Abstract
Paraspeckles are unique subnuclear structures that are built around a specific long non-coding RNA (lncRNA), NEAT1, which is comprised of two isoforms (NEAT1_1 and NEAT1_2) that are produced by alternative 3′-end processing. NEAT1 lncRNAs are unusual RNA polymerase II transcripts that lack introns. The non-polyadenylated 3′-end of NEAT1_2 is non-canonically processed by RNase P. NEAT1_2 is an essential component for paraspeckle formation. Paraspeckles form during the NEAT1_2 lncRNA biogenesis process, which encompasses transcription from its own chromosome locus through lncRNA processing and accumulation. Recent RNAi analyses of 40 paraspeckle proteins (PSPs) identified four PSPs that are required for paraspeckle formation by mediating NEAT1 processing and accumulation. In particular, HNRNPK was shown to arrest CFIm-dependent NEAT1_1 polyadenylation, leading to NEAT1_2 synthesis. The other three PSPs were required for paraspeckle formation, but did not affect NEAT1_2 expression. This observation suggests that NEAT1_2 accumulation is necessary but not sufficient for paraspeckle formation. An additional step, presumably the bundling of NEAT1 ribonucleoprotein sub-complexes, may be required for construction of the intact paraspeckle structure. NEAT1 expression is likely regulated at transcriptional and post-transcriptional steps under certain stress conditions, suggesting roles for paraspeckles in the lncRNA-mediated regulation of gene expression, such as the nucleocytoplasmic transport of mRNA in response to certain stimuli.
Keywords: 3′-end processing, RNA-binding protein, RNA-protein interaction, long non-coding RNA, nuclear bodies
Introduction
Mammalian cells contain a highly organized nucleus composed of distinct subnuclear structures called nuclear bodies. These membraneless organelles contain specific proteins or RNAs characteristic of particular nuclear processes. Nuclear bodies are the sites of the regulation of the expression of specific genes and the biogenesis of macromolecular ribonucleoprotein (RNP) machineries, such as ribosomes and spliceosomes.1 Many nuclear bodies have been characterized, and numerous relatively abundant long non-coding RNAs (lncRNAs) were recently found to localize to specific nuclear bodies.1,2
Paraspeckles are nuclear bodies that usually occur in cultured cell lines as 2~20 foci in close proximity to nuclear speckles (Fig. 1B).3 Paraspeckles were initially defined as foci enriched in characteristic RNA-binding proteins, including three mammalian Drosophila melanogaster behavior and human splicing (DBHS) proteins, Paraspeckle component 1 (PSPC1), a non-POU domain containing, octamer-binding (NONO) (p54nrb) protein and a splicing factor proline/glutamine-rich (SFPQ) (PSF) protein.4 Paraspeckle diameter was estimated to be ~360 nm.5 According to their sizes, shapes and electron density, the paraspeckle was confirmed to be equivalent to the interchromatin granule-associated zone (IGAZ) that had been observed by electron microscopy.4-6
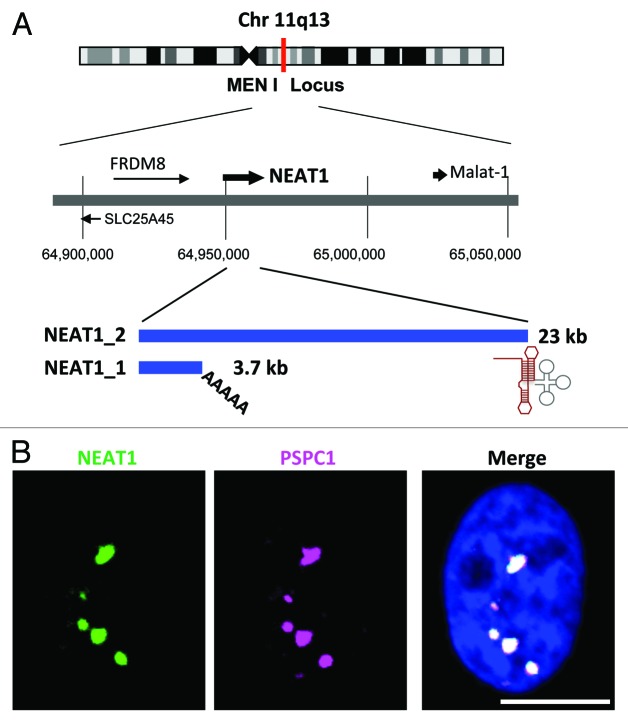
Figure 1. (A) The NEAT1 long non-coding RNA (lncRNA) is an RNA polymerase II transcript with unusual features. Schematics of the NEAT1 genomic locus and NEAT1 transcripts are shown. Chromosome 11 is shown with the chromosome bands seen on Giemsa-stained chromosomes. The position of the chromosome locus can be estimated according to the numbers below. FRDM8 and SLC25A45 are the protein-coding genes located adjacent to NEAT1. The arrows indicate the directions of transcription. NEAT1_1 and NEAT1_2 possess distinct 3′-terminal structures. The triple-helix structure (red line) stabilizes NEAT1_2. The tRNA-like structure (gray line) is recognized by RNase P to create the 3′-end of NEAT1_2. (B) NEAT1 lncRNA (green) and PSPC1 (magenta) are localized to nuclear paraspeckles, which appear as bright nuclear foci in the right photograph. Nuclear DNA was stained with DAPI (blue). Scale bar is 10 μm.
Prasanth et al. reported that CAT2 transcribed nuclear-RNA (CTN-RNA), an isoform of mouse cationic amino acid transporter 2 (mCAT2) mRNA, is retained specifically in the paraspeckle.7 Intriguingly, the long 3′ untranslated region (UTR) of CTN-RNA is cleaved by an unidentified endoribonuclease upon exposure to certain stresses. This process leads to the export of processed mCAT2 mRNA as a 5′ part of CTN-RNA for cytoplasmic translation.7 The CTN-RNA 3′-UTR contains a long inverted-repeat sequence that is capable of forming intramolecular double-stranded RNAs that are A-to-I edited. The hyperedited CTN-RNAs are enriched in paraspeckles. Thus, paraspeckles are thought to suppress the protein synthesis of hyperedited transcripts through nuclear retention.7
It was determined that paraspeckles are sensitive to RNase treatment after cell permeabilization.7,8 They suggested that an as-yet-unidentified RNA molecule was required for their structural maintenance. The discovery of the specific paraspeckle localization of nuclear paraspeckle assembly transcript 1 (NEAT1) lncRNA opened a new window in paraspeckle research.9-12 NEAT1 lncRNAs are transcribed from a genetic locus, called familial tumor syndrome multiple endocrine neoplasia (MEN) type I, on human chromosome 11 (Fig. 1A).13 NEAT1 is comprised of two isoform transcripts, 3.7-kb NEAT1_1 (MENε) and 23-kb NEAT1_2 (MENβ). Both RNAs are produced from the same promoter as explained below (Fig. 1A). Importantly, the knockdown of both NEAT1 lncRNA isoforms leads to the disintegration of paraspeckles, suggesting that these lncRNAs serve as a core structural component.9-12
Paraspeckles can be thought of as tremendously large RNP complexes, because their structural integrity is maintained by the interactions of NEAT1_2 lncRNA with at least two DBHS proteins.11 This article summarizes how the paraspeckle structure is organized, focusing on the essential components and steps required for paraspeckle formation.
The unique features of NEAT1 lncRNA required for paraspeckle formation
The NEAT1 lncRNA biogenesis process has some unusual features. NEAT1_1/2 are exceptionally abundant lncRNAs and are as abundant as most moderately abundant mRNAs in mammalian cultured cells.11,14 They are transcribed by RNA polymerase II (RNAPII) as protein-coding pre-mRNAs; however, the NEAT1_1/2 biogenesis process has distinct features compared with the mRNA biogenesis process. First, NEAT1_1/2 lacks introns. Therefore, both NEAT1 isoforms are transcribed as single exon transcripts, even though the size of NEAT1_2 reaches ~23 kb (Fig. 1A). This feature is unusual for pre-mRNAs; other RNAPII transcripts in humans have an average exon size of 145 bp.15 Second, NEAT1 is processed at its 3′-end to produce the canonically polyadenylated NEAT1_1 and the non-canonically processed NEAT1_2. RNase P recognizes the tRNA-like structure and cleaves it to form the non-polyadenylated 3′-end of NEAT1_2.12 It was recently reported that the non-polyadenylated 3′-end of NEAT1_2 forms a characteristic triple-helix structure that is critical for its stabilization (Fig. 1A),16,17 although the significance of the non-canonical 3′-end processing remains uncertain. Third, both NEAT1 isoforms are never transported to the cytoplasm; instead, it is retained in the nucleus as a core molecule of the paraspeckle. During mRNA biogenesis, the mRNA export factors are recruited via the 5′ cap structure.18,19 The mechanism to arrest export factor recruitment may act during NEAT1 lncRNA biogenesis. The association of nuclear RNA export factor 1 (NXF1) with the X (inactive)-specific transcript (XIST) lncRNA, which is a nuclear-retained lncRNA, is weaker than its association with a protein-coding mRNA,20 suggesting a common mechanism for various nuclear-retained lncRNAs. Because the NEAT1_1/2 5′ cap structure remains uncharacterized, we cannot rule out the possibility that NEAT1 may possess an unusual cap structure at its 5′ terminus. Alternatively, NEAT1_1/2 may possess a nuclear retention element within its body, as has been reported for metastasis-associated lung adenocarcinoma transcript 1 (Malat-1) lncRNA.21 Together, these unusual features in NEAT1 biogenesis cause it to act as “architectural RNA” for paraspeckle formation in the nucleus.
The significance of the two NEAT1 isoforms for paraspeckle formation has only just begun to be understood. Elimination of either of the two DBHS proteins, NONO or SFPQ, markedly reduces the NEAT1_2 level and leads to paraspeckle disintegration11 but does not affect the NEAT1_1 level. These results strongly suggest that NEAT1_2 is the essential paraspeckle RNA component. This argument is supported by observations in mouse tissues, in which the expression of NEAT1_2 but not NEAT1_1 was highly correlated with paraspeckle appearance.22 Other studies showed that overexpressed NEAT1_1 is capable of increasing the number of paraspeckles,10 and that the artificial tethering of MS2-tagged NEAT1_1 RNA to the LacO array using the co-expressed GFP-NLS-LacI-MS2 coat protein triggers paraspeckle formation at the corresponding site.23 These findings suggest that NEAT1_1 may be the functional isoform for paraspeckle formation. Recent rescue experiments using plasmids expressing either NEAT1_1 or both NEAT1_1 and NEAT1_2 in embryonic fibroblasts from NEAT1_1/2-knockout mice confirmed that NEAT1_2 is an authentic RNA component that is capable of de novo paraspeckle formation.24
The organization of NEAT1_1/2 lncRNAs within paraspeckles has been precisely delineated by electron microscopy.5 These analyses revealed that the 5′-region of NEAT1_1/2, which is common to both isoforms, and the 3′-end of NEAT1_2 are located at the paraspeckle periphery, whereas the NEAT1_2 middle region is located in the paraspeckle interior (Fig. 2). These observations support the importance of NEAT1_2 for paraspeckle formation. Locally concentrated NEAT1_1 synthesized from the transfected plasmid likely captures the preexisting paraspeckles or their sub-particles containing NEAT1_2, which results in the accelerated formation of the paraspeckles containing exogenous NEAT1_1. The overexpressed NEAT1_1 is efficiently incorporated into the peripheral area of paraspeckles, whose core is constructed around endogenous NEAT1_2.
Figure 2. Paraspeckle formation proceeds during NEAT1 lncRNA biogenesis. NEAT1_1 and NEAT1_2 are shown by blue and red lines, respectively. The steps in paraspeckle formation in which essential PSPs (categories 1A and 1B) participate are shown. SFPQ (blue oval) and NONO (purple oval) preferentially bind to NEAT1_2. HNRNPK (black oval) binds to the pyrimidine stretch (open square) and arrests CFIm (pink and orange ovals)-dependent NEAT1_1 polyadenylation (CFIm-binding site cluster is shown by dark blue square).
The significant roles of paraspeckle proteins
Researchers initially classified four RNA-binding proteins, including three DBHS proteins (NONO, PSPC1 and SFPQ) and RNA-binding motif protein 14 (RBM14), as “classical” PSP components.3,4,7 Cleavage and polyadenylation-specific factor subunit 6 (CPSF6) was later added to this list.25 Recently, 35 additional PSPs were identified by colocalization screening with a fluorescent protein-tagged human full-length cDNA library.24 Among the 40 compiled PSPs, 30 possess putative RNA-binding domains, such as RRM, KH, RGG box and Zinc finger motifs.24 Most of the newly identified PSPs possess various putative RNA-binding domains, and these PSPs may interact with NEAT1_1 and/or 1_2 lncRNAs as the classical PSPs11 or unidentified RNAs localized in paraspeckles. Thus, it may be that paraspeckles form by gathering the PSPs that NEAT1_1/2 lncRNAs capture. PSPs are not exclusively localized in paraspeckles, but are broadly distributed in the nucleoplasm and enriched in paraspeckles.24 A study showed that PSPC1 dynamically moves in and out of paraspeckles,3 suggesting the dynamic movement of other PSPs around the paraspeckles. These movements may be sustained by RNA-protein interactions with NEAT1_1/2 lncRNAs and interactions between the PSPs.11,26
The results of RNAi knockdown and RNA immunoprecipitation of classical PSPs revealed that NONO and SFPQ but not PSPC1 are required for paraspeckle formation, via their interaction with NEAT1_2 lncRNA.11 This finding indicates distinct roles for each PSP in paraspeckle formation. Further extensive RNAi analyses allowed the categorization of the identified PSPs into three categories (category 1~3) based on the paraspeckle phenotype, and category 1 and category 3 were further grouped into subcategories (1A and 1B, 3A and 3B) based on their influence on the accumulation of NEAT1 isoforms.24 We found seven “category 1” PSPs that are essential for paraspeckle formation (Table 1 and Fig. 2).24 Ten other PSPs whose elimination leads to moderately decreased paraspeckle numbers and sizes were characterized as category 2 proteins. The remaining 16 “category 3” PSPs do not affect paraspeckle structure.24 Thus, each PSP is classified according to its role in building the paraspeckle structure.
Table 1. The paraspeckle proteins essential to paraspeckle formation.
| PSP | The steps involved in PS formation | Category |
|---|---|---|
| HNRNPK |
Alternative 3′-end processing for NEAT1_2 |
1A |
| NONO |
Stabilization of NEAT1_2 |
1A |
| RBM14 |
Stabilization of NEAT1_2 |
1A |
| SFPQ |
Stabilization of NEAT1_2 |
1A |
| DAZAP1 |
Assembly of NEAT1 RNP sub-complexes (?) |
1B |
| FUS |
Assembly of NEAT1 RNP sub-complexes (?) |
1B |
| HNRNPH3 | Assembly of NEAT1 RNP sub-complexes (?) | 1B |
Reference: Naganuma et al., EMBO J 2012, Sasaki et al., PNAS 2009.
Paraspeckle formation requires the ongoing transcription of NEAT1_1/2 lncRNAs (Fig. 2).9,27 The paraspeckles are usually assembled co-transcriptionally at the NEAT1 transcription site, and the formed paraspeckles rarely left from this site.27 NEAT1_1 and NEAT1_2 are synthesized by alternative 3′-end processing (Fig. 2), which likely occurs co-transcriptionally. Given the essential role of the NEAT1_2 isoform, the alternative 3′-end processing step that synthesizes this isoform is the prerequisite molecular event for paraspeckle formation.
The alternative 3′-end processing of NEAT1 is unique, because two 3′ ends are processed by distinct 3′-end processing mechanisms: canonical polyadenylation for NEAT1_1 and RNase P-mediated cleavage for NEAT1_2 (Fig. 2).12 The aforementioned extensive RNAi experiments identified PSPs that mediate the alternative 3′-processing of NEAT1.24 Two PSPs, CPSF6 and Nudix (nucleoside diphosphate linked moiety X)-type motif 21 (NUDT21), form a functional heterodimer [Cleavage factor Im (CFIm) complex] to facilitate the canonical 3′-end processing of NEAT1_1 (Fig. 2). By contrast, the category 1A protein heterogeneous nuclear ribonucleoprotein K (HNRNPK) represses the 3′-end processing of NEAT1_1 (Fig. 2), which may enhance the 3′-end processing of NEAT1_2. The molecular mechanism of the HNRNPK-mediated 3′-end processing of NEAT1_1 was dissected in a recapitulated in vitro system.24 HNRNPK binds to the short pyrimidine stretch located between the canonical polyadenylation signal for NEAT1_1 and the upstream CFIm binding cluster (Fig. 2), where HNRNPK displaces NUDT21 from the functional CFIm complex, resulting in the arrest of the 3′-end processing of the NEAT1_1 isoform.
The NEAT1_1/2 isoform ratio dynamically changes in a cell type-specific manner. In adult mouse tissues, NEAT1_2 is expressed in limited cell types, such as the epithelial layers of digestive tissues. By contrast, NEAT1_1 is expressed in a broader range of cell types.22 These observations suggest that the 3′-end processing site for NEAT1_1 is selected in numerous cell types, whereas the 3′-end processing site for NEAT1_2 is only selected in specific cell types. Because all of the identified PSPs involved in the alternative 3′-end processing of NEAT1 (HNRNPK, CPSF6 and NUDT21) are broadly expressed in multiple tissues. Although we cannot rule out the possibility that the protein levels of the above factors vary in different cell types, the cell type-specific regulation of NEAT1_1/2 3′-end processing is thought to be controlled by additional factors that modulate the roles of the factors above. Alternatively, the 3′-end processing could be regulated by cell type-specific post-translational modifications of HNRNPK or CFIm components. Indeed, HNRNPK is the target of multiple protein kinases, methylases and ubiquitin/SUMO ligases that act under certain physiological conditions.28-31
Among the seven category 1 proteins, three proteins (category 1A proteins, including NONO, RBM14 and SFPQ) are required for the accumulation of NEAT1_2 but not NEAT1_1 (Table 1), suggesting they are unlikely to act on transcription from the promoter common to both NEAT1 isoforms but may affect the stability of the NEAT1_2 isoform (Fig. 1A). NONO and SFPQ interact with NEAT1_2. This interaction strongly suggests that they associate with NEAT1_2 to maintain its stability (Fig. 2). NEAT1_1/2 lncRNAs were recently shown to have a relatively short half-life (< 4 h),32,33 suggesting that the association of NONO and SFPQ may control the RNA degradation rate of NEAT1_2 lncRNA. An intriguing possibility is that the rapid turnover of NEAT1_1/2 lncRNA affects the dynamics of the PSPs in paraspeckles. The precise mapping of binding sites of these category 1A proteins onto NEAT1_2 will provide important information regarding the lncRNA sequences responsible for its architectural function.
Construction of the higher-order paraspeckle structure from NEAT1 RNP sub-complexes
The aforementioned extensive RNAi experiments revealed that NEAT1_2 accumulation was insufficient for paraspeckle formation. The knockdown of category 1B proteins leads to the disintegration of paraspeckles, without affecting the accumulation of NEAT1_2 (Table 1B). Accordingly, the NEAT1_2 accumulation that is associated with category 1A proteins appears not to be sufficient for intact paraspeckle formation. Category 1B proteins may help to build the paraspeckle structure, by bundling multiple copies of the NEAT1_2 RNP sub-complexes (Table 1 and Fig. 2). All three of the category 1B proteins possess canonical RRMs. Thus, category 1B proteins may associate with the NEAT1_1 and/or 1_2 lncRNAs without affecting their stability. Additionally, the protein-protein interactions between NEAT1-bound PSPs seem to be critical for the construction of the higher-order paraspeckle structure from NEAT1_1/2 sub-complexes, which are formed to stabilize each NEAT1 isoform.
Low-complexity regions in various RNA-binding proteins, including the category 1B proteins DAZ-associated protein 1 (DAZAP1) and fused in sarcoma (FUS), are thought to be critical for the formation of membraneless subcellular structures and mediate the subsequent recruitment of additional proteins and RNAs to these structures.34-36 Indeed, category 1B proteins reportedly interact with category 1A proteins,37 and Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) analysis showed that one of category 1B proteins (FUS) binds to both NEAT1_1 and NEAT1_2.38 Further analyses to investigate roles of category 1B proteins would be extremely important for understanding how the higher-order structure of the paraspeckle is organized.
Optical fluorescent microscopy results showed that NEAT1_1 itself does not contribute to the formation of paraspeckle foci. Extensive RNAi analyses revealed that three PSPs, CPSF6, NUDT21 and Ubiquitin-associated protein 2-like (UBAP2L), which belong to category 3A, are required for NEAT1_1 accumulation.24 These data are consistent with the electron microscopic observation that NEAT1_1 is located in the peripheral parts of paraspeckles.5 However, it remains unknown how the NEAT1_1 sub-complexes join to form the core complex around NEAT1_2.
What is the purpose of the paraspeckle formation?
The paraspeckle is a tremendously large RNP particle whose formation is tightly coupled with NEAT1 lncRNA biogenesis and involves the subsequent bundling of the NEAT1_1/2 RNP complexes. In contrast to the research progress in paraspeckle formation, functional explorations of the paraspeckle have been limited. Studies in NEAT1_1/2-knockout mice revealed that paraspeckles were not essential for viability and development in a mouse model under normal conditions.22 Paraspeckles likely play roles under certain stress conditions, since NEAT1_1 levels are increased by various internal and external stimuli, including Japanese encephalitis virus, Rabies virus39 and differentiation from embryonic stem cells to trophoblasts or from myoblasts to myotubules.9,12 The analysis of the RNA binding of TAR DNA-binding protein (TARDBP) (TDP-43), which is involved in the regulation of pre-mRNA splicing in brains from subjects with frontotemporal lobar degeneration, revealed that one of the largest increases in binding was to NEAT1_1/2 lncRNAs.40 The cellular levels of NEAT1_1/2 were downregulated in a particular set of cells in Malat1-knockout mice, suggesting that Malat1 transcribed from the adjacent region (Fig. 1A) affects the expression of NEAT1_1/2 lncRNAs in specific cells.41 Paraspeckles presumably regulate the nuclear retention of specific mRNAs that possess a long inverted repeat (IR) in their 3′-UTRs, since nuclear-retained IR-containing mRNAs were partially transferred to the cytoplasm when NEAT1_1/2 were knocked down.7,9,42 These facts raise the possibility that the induction of NEAT1_1/2 lncRNAs that leads to paraspeckle formation also controls the nucleocytoplasmic transport of specific mRNAs under certain physiological conditions.
Paraspeckles may be involved in other regulatory events in gene expression, because PSPs regulate the transcription and pre-mRNA splicing events that are associated with important physiological events and diseases.43,44 NEAT1 could control these events by modulating the functions of PSPs upon exposure to specific stresses. Currently, the paraspeckle is considered to be a unique lncRNA-directed nuclear body that is potentially involved in stress response. A precise understanding of NEAT1-PSP interactions and their dynamics could elucidate the molecular mechanisms underlying the dynamic nature of this enigmatic RNP nuclear body.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23547
References
- 1.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caudron-Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev. 2012;22:179–87. doi: 10.1016/j.gde.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, et al. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/S0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 4.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–44. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–7. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visa N, Puvion-Dutilleul F, Bachellerie JP, Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur J Cell Biol. 1993;60:308–21. [PubMed] [Google Scholar]
- 7.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–63. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 8.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–15. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–78. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, et al. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997;7:725–35. doi: 10.1101/gr.7.7.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Wilusz JE, Jnbaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci USA. 2012;109:19202–7. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 19.Nojima T, Hirose T, Kimura H, Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J Biol Chem. 2007;282:15645–51. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- 20.Cohen HR, Panning B. XIST RNA exhibits nuclear retention and exhibits reduced association with the export factor TAP/NXF1. Chromosoma. 2007;116:373–83. doi: 10.1007/s00412-007-0100-1. [DOI] [PubMed] [Google Scholar]
- 21.Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–51. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–9. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–73. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 24.Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–34. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–97. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 26.Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A, et al. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci USA. 2012;109:4846–50. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–38. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Zhou X, Liu N, Wang C, Zhang L, Mo W, et al. Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett. 2008;582:1761–5. doi: 10.1016/j.febslet.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 30.Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–78. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 31.Pelisch F, Pozzi B, Risso G, Muñoz MJ, Srebrow A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J Biol Chem. 2012;287:30789–99. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–56. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–79. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 37.Yang HT, Peggie M, Cohen P, Rousseau S. DAZAP1 interacts via its RNA-recognition motifs with the C-termini of other RNA-binding proteins. Biochem Biophys Res Commun. 2009;380:705–9. doi: 10.1016/j.bbrc.2009.01.166. [DOI] [PubMed] [Google Scholar]
- 38.Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, Farazi TA, et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18:1428–31. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol. 2006;87:1991–5. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 40.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–8. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–99. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002;531:109–14. doi: 10.1016/S0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 44.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–4. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]