitating parasite clearance or infection.
C. parvum can repress miR-513 transcription and simultaneously activate the expression of B7-H1, residing a binding site for miR-513. This leads to an increase of B7-H1expressed on the cell surface that in vitro induces the apoptosis of activated T cells.73 This is of particular interest because it illustrates the feasibility that parasites take advantage of host miRNA pathways as a method of defense against the host. The in vivo events following increased B7-H1 will be of interest to be further investigated. Parasite-induced host miRNA expression alterations were also observed in T. gondii, though the relationship between elevating miRNA level and the infection is not clear.74 In contrast to the unaltered level in response to Neospora caninum infection, the expression of mature miR-17 family was upregulated in the cells infected by T. gondii, illustrating the elevated abundance specific to T. gondii infection. Collectively, host miRNA-induced silencing networks take part in interplay between host and pathogen via modulation of key players that are active in a course of infection and immunology.
Concluding Remarks
Although the vast majority of miRNAs are enigmatic in function, it is clear that the miRNA-induced silencing machinery directly or indirectly affects many processes of organisms and their responses to environments. Parasite and host miRNA profiles can be served as a probe to investigate underlying mechanisms and, thus, deeply understand pathogen-host interplay. Likewise, miRNA silencing network will be an alternative to help us understand global deleterious drug resistance in parasitic nematodes.75 The intervention of miRNA pathways to control diseases is in infancy, but successful attempts to control viral infection in chimpanzees or tumorigenesis in animal models76,77 have shed light on the potential of miRNA pathways as the therapeutical targets.
Acknowledgments
This study was financially supported by National Natural Science Foundation of China (31201900), State Public-interest Institution Basal Research Fund, Chinese Academy of Agricultural Sciences (0032012037), the Open Fund of the Key Laboratory of the New Animal Drug Project of Gansu Province and the Key Laboratory of Veterinary Pharmaceutical Development of the Ministry of Agriculture (1610322011011) and by the Science Fund for Creative Research Groups of Gansu Province (Grant No.1210RJIA006). The part of analyses in this study was conducted in the University of Nottingham under the financial support of Overseas Research Students Awards (Y.Z.), UK and the University of Nottingham scholarship (Y.Z.). The authors would like to give thanks to reviewers for their constructive suggestions. We also thank institutes or sponsors for free use of unpublished genomic data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23716
References
- 1.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–7. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–8. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 3.Militello KT, Refour P, Comeaux CA, Duraisingh MT. Antisense RNA and RNAi in protozoan parasites: working hard or hardly working? Mol Biochem Parasitol. 2008;157:117–26. doi: 10.1016/j.molbiopara.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 5.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–82. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–8. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–56. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Winter F, Edaye S, Hüttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–62. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prucca CG, Slavin I, Quiroga R, Elías EV, Rivero FD, Saura A, et al. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456:750–4. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- 15.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–64. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–42. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherer LJ, Frank R, Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–8. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–9. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 24.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–22. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–6. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaubert-Possamai S, Rispe C, Tanguy S, Gordon K, Walsh T, Edwards O, et al. Expansion of the miRNA pathway in the hemipteran insect Acyrthosiphon pisum. Mol Biol Evol. 2010;27:979–87. doi: 10.1093/molbev/msp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia Silva MR, Tosar JP, Frugier M, Pantano S, Bonilla B, Esteban L, et al. Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene. 2010;466:26–35. doi: 10.1016/j.gene.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, et al. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol. 2010;171:64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–9. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 30.Braun L, Cannella D, Ortet P, Barakat M, Sautel CF, Kieffer S, et al. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog. 2010;6:e1000920. doi: 10.1371/journal.ppat.1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–9. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu S, Sun YH, Chiang VL. Adenylation of plant miRNAs. Nucleic Acids Res. 2009;37:1878–85. doi: 10.1093/nar/gkp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 35.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–86. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–40. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, Zavolan M, et al. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009;5:e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–86. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–6. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Piriyapongsa J, Mariño-Ramírez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–37. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–7. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–60. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoang C, Ferré-D’Amaré AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–39. doi: 10.1016/S0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 45.Raabe CA, Sanchez CP, Randau G, Robeck T, Skryabin BV, Chinni SV, et al. A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res. 2010;38:608–17. doi: 10.1093/nar/gkp895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakrabarti K, Pearson M, Grate L, Sterne-Weiler T, Deans J, Donohue JP, et al. Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA. 2007;13:1923–39. doi: 10.1261/rna.751807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uliel S, Liang XH, Unger R, Michaeli S. Small nucleolar RNAs that guide modification in trypanosomatids: repertoire, targets, genome organisation, and unique functions. Int J Parasitol. 2004;34:445–54. doi: 10.1016/j.ijpara.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Morales L, Romero I, Diez H, Del Portillo P, Montilla M, Nicholls S, et al. Characterization of a candidate Trypanosoma rangeli small nucleolar RNA gene and its application in a PCR-based parasite detection. Exp Parasitol. 2002;102:72–80. doi: 10.1016/S0014-4894(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 49.Chen XS, Collins LJ, Biggs PJ, Penny D. High throughput genome-wide survey of small RNAs from the parasitic protists Giardia intestinalis and Trichomonas vaginalis. Genome Biol Evol. 2009;1:165–75. doi: 10.1093/gbe/evp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Copeland CS, Marz M, Rose D, Hertel J, Brindley PJ, Santana CB, et al. Homology-based annotation of non-coding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum. BMC Genomics. 2009;10:464. doi: 10.1186/1471-2164-10-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the Planarian Schmidtea mediterranea: a model system for stem cell biology. RNA. 2006;12:1640–9. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spiliotis M, Lechner S, Tappe D, Scheller C, Krohne G, Brehm K. Transient transfection of Echinococcus multilocularis primary cells and complete in vitro regeneration of metacestode vesicles. Int J Parasitol. 2008;38:1025–39. doi: 10.1016/j.ijpara.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Lu YC, Smielewska M, Palakodeti D, Lovci MT, Aigner S, Yeo GW, et al. Deep sequencing identifies new and regulated microRNAs in Schmidtea mediterranea. RNA. 2009;15:1483–91. doi: 10.1261/rna.1702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, et al. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci USA. 2009;106:11546–51. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.González-Estévez C, Arseni V, Thambyrajah RS, Felix DA, Aboobaker AA. Diverse miRNA spatial expression patterns suggest important roles in homeostasis and regeneration in planarians. Int J Dev Biol. 2009;53:493–505. doi: 10.1387/ijdb.082825cg. [DOI] [PubMed] [Google Scholar]
- 56.Huang J, Hao P, Chen H, Hu W, Yan Q, Liu F, et al. Genome-wide identification of Schistosoma japonicum microRNAs using a deep-sequencing approach. PLoS One. 2009;4:e8206. doi: 10.1371/journal.pone.0008206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–8. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomes MS, Cabral FJ, Jannotti-Passos LK, Carvalho O, Rodrigues V, Baba EH, et al. Preliminary analysis of miRNA pathway in Schistosoma mansoni. Parasitol Int. 2009;58:61–8. doi: 10.1016/j.parint.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 59.Xue X, Sun J, Zhang Q, Wang Z, Huang Y, Pan W. Identification and characterization of novel microRNAs from Schistosoma japonicum. PLoS One. 2008;3:e4034. doi: 10.1371/journal.pone.0004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao L, Cai P, Jiang N, Wang H, Chen Q. Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum. BMC Genomics. 2010;11:55. doi: 10.1186/1471-2164-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang QX, Cheng XY, Mao ZC, Wang YS, Zhao LL, Yan X, et al. MicroRNA discovery and analysis of pinewood nematode Bursaphelenchus xylophilus by deep sequencing. PLoS One. 2010;5:e13271. doi: 10.1371/journal.pone.0013271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 63.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 64.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–82. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 65.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 66.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–85. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035–48. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nash TE. Surface antigenic variation in Giardia lamblia. Mol Microbiol. 2002;45:585–90. doi: 10.1046/j.1365-2958.2002.03029.x. [DOI] [PubMed] [Google Scholar]
- 69.Mallick B, Ghosh Z, Chakrabarti J. MicroRNA switches in Trypanosoma brucei. Biochem Biophys Res Commun. 2008;372:459–63. doi: 10.1016/j.bbrc.2008.05.084. [DOI] [PubMed] [Google Scholar]
- 70.Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–38. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Hara SP, Splinter PL, Gajdos GB, Trussoni CE, Fernandez-Zapico ME, Chen XM, et al. NFkappaB p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated transcriptional repression of microRNA let-7i following microbial infection. J Biol Chem. 2010;285:216–25. doi: 10.1074/jbc.M109.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gong AY, Zhou R, Hu G, Liu J, Sosnowska D, Drescher KM, et al. Cryptosporidium parvum induces B7-H1 expression in cholangiocytes by down-regulating microRNA-513. J Infect Dis. 2010;201:160–9. doi: 10.1086/648589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeiner GM, Norman KL, Thomson JM, Hammond SM, Boothroyd JC. Toxoplasma gondii infection specifically increases the levels of key host microRNAs. PLoS One. 2010;5:e8742. doi: 10.1371/journal.pone.0008742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Devaney E, Winter AD, Britton C. microRNAs: a role in drug resistance in parasitic nematodes? Trends Parasitol. 2010;26:428–33. doi: 10.1016/j.pt.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De S, Pal D, Ghosh SK. Entamoeba histolytica: computational identification of putative microRNA candidates. Exp Parasitol. 2006;113:239–43. doi: 10.1016/j.exppara.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 79.Xu MJ, Liu Q, Nisbet AJ, Cai XQ, Yan C, Lin RQ, et al. Identification and characterization of microRNAs in Clonorchis sinensis of human health significance. BMC Genomics. 2010;11:521. doi: 10.1186/1471-2164-11-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, Xue X, Sun J, Luo R, Xu X, Jiang Y, et al. An “in-depth” description of the small non-coding RNA population of Schistosoma japonicum schistosomulum. PLoS Negl Trop Dis. 2010;4:e596. doi: 10.1371/journal.pntd.0000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Poole CB, Davis PJ, Jin J, McReynolds LA. Cloning and bioinformatic identification of small RNAs in the filarial nematode, Brugia malayi. Mol Biochem Parasitol. 2010;169:87–94. doi: 10.1016/j.molbiopara.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 82.Cucher M, Prada L, Mourglia-Ettlin G, Dematteis S, Camicia F, Asurmendi S, et al. Identification of Echinococcus granulosus microRNAs and their expression in different life cycle stages and parasite genotypes. Int J Parasitol. 2011;41:439–48. doi: 10.1016/j.ijpara.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 83.Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci USA. 2010;107:12168–73. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]