Abstract
Autophagy is a highly conserved cellular process by which cytoplasmic components are sequestered in autophagosomes and delivered to lysosomes for degradation. As a major intracellular degradation and recycling pathway, autophagy is crucial for maintaining cellular homeostasis as well as remodeling during normal development, and dysfunctions in autophagy have been associated with a variety of pathologies including cancer, inflammatory bowel disease and neurodegenerative disease. Stem cells are unique in their ability to self-renew and differentiate into various cells in the body, which are important in development, tissue renewal and a range of disease processes. Therefore, it is predicted that autophagy would be crucial for the quality control mechanisms and maintenance of cellular homeostasis in various stem cells given their relatively long life in the organisms. In contrast to the extensive body of knowledge available for somatic cells, the role of autophagy in the maintenance and function of stem cells is only beginning to be revealed as a result of recent studies. Here we provide a comprehensive review of the current understanding of the mechanisms and regulation of autophagy in embryonic stem cells, several tissue stem cells (particularly hematopoietic stem cells), as well as a number of cancer stem cells. We discuss how recent studies of different knockout mice models have defined the roles of various autophagy genes and related pathways in the regulation of the maintenance, expansion and differentiation of various stem cells. We also highlight the many unanswered questions that will help to drive further research at the intersection of autophagy and stem cell biology in the near future.
Keywords: autophagy, embryonic stem cells, tissue stem cells, cancer stem cells
Introduction
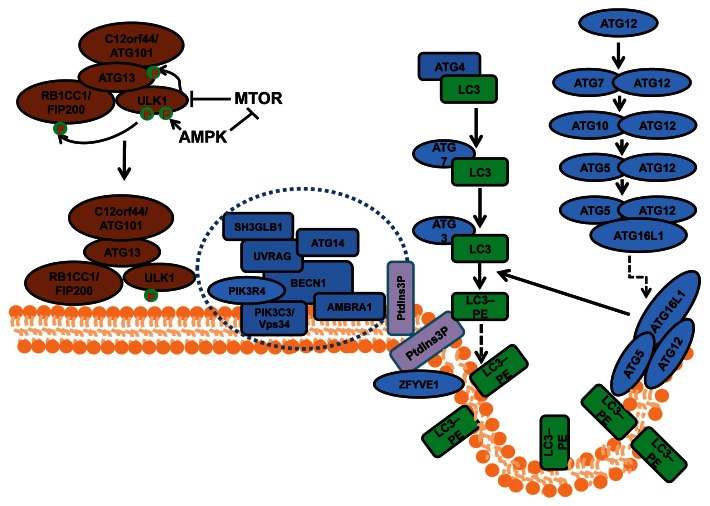
Macroautophagy (herein referred to as autophagy) is an evolutionarily conserved cellular process by which cells sequester a portion of their cytoplasm and organelles into double-membraned vesicles called autophagosomes that subsequently fuse with lysosomes for degradation of the enclosed materials.1 Autophagosome formation is controlled by a series of protein complexes acting sequentially, including the ULK1-ATG13-RB1CC1/FIP200-C12orf44/ATG101 complex for autophagy induction, the class III phosphatidylinositol (PtdIns) 3-kinase complex (including BECN1, ATG14/ATG14L/Barkor, PIK3R4/VPS15, PIK3C3/VPS34 and AMBRA1) for initiation of autophagosomes, and the ATG12–ATG5-ATG16L1 complex and the MAP1LC3A/LC3 (ATG8 homolog)–phosphatidylethanolamine (PE) complex (both ubiquitin-like conjugation systems) for the extension and closure of the autophagosome double membranes (Fig. 1). Besides these core components, a number of other autophagy-related (ATG) genes characterized in yeast, and their mammalian orthologs, are also required for autophagy.2 In addition, recent studies using proteomics approaches identified many other proteins in extensive networks that connect various cellular processes and signaling pathways to the autophagy machinery.3
Figure 1. The process and regulation of autophagosome formation in mammalian cells. Mammalian autophagosome formation is induced by the activation of the ULK1 complex (comprising ULK1, ATG13, RB1CC1/FIP200 and C12orf44/ATG101), which is promoted by suppression of MTOR complex 1 (MTORC1) under starvation conditions or AMPK (both directly as well as by repressing MTORC1) in low-energy states. Once the activated ULK1 complex translocates to part of the endoplasmic reticulum (or possibly other intracellular membranes), it coordinates with the class III phosphatidylinositol-3-phosphate (PtdIns3P) kinase complex (including PIK3C3, BECN1, PIK3R4/p150, AMBRA1, UVRAG, ATG14 and SH3GLB1) for the initiation of autophagosomes upon formation of PtdIns3P and recruitment of double ZFYVE 1. Two ubiquitin-like systems including the ATG12–ATG5-ATG16L1 complex (containing ATG5, ATG7, ATG10, ATG12 and ATG16L1) and the LC3–phosphatidylethanolamine (PE) conjugation system (including ATG3, ATG4, ATG7 and MAP1LC3) will then promote the elongation of phagophore membranes for the formation of autophagosomes.
Upon autophagosome maturation and the fusion of its outer membrane with the lysosome membrane, the contents as well as the inner membrane of autophagosomes are degraded to generate amino acids and other cellular building blocks for recycling by the cell. Besides this recycling function in response to nutrient or energy starvation, autophagy is increasingly recognized as a quality control mechanism for both proteins and organelles.2,4,5 Autophagy is induced as a result of various intrinsic and extrinsic cellular stress conditions, such as endoplasmic reticulum stress, reactive oxygen species (ROS) accumulation, hypoxia and bacterial infections. Activation of autophagy serves to clear the damaged protein aggregates, and impaired organelles as well as intracellular pathogens. Therefore, autophagy is crucial for maintaining cellular homeostasis as well as remodeling during normal development, and dysfunctions in autophagy have been associated with a variety of diseases such as cancer, inflammatory bowel diseases and neurodegenerative diseases.4,5 A hallmark of cellular defects in neurodegenerative diseases is the accumulation of ubiquitinated protein aggregates (or inclusion bodies), which is normally cleared by the ubiquitin-proteasome degradation system as well as autophagy for larger aggregates.6-8 The accumulation of large protein aggregates has been linked to increased neuronal apoptosis and axonal degeneration, accounting for the phenotypes in neurodegenerative diseases. In cancer, defective autophagy has been linked to increased DNA damage and gene mutations, leading to increased tumorigenesis,9 as well as to the reduced proliferation of tumor cells during cancer progression and metastasis.10,11
In contrast to the body of data derived from studies of somatic cells and disease models, the role of autophagy in the maintenance and function of stem cells is poorly understood. Both embryonic and various tissue stem cells play essential roles in development, tissue renewal and certain disease processes. Many stem cells are also long lived and persist throughout the adult life of an organism, thus the quality control mechanisms and maintenance of cellular homeostasis would be crucial for the maintenance of these cells. Thus, autophagy is expected to play an important role in the normal function of stem cells and associated diseases. Indeed, consideration of the available data suggests that the unique properties of stem cells (self-renewal, pluripotency, differentiation and quiescence) are dependent on the activation of the autophagic process.12,13 First, the processes of self-renewal and differentiation both require a strict control of cellular remodeling that involves protein turnover and lysosomal degradation of organelles.14 Second, the elimination and turnover of damaged macromolecules and organelles are essential to preserve the pluripotency of long-lived stem cells during their lengthy periods of quiescence.15,16 Third, basal autophagy is a homeostatic process that mediates quality control, clearance of altered and damaged intracellular proteins and organelles, and cellular remodeling through degradation of structural components. Not surprisingly, there is now accumulating evidence for the active role of autophagy in the regulation of embryonic stem cells (ESCs), several tissue stem cells, in particular hematopoietic stem cells (HSCs), as well as a number of cancer stem cells (CSCs). In addition to serving as an important mechanism for stem cell maintenance and self-renewal, autophagy has been implicated in the regulation of differentiation of various stem cells and their progenies, such as clearance of mitochondria and other organelles during erythrocyte maturation reported decades ago17,18 and recently validated by gene knockout studies.19
In this review, we describe a growing body of knowledge encompassing a range of stem cell systems that have significantly advanced our understanding of autophagy in stem cell biology. We discuss how recent studies of multiple knockout mice models have defined the functions of the autophagy pathway and specific autophagy proteins in ESCs, HSCs and other stem cells. In each system, autophagy has been suggested to regulate the maintenance, expansion and differentiation of stem cells, although different cellular mechanisms may be involved. Given the special capacity of stem cells in perpetuating themselves through self-renewal and generating mature cells through differentiation, the recent advances suggest the emerging concepts that autophagy may function to balance the quiescence, self-renewal and differentiation of stem cells in normal physiological and various stress conditions.
Autophagy in Embryonic Stem Cells
ESCs are pluripotent stem cells present in the early embryo with the capacity to undergo long-term renewal and differentiate into the primary germ layers: ectoderm, endoderm and mesoderm. Investigations using this cell type will allow us to acquire an understanding of the role that autophagy plays in early stages of mammalian development. Much of our knowledge concerning the molecular aspects of autophagy is founded on studies in the yeast Saccharomyces cerevisiae that led to the identification of the autophagy-related (ATG) genes (see ref. 1 for review). These genes provided the essential tools for investigating the mechanism, regulation and role of autophagy. The prediction that a number of these ATG genes existed as homologs in higher eukaryotes prompted molecular studies in mammalian cells. The first detailed molecular study into autophagy in a mammalian cell setting was performed using mouse embryonic stem cells.20 This study showed that bulk turnover of proteins labeled with [14C] amino acids can be induced by subjecting wild-type mouse ESCs to amino acid starvation. This bulk protein turnover is significantly reduced (> 50%) in mouse ESC (mESC) atg5−/− cells indicating the importance of autophagy in protein turnover. However, the absence of the ATG5 protein in these cells does not appear to affect their growth rate or colony morphology under replete culture conditions. Similarly, mES becn1−/− cells, lacking expression of the homolog of VPS30/ATG6, show no evidence of a growth defect under nonstarvation conditions,21 although in a later study it was reported that these cells are unable to form embryoid bodies (see below).22 Mice lacking expression of other individual autophagy-related genes including Atg7,23 Atg9,24 Atg3,25 Atg16l1,26 Becn1,21 and Ambra127 have been generated, but investigations of ESCs isolated from these mice have not been reported in any detail.
Autophagy is essential for early but not late differentiation and development of embryos
Development and differentiation processes are often accompanied by considerable remodeling of cells and tissues, and it has been suggested that autophagy plays a key role in this process providing both recycled material for building new structures while removing the old material.28 Since ESCs give rise to each of the three germ layers, it might be reasonable to expect that autophagic activity would be indispensable for normal differentiation and development of the embryo. However, it is not clear from the available evidence that this is the case.
It has been established that autophagy is essential for the very early stages of embryogenesis. Fertilized mouse oocytes lacking ATG5 (by oocyte-specific conditional knockout of the Atg5 gene, thus also removing maternal ATG5 protein) do not proceed beyond the 4- to 8-cell stage if they were fertilized by Atg5-null sperm, and therefore fail to form the blastocysts and the inner cell mass.29 Using GFP-LC3 as a marker, autophagy was found to be significantly upregulated in wild-type fertilized rat oocytes immediately (4 h) after fertilization of the oocyte by the spermatoazoa.29 Autophagic levels were subsequently suppressed between the 1- and 2-cell stage but elevated at the 4-cell stage. The levels of autophagy in cells in the later stages of development including the blastocyst stage containing the inner-cell mass from which the ESCs were derived were not reported.29 It is not clear why the very early stages are dependent on autophagic activity for survival. Presumably autophagy is required for controlling levels of key regulatory protein complexes or perhaps to provide substrates for cellular energy homeostasis prior to pre-implantation, after which cells have access to trans-placental nutrients.
The role of autophagy in ESC function in later stages of embryo development is less clear according to experimental observations of mice lacking expression of a number of different ATGs. atg3−/−,25 atg5−/−,30 atg7−/−,23 atg9−/−24 or atg16l1−/−26 mice give birth to phenotypically normal litters at Mendelian frequencies without any apparent anatomical abnormalities, suggesting that autophagy does not appear to play a pivotal role in the timing and coordination of differentiation in the developing embryo. These autophagy-deficient embryos are able to develop beyond the 8-cell stage (and indeed give rise to birth of live pups) due to the presence of maternally inherited ATG proteins in the oocyte cytoplasm. However, compared with pups of wild-type litters, autophagy-deficient pups have reduced birth weight and typically die within 1–2 d of birth possibly due to suckling defects caused by deficient neurological development.
Although the results of investigations using autophagy-deficient mice suggest that autophagy is dispensable for embryogenesis, it is possible that cellular and tissue defects arising from autophagic deficiency are more subtle, and have yet to be identified in the embryo. For example, atg5−/− mice develop neuronal inclusions and have decreased adipose mass. Autophagy plays an important role in quality control of cell components. It is possible that the lack of autophagy in ESCs contributes to the accumulation of damaged cellular components during embryogenesis, the implications of which are manifested later in life, and in terminally differentiated cells. Tissue-specific knockouts of different ATG genes give rise to a range of phenotypes, many of which relate to the unwanted accumulation of aggregates and damaged organelles such as mitochondria (reviewed in ref. 2) .
It is possible that other quality control pathways such as the ubiquitin-proteasome system (UPS) are to some extent able to compensate for the absence of autophagic activity in ESCs. Human ESCs (hESCs) exhibit high proteasome activity that is downregulated upon differentiation, suggesting that high proteasome activity is an intrinsic characteristic of hESC identity.31 Furthermore, hESCs lose their high proteasome activity in a continuous and progressive manner during the differentiation process, and differentiated cells showed increased levels of polyubiquitinated proteins. However, in another study it was reported that proteins damaged by carbonylation or formation of advanced glycation end products accumulate in murine ESCs but are cleared upon differentiation, an event that correlates with increased proteasome activity.32 It is possible that increased autophagic activity observed upon differentiation contributes to the removal of such damaged proteins. Further studies are required to investigate the relationship between the UPS and autophagy in ESCs.
In contrast to other ATG genes, Becn1−/− mice exhibit a profound developmental delay at E6.5 with the amniotic fold failing to develop33 and consequently die at an early embryonic stage. Widespread cell death is present in null embryos at E7.5. It has been suggested that BECN1 through its binding partners and control of the PIK3C3 lipid kinase is involved in membrane trafficking events other than autophagy, thereby explaining the more severe phenotype of becn1−/− mice34 compared with other ATG knockout mice. Becn1+/− mice survive, but develop tumors indicating that Becn1 can also function as a haploinsufficient tumor suppressor gene. AMBRA1 is a positive regulator of BECN1-dependent autophagy. However, a functional deficiency of AMBRA1 in mouse embryos does not phenocopy BECN1 deficiency, but rather leads to severe neural tube defects, accumulation of ubiquitinated proteins, unbalanced cell proliferation and excessive apoptotic cell death, suggesting that AMBRA1 may regulate target genes other than Becn1 or that BECN1 may have additional roles at later developmental stages.
Autophagy is required for embryoid body formation
mESCs deficient in ATG5 progress normally through embryonic development. However, there is some evidence from studies using an in vitro model of development that suggests autophagy may be important under particular circumstances. In one study it was reported that when compared with wild-type mESCs, autophagy-deficient mESCs cultured outside of the blastocyst exhibit altered behavior.22 Wild-type mESCs cultured in the absence of feeder cells and leukemia inhibitory factor (LIF) are able to form undifferentiated cell aggregates that develop into simple embryoid bodies (EBs) that contain an outer layer of primitive endoderm cells and an inner solid core of ectodermal cells. Cystic EBs are formed when the inner ectodermal cells undergo programmed cell death. These events mimic cavitation in early embryo development (PCD).
In this model of early embryo development, autophagy in wild-type ESCs is detectable throughout development, but is greatest at times when programmed cell death is maximal during cavitation. Compared with wild-type ESCs, atg5−/− and becn1−/− ES cells cultures in vitro fail to undergo cavitation. The results of further experiments indicated that dying cells do not express the engulfment signals for phagocytosis including phosphatidyl or lysophosphatdylcholine. EBs derived from these atg5−/− and becn1−/− cells have a defect in ATP production which, when reversed using methylpyruvate, restores cavitation and removal of apoptotic corpses. Methylpyruvate is a cell-permeable form of pyruvate that can feed in to the mitochondrial tricarboxylic acid cycle. These results suggest that the cells in EBs rely on autophagy for energy homeostasis presumably through the production of amino acids.
The role of autophagy for energy homeostasis in normal mammalian post-implantation embryo development may be limited, given the reliable supply of trans-placental nutrients. This notion is supported by the results of experiments investigating development in other organisms including insects such as the fruit fly Drosophila melanogaster, which do not have a maternal nutrient supply. In these species autophagy-deficient embryos suffer severe developmental defects.35 Nevertheless, the lowered birth weight of autophagy-deficient mice suggests a disturbance in their metabolism.
The majority of studies investigating autophagy in ESCs have been performed using mice. However, hESC lines that constitutively express GFP-LC3 (HES3-GFP-LC3) have been developed and validated for monitoring autophagic events.36 Since GFP-LC3 expression is maintained after teratoma formation, these cell lines represent a useful tool for investigating autophagy during early human embryogenesis. The results of studies using these cells suggest links between autophagy and differentiation. HES3-GFP-LC3 cells deprived of mouse embryonic fibroblast (MEF)-secreted maintenance factors by culturing in unconditioned medium for 3 d undergo spontaneous differentiation, and exhibit a significant increase in the number of fluorescent puncta compared with cells maintained in conditioned medium.36 Acute induction of differentiation using the TGFβ/TGFβ receptor II inhibitor SB431542 leads to a rapid increase in fluorescent puncta as early as 2 h after addition of the inhibitor. The inhibitor does not induce autophagy in differentiated cell types. Although unclear as to whether autophagy acts upstream or concomitant with differentiation, acute induction by SB431542 suggests a regulatory upstream role for degradation of pluripotency-regulating protein complexes.
The available evidence suggests that MTOR plays an important role in the regulation of pluripotency and self-renewal in hESCs. MTOR is also a central player in the regulation of autophagy. Inhibition of MTOR with rapamycin or depletion of MTOR transcript in hESCs leads to a significant reduction in levels of the pluripotency regulation transcription factors (POU5F1/Oct4 and SOX2), promotion of mesoderm and endoderm activities and decreased proliferation.37 Disrupting the kinase activity of MTOR in mESCs results in reduced cell size and a proliferation arrest.38 The regulation of MTOR appears to differ according to the differentiation pathway. A transcriptome analysis of hESCs differentiating into neural cells indicated that transcripts associated with MTOR are upregulated.39 Although not reported in these studies, autophagic activity would be expected to be significantly upregulated upon MTOR inhibition. It is possible that autophagy plays a role, albeit indispensable, in the loss of pluripotency and self-renewal properties of ESCs. The targets for degradation by autophagy remain to be identified.
Mitochondria and mitophagy in ESCs
Macroautophagy was originally thought to be restricted to the bulk turnover of cellular components. However, it is now apparent that a number of different cellular components can be targeted by the autophagic machinery in a selective manner using different receptor complexes.40 Studies in ESCs on the different selective forms of autophagy are limited to a few reports, and include the elimination of mitochondria (mitophagy)41,42 and midbodies (midbophagy).43 It is likely that other selective forms of autophagy occur in ESCs but have yet to be reported.
In mammalian cells the mitochondrion is responsible for satisfying the majority of ATP requirements. Our knowledge of mitochondria in stem cells is limited.44 Observations of mitochondria in a number of human and mouse ESC lines using electron microscopy show the presence of few mitochondria that also have poorly developed cristae, the site of oxidative phosphorylation, and ATP production. Fluorescence microscopy shows mitochondria to be located in small perinuclear groups. The energy demands of stem cells are thought to be largely met by the action of glycolysis.45 The proliferative capacity of mouse ESCs indeed closely correlates with high activity of different glycolytic enzymes, elevated glycolytic flux and low mitochondrial oxygen consumption. However, Zhou et al. showed that hESCs rather exhibit aerobic glycolysis,46 a phenomenon often seen in highly proliferative cells such as cancer cells and during embryogenesis,47,48 and that the uncoupling of glycolysis and oxidative phosphorylation in hESCs is caused by the expression of UCP2, presumably inhibiting the entry of pyruvate into the TCA cycle via pyruvate dehydrogenase. These data challenge the commonly held belief that mitochondria in hESCs are not metabolically active because they exhibit only few and underdeveloped cristae.49-51
Maintaining integrity, function and regulation of mitochondrial dynamics is important for all cells, and mitophagy contributes to their quality control. Mitophagy is tightly controlled in ESCs. BNIP3L/NIX, a BCL2 family member, is required for the elimination of mitochondria by mitophagy during erythroid maturation. Although mESC bnip3l−/− cells appear normal, mice derived from the cell are anemic.52 The results of a recent study provide evidence that mitophagy can be induced under particular circumstances in mESCs, and may play an important role in quality control in stem cells. In this study it was found that downregulation of the growth factor, augmentation of liver regeneration (GFER) protein, a portion of which is located in the inter-membrane space of the mitochondrion, results in loss of mitochondrial membrane potential, excessive fragmentation and elimination of damaged mitochondria by mitophagy.41,42 These ESCs also have significantly reduced levels of pluripotency marker gene expression. It is suggested that GFER modulates the mitochondrial fission GTPase DNMIL/DRP1, a protein responsible for promoting mitochondrial fragmentation. Depletion of GFER in differentiated cells does not affect mitochondrial function42 suggesting that the role of GFER is restricted to ESCs. The organization and function of mitochondria in ESCs are distinct from their differentiated counterparts, and maintaining the “status quo” appears to be vital for their survival. Mitophagy could also be induced in ESCs by inhibition of complex I of the respiratory chain using rotenone, and mild inhibition of complex III enhances pluripotency of ESC, suggesting that maintenance of mitochondrial integrity is important in ESCs.53
The trigger for loss of pluripotency markers in cells deficient in GFER is not clear, but it is possibly related to changes in mitochondrial dynamics. Mitochondria in normal ESCs undergo dramatic changes in appearance and function during differentiation, and mitophagy may serve in a surveillance role ensuring alterations in mitochondrial behavior is suppressed in the absence of developmental signals.
It has been suggested that the low numbers of mitochondria and the immature morphology of mitochondria in ESCs may be required to maintain the “stemness” of ESCs.41,42 Mitochondrial mass, mitochondrial DNA and ATP content of cells increase significantly when hESCs undergo differentiation.54 Increased mitochondrial activity is accompanied by increased production of ROS as a by-product of oxidative phosphorylation. Antioxidant enzyme production including mitochondrial and cytosolic dismutases, catalase and peroxiredoxins are also increased in differentiating cells.54 Nevertheless, ROS is elevated suggesting these cells are at risk from damage by ROS. Since autophagy is a mechanism for removing cellular constituents that have sustained damage, it is likely that autophagy would be upregulated in ESCs in response to increased mitochondrial activity and ROS production. It is also possible that increased ROS is required for signaling and control of events during differentiation.55 Accordingly, in addition to a role for energy homeostasis, autophagy (mitophagy) is likely to be responsible for quality control of mitochondria in ESCs. Given the pluripotent nature of ESCs, such quality control would be essential, which needs to be further investigated during ESC renewal and differentiation if these cells are to be used for therapeutics.
Midbophagy
The existence of multiple forms of autophagy (including nonspecific autophagy) implies that each is separately regulated in a temporal and spatial manner. Accordingly, mitophagy is activated to maintain the quality of the mitochondrial population, and other forms of autophagy may be activated under different conditions. The removal of midbodies by midbophagy occurs as ESCs undergo differentiation.43 Midbodies are organelles that form between daughter cells during cytokinesis and are required for their final separation. They selectively accumulate in stem cells and induced pluripotent stem cells (iPSCs) in vitro and in vivo. It is suggested that their accumulation may function to maintain or enhance the pluripotency of stem cells. Correct control of midbophagy would be essential to maintain the pluripotent state of ESCs. It is possible that induction of bulk autophagy by starvation or treatment with rapamycin stimulates differentiation through removal of midbodies.
Role of Autophagy in Hematopoietic Stem Cells
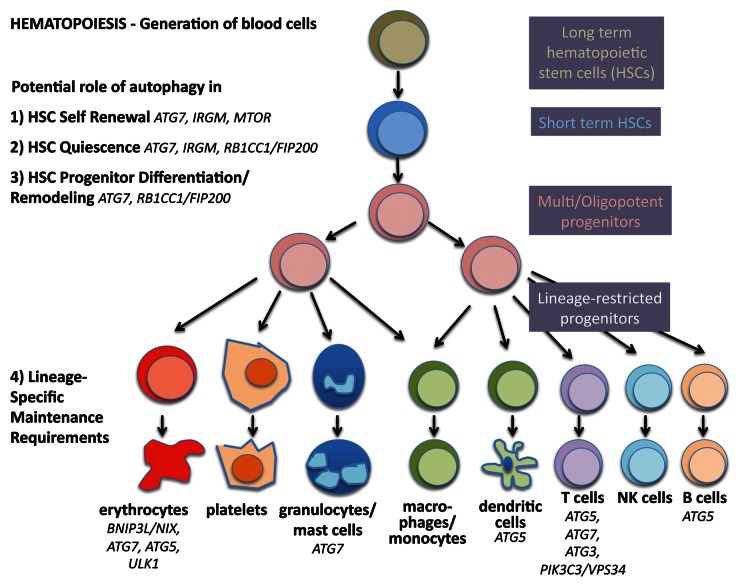
Hematopoietic stem cells are perhaps the best studied stem cells in the body due to relatively easy access to populations in the bone marrow and blood, combined with well-established assays in vitro and vivo. The hematopoietic system develops in a hierarchical fashion; a small number of long-term stem cells that are relatively quiescent divide to become progenitors that proliferate and differentiate into mature blood cell lineages that are produced in large numbers every day (Fig. 2). The lympho-hematopoietic system is composed largely of cells with short life spans and therefore requires continuous replenishment. In contrast, the cellular source of these blood cell lineages, the adult hematopoietic stem cells (HSCs) are long-lived and not depleted during the lifetime of the organism.
Figure 2. Autophagy in stemness of hematopoietic stem cells. During hematopoiesis, differentiated blood cells are generated from hematopoietic stem cells (HSCs). Long-term HSCs develop into short-term HSCs, multi- and then oligopotent progenitors and lineage-restricted progenitors, which give rise to the differentiated blood cells. These include erythrocytes, platelets, granulocytes, macrophages, dendritic cells and the lymphocytes T, B and NK cells. Autophagy is thought to play a role in self-renewal and quiescence in HSCs, the two hallmarks of stemness. Moreover, during differentiation autophagy is hypothesized to help multipotency and remodeling. Last, evidence is increasing that autophagy is required to maintain the healthy differentiated lineages.
Over the last decade, much progress has been made toward murine and human HSC isolation and identification, based mainly on their characteristic cell surface proteins that are either present (ATXN1/Sca-1 and KIT/c-Kit) or absent (lineage markers such as ITGAM/MAC-1 for mature myeloid cells, CD8 for T cells, PTPRC/B220 for B cells) on immature cells. True long-term murine HSCs are lin-, KIT+, ATXN1+, and further identified by SLAM family markers (CD48- and SLAMF1/CD150+), while lin- CD34+ CD38- enriches them in humans. HSCs reside in a specific hypoxic microenvironment or niche within the bone marrow, the majority in a quiescent state (reviewed in ref. 56). In response to intrinsic or extrinsic cues regarding the need for mature blood cells, HSCs can be stimulated to regenerate another HSC (self-renewal) or in asymmetric cell division generate a progenitor cell that can then produce the mature hematopoietic cell lineages (differentiation/multipotency). In summary, and like other stem cells, true HSCs strike a fine balance between quiescence, self-renewal and differentiation. When this balance is not executed properly, the consequences are severe, including either biased differentiation and thereby depletion or overrepresentation of some lineages (e.g., anemia, lymphopenia, myeloproliferation,) and/or hematopoietic malignancies (e.g., leukemias); it is well accepted that transformation events may occur as early as the stem or progenitor cell stage.
There are very few studies that have investigated autophagy in HSCs, but at least one of them has indicated that autophagy is highly active in HSCs in humans.57 Furthermore, murine HSCs, in contrast to their short-lived myeloid progeny, robustly induce autophagy after ex vivo cytokine withdrawal and in vivo calorie restriction, driven by the transcription regulator FOXO3.58 For this review, our prevailing hypothesis is that autophagy is essential to strike the balance between quiescence, self-renewal and differentiation in HSCs.
Autophagy in quiescence of HSCs
The majority of HSCs reside in the G0/G1 phase of the cell cycle59 and are in a state of reversible cell cycle arrest known as quiescence. Quiescent cells are particularly hardy and able to survive long periods of nutrient starvation; entry into quiescence is often associated with metabolic changes (reviewed in ref. 60). While proliferating cells devote much of their metabolic capacity to biosynthesis in order to create the material necessary to form a new cell, quiescent cells are relieved of this expensive metabolic requirement. HSCs downregulate protein synthesis and activate pathways that sustain them during periods of nondivision. In addition to cyclin-dependent kinase inhibitors, regulation of quiescence is achieved through the PtdIns3K-MTOR pathway.61-64 Indeed, increased MTOR activity has deleterious effects on HSCs, including excessive HSC proliferation leading to leukemias, followed by stem cell depletion and failure to repopulate the hematopoietic lineages.
The following indirect evidence points toward the idea that autophagy is required for the maintenance of quiescence in HSCs:
First, the slow metabolic turnover of quiescent HSCs means that damaged mitochondria and proteins cannot be easily passed on to daughter cells and thereby diluted, so autophagy may be essential for the required increase in catabolic rate. Interestingly, the UPS, the other major degradation system in the cell, has also been implicated in leukemogenesis. CBL/C-CBL, FBXW7/FBW-7, VHL and MDM2 are molecules from the UPS pathway that are often modified in hematopoietic malignancies. The deletion of genes encoding these protens in mice leads to deleterious HSC phenotypes (reviewed in ref. 65).
Second, quiescence is accompanied by changes in metabolism, in which autophagy could play a role due to its role in the clearance of mitochondria (mitophagy). Only a few studies have addressed how HSCs generate ATP.66 However, high levels of autophagy,57,67 low mitochondrial content in HSCs68 and the fact that hypoxia, common in the oxygen-limited niche environment, switches cells to a glycolytic metabolism all point toward the idea that quiescent HSCs primarily use glycolysis rather than oxidative phosphorylation to meet energy requirements.66,69
Third, we know from other cell types that mitophagy controls ROS levels. Increased ROS promote HSCs’ exit out of the quiescent state69 and mice with loss of autophagy in HSCs develop myeloproliferation,67 a sign of loss of quiescence. Interestingly, ATG7 has recently been shown to bind and modulate TP53/p53, thereby controlling exit out of the cell cycle in response to metabolic stress. Indeed, starved murine fibroblasts lacking ATG7 fail to undergo cell cycle arrest.70
Induction of quiescence and slow cell cycle turnover help ensure maintenance and preservation of long-lived stem cells. This also ensures that HSCs age more slowly, undergoing fewer cycles of replication leading to a minimum of telomere shortening.71 However, cell damage and telomere erosion is not completely prevented in HSCs, despite the slow turnover and the fact that stem cells are one of the few cell types in the body to express telomerase.72 Similar to other cells, entering senescence is important for stem cells to avoid accumulation of damaged proteins, lipids and nucleic acids, and may prevent transformation. Clearly, maintaining cell health and preventing stem cell aging is vital for hematopoietic health, and autophagy’s role in degrading damaged molecules and organelles may be essential to keep cells young. Importantly, autophagy has been inextricably linked to aging: (1) autophagy levels generally decrease with age in multiple cell types including hematopoietic cells,73 (2) loss of function mutations in ATG genes decreases life span in yeast, (3) calorie restriction and rapamycin increases life span in C. elegans and rats, and (4) tissue-specific loss of autophagy genes in mice leads to age-related diseases such as neurodegeneration, diabetes and cancer.5 HSCs from aged mice show a reduced repopulating activity per cell, attenuated self-renewal and homing abilities, myeloid skewing of differentiation, and an increased level of stress-induced apoptosis.74 This phenotype is largely reminiscent of the one found in HSCs from mice deficient for ATG7 or RB1CC1/FIP200.67,75,76 Furthermore, Chen et al. showed that HSCs can be rejuvenated through inhibition of MTOR signaling, restoring self-renewal and hematopoiesis in aged mice and enabling effective vaccination against flu.77
Although it has not been shown directly that autophagy plays a role in HSC senescence, links exist and it is clear that prevention of senescence holds the key to prolonged HSC self-renewal capacity and maintaining hematopoietic homeostasis during the lifetime of an animal or human.
Autophagy in self-renewal of HSCs
Life-long maintenance of HSCs is achieved by ensuring a balance between differentiation and self-renewal. Not much is known about the molecular mechanisms of self-renewal except that control is exerted by the WNT pathway and BMI1, a member of the polycomb protein group. In bmi1−/− mice, HSC numbers are markedly reduced, and stem cell-associated survival gene and proliferation-modulating gene Cdkn2a (encoding P16INK4A and P19ARF in overlapping reading frames) is controlled by BMI1.78 Moreover, recent evidence suggests that accumulation of DNA damage restricts the self-renewal capacity of human HSCs79 with some molecular aspects of the pathway just being elucidated in mice.80 Interestingly, according to this recent study, by committing to differentiate, the lymphoid-biased HSCs forgo self-renewal.80
To date, serial transplantation is the best way to test the ability of HSCs to undergo self-renewal. According to this assay, HSCs from the original donor have self-renewal ability if the recipient of a bone-marrow transplant, Recipient 1 (whose own HSCs were completely ablated before the transplant), serves as a successful donor for subsequent (serial) transplants (Recipient 2, etc.). The number of serial transplants that the original donor's bone marrow can perform successfully is a measure of the self-renewal capacity of the donor’s HSCs.81 To ascertain a role for autophagy in self-renewal, this assay should ideally be performed on autophagy-depleted HSCs. The fact that HSCs without essential autophagy genes fail to survive the first transplantation suggests that self-renewal requires autophagy.67,75 In a surrogate in vitro assay, the serial colony-forming assay, atg7−/− HSCs show a serious loss of colony formation on replating.67 Similarly, human adult HSCs fail to form colonies in colony-forming assays when autophagy is inhibited by 3-methyladenine (3-MA) or ATG5 siRNA.57 The authors of this study conclude diminished self-renewal capacity of HSCs in the absence of autophagy; however, without replating, it is not clear whether colonies formed from self-renewed stem cells, so strictly speaking replating would have been necessary to draw this conclusion.
In summary, as a self-renewing population that may have prolonged periods of quiescence, HSCs must possess robust defense and repair mechanisms in order to avoid damage. By contrast, differentiation of hematopoietic cells leads within days or weeks to cell death (i.e., in RBCs, neutrophils, platelets, and some types of myeloid cells), thereby eliminating long-term damage effects. In long-term self-renewing HSCs, such mechanisms have been shown to include senescence, apoptosis and premature differentiation.69 Increasing evidence suggests that autophagy should be added to this list.
Autophagy in differentiation and multipotency of HSCs
As opposed to the UPS, autophagy can degrade cytoplasm in bulk, including damaged organelles such as mitochondria, ER and ribosomes. One of the best-studied examples for autophagy’s role in differentiation is the clearance of mitochondria in the developing red blood cell (RBC). As RBCs mature they degrade organelles and proteins, leaving only hemoglobin behind, allowing cells to shrink and squeeze through the smallest capillaries. While the nucleus is expelled from differentiating RBCs, clearance of mitochondria is mediated by autophagy. Mitophagy impairment via loss of either the autophagosome-targeting molecule BNIP3L,52 ULK1 (a homolog of yeast Atg1, part of the initial signaling complex for autophagy19) or ATG7, an essential molecule for the elongation of the autophagosome, results in serious RBC developmental and functional defects, including mitochondrial retention.82,83 This is the first convincing example in which autophagy aids execution of cellular differentiation requiring massive remodeling to accommodate specialized cellular functions. Interestingly, GATA1, the master regulator of hematopoiesis and erythropoiesis, was found to control the expression of several autophagy genes (MAP1LC3B and its homologs) as well as genes that control lysosomal biogenesis.84
The bone marrow niche where HSCs reside is usually low in oxygen, and HSCs appear to avoid using oxidative phosphorylation for their energy supply, keeping the niche low in ROS.85 Accordingly, normal HSCs have few mitochondria, and increased mitochondrial biogenesis results in defective HSC maintenance. The transition from quiescence to proliferation/differentiation is accompanied by increased MTOR activity, resulting in increased metabolic rate and ROS.85,86 Antioxidants or rapamycin restore the self-renewal ability of ROShigh HSCs.77,86 Interestingly, HSCs can be identified by their low mitochondrial metabolism using Rhodamine123, a vital dye based on mitochondrial membrane potential.87 The migration of HSCs from the hypoxic niche to a microenvironment that is rich in oxygen and increased ROS is thought to promote their differentiation toward the myeloid lineage.88 Interestingly, increased differentiation toward the myeloid lineage is a hallmark of both the aged and autophagy-deficient hematopoietic system.
Indeed, ROS accumulate in ATG7-deficient hematopoietic stem cells, possibly due to the generation of mitochondrial superoxides produced by spent mitochondria that are not being cleared away. Mice develop myelodysplastic syndrome with accumulation of myeloid cells; this may be a consequence of increased ROS levels biasing differentiation toward the myeloid lineage.67 Similarly, a defect in RB1CC1/FIP200, a component of the ULK1-ATG13-RB1CC1/FIP200-C12orf44/ATG101 complex, leads to increased myeloid lineage cells.75 In both of these models autophagy was deleted in the hematopoietic lineage alone (using the promoter Vav or Tie-2, respectively). Whether osteoclasts or macrophages or the HSC themselves (all present in the bone marrow niche) are responsible for providing a ROS rich environment remains to be shown. It may turn out to be similar, however, to the metabolic interactions between breast tumor cells and stroma demonstrated by Lisanti et al., where levels of autophagy and ROS in the tumor stroma have an impact on tumor progression (reverse Warburg effect).89
Last, many publications point toward the idea that mouse models with modified signaling upstream of the MTOR pathway (constitutively active AKT1 or PTEN deficient) and in theory with decreased autophagy develop myeloid proliferation with increased LY6G+/Gr1+ ITGAM+/MAC-1+ myeloid cells,63,90 similar to the hematopoietic Atg7 knockout model. Conversely, the deletion of the MTORC1 component RAPTOR, therefore theoretically causing an increase in autophagy, results in a decrease of this myeloid population.91 However, it remains to be shown definitively that loss or gain of autophagy contributes to this phenotype, as MTOR inhibition signals for many other important cellular functions such as inhibition of protein translation, mitochondrial biogenesis, cell growth, motility and proliferation.
In summary, there is strong evidence that autophagy plays a role in remodeling during hematopoietic cell differentiation and in the multipotency of HSCs.
Autophagy in Neural Stem Cells
Although it was initially described as a cellular response to starvation conditions in many organisms, autophagy at basal levels (i.e., independent of nutrient stress) has been increasingly recognized as an important mechanism to maintain the cellular homeostasis, particularly in postmitotic cells such as neurons. Indeed, the first observation of the autophagy process, that is, self-eating of the cytoplasm by lysosomes, was made by electron microscopy in neurons almost half a century ago.92,93 Along with ubiquitin-proteasome degradation machinery, autophagy has been proposed to play a protective role against the development of a number of neurodegenerative diseases.6-8 Indeed, neural-specific conditional knockout of essential autophagy genes such as Atg5, Atg7 or Rb1cc1/Fip200 result in abnormal accumulation of ubiquitinated protein aggregates, SQSTM1/p62 and damaged mitochondria, increased apoptosis and neurodegeneration,94-96 providing direct support for a role of basal autophagy in protecting against neurodegenerative diseases.
Despite extensive studies of autophagy in neurodegenerative diseases, surprisingly little is known about the potential role of autophagy in the regulation of neuronal stem cells (NSCs), which, unlike terminally differentiated and quiescent neurons, are capable of expansion through self-renewal and proliferation during neurogenesis as well as differentiation into other neural lineages. Like HSCs, NSCs have been extensively characterized by the use of a combination of markers as well as other approaches such as long-term label retention by the relatively quiescent NSCs residing within the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone of the hippocampal dentate gyrus in adult brain.97,98 Similar to HSCs, niches for NSCs are also hypoxic (e.g., the subgranular zone of the dentate gyrus99) perhaps in order to maintain the relatively slow cycling property of these cells. Therefore, one hint of possible roles of autophagy in NSCs could be through regulation of ROS level and oxidative damage, which might otherwise be elevated in the hypoxic environment. Consistent with such an idea, deletion of the gene encoding the transcription factor PRDM16 results in an increased ROS production and abrogated NSC self-renewal, which can be rescued upon treatment with the antioxidant N-acetyl cysteine.100 Moreover, although in other cells, recent studies suggested that hypoxia inducible factor 1, α subunit (basic helix-loop-helix transcription factor; HIF1A)-dependent expression of BNIP3 promotes mitophagy to control excess ROS production and ROS-induced cell death under conditions of prolonged hypoxia.101,102 In addition, inactivation of transcription factors FOXO1, FOXO3 and FOXO4 (either in combination103 or FOXO3 only104) leads to defective self-renewal and differentiation of NSCs, accompanied with increased ROS, and such phenotypes are rescued by N-acetyl cysteine.103 As recent studies suggested that FOXO1 can control autophagy in cancer cells105 and that FOXO3 regulates autophagy in muscles,106,107 these findings raised the interesting possibility that possible autophagy defects in these mice might contribute to the ROS elevation and NSC phenotypes caused by FOXO-deficiency.
Although autophagy may regulate NSCs in a similar manner as HSCs by controlling ROS levels, it is not clear whether the elevated ROS upon autophagy inhibition affect self-renewal of NSCs and HSCs through similar mechanisms. Whereas the increased ROS has been shown to deplete HSCs through promoting exit of the quiescent state, a recent study suggested that a high level of ROS is required for self-renewal of NSCs based on the observation of higher endogenous ROS levels in NSCs within the SVZ.108 Nevertheless, there is considerable heterogeneity of the level of ROS in the SVZ, raising the interesting possibility that NSCs with the relatively higher level of ROS are in the proliferative or “activated” state whereas those with lower levels are the quiescent NSCs. It will be important to examine directly the potential role of autophagy in the self-renewal of NSCs by deletion of essential autophagy genes in these cells, and assuming that elevated ROS will be observed as in the case of HSCs, also to resolve the potentially complex regulation of NSCs and their progenies by ROS in both positive and negative manners.
In contrast to the lack of direct studies on self-renewal of NSCs, a recent study showed increased expression of Atg7, Becn1, Map1lc3a and Ambra1, all involved in autophagy, during neurogenesis in mouse olfactory bulbs, implicating a role for autophagy in NSC differentiation.109 Indeed, neuronal differentiation was found to be impaired in Ambra1 knockout mice as shown by decreased expression of several neural markers as well as the previous observation of severe neural tube defects during embryogenesis.27 Consistent with these findings in vivo, increased autophagy was also observed in neuronal differentiation in cultured NSC/progenitors in vitro, which could be markedly inhibited by 3-MA or wortmannin. Moreover, both Ambra1 haploinsufficient and Atg5-null olfactory bulb cells exhibit decreased neuronal differentiation in vitro. Another recent report showed that treatment of chick embryos with 3-MA significantly changes the spatiotemporal expression pattern of several neural markers and decreases the size of the acoustic-vestibular ganglion, suggesting a potential role of autophagy in chicken otic neurogenesis. In both of these studies, restoration of ATP levels by addition of methylpyruvate, a permeable analog for the citric acid cycle, partially rescued the differentiation defects, suggesting a role for autophagy in energy supply to promote neuronal differentiation. It was also reported that induction of autophagy by MTORC1 inhibition with rapamycin potentiates dbcAMP-induced differentiation of NG108–15 neuroblastoma cells, which could be abrogated by inhibition of the autophagy pathway by 3-MA or silencing Becn1, providing further support that autophagy may promote neuronal differentiation.110 Therefore, similar to observations showing altered differentiation in HSCs with loss of RB1CC1/FIP200 or Atg767,75 or preadipocytes lacking Atg5 or Atg7111,112 as well as reduced terminal differentiation of autophagy-deficient reticulocytes,19,52,113 autophagy plays a critical role in the promotion of neuronal differentiation of NSCs.
Autophagy in Cardiac Stem Cells
A role for autophagy in cardiac stem cell differentiation was initially recognized by Fen Wang and his colleagues.114 Tissue-specific ablation of FGF receptors and FGF receptor substrate 2α (FRS2) in heart progenitor cells leads to cardiac premature differentiation in mice. The differentiation defect is associated with abnormal autophagy activity, indicating that autophagy is required for FGF to regulate cardiac differentiation. Whole embryo and embryoid body culture were performed to investigate the underlying mechanisms. The group discovered that inhibiting autophagy either by small molecules or siRNA suppressed cardiac differentiation of embryoid body and embryo explants. In addition, suppressing autophagy activity blocked the enhancement of differentiation by FGFR inhibitors, while increasing autophagy activity abolished the suppression activity of FGF2 treatment. Thus, it is concluded that FGF signaling inhibits cardiac stem cell differentiation via suppressing autophagy activity. More detailed analysis indicates that FGF signaling suppresses autophagy activity through promoting AKT and MAPK pathways. Further study from whole-heart culture shows autophagy also inhibits cardiomyocyte maturation. These findings raise a number of interesting questions:
First, how does autophagy promote cardiac stem cell differentiation? Autophagy has been demonstrated to mediate the elimination of organelles during cell differentiation, such as erythrocyte and T cell maturation. Whether cardiac stem cells use the same mechanism remains to be answered.
Second, are there any other signal pathways involved in autophagy regulation during cardiac stem cell differentiation? WNT signaling has been proposed to be the upstream regulator of FGF signaling during cardiac development and it also inhibits cardiac differentiation.115,116 GSK3 is a part of the canonical CTNNB1/β-catenin-WNT pathway. One recent study demonstrated that a GSK3-KAT5/TIP60-ULK1 signal directly regulates autophagy.117 Thus, it is possible that autophagy mediates WNT signaling during cardiac development.
This question can be posed in more general terms: What are the mechanisms underlying the transcriptional and epigenetic regulation of autophagy and whether the regulation is organ- or cell-type specific? Indeed, accumulating data implicate an important role of epigenetic factors in regulating autophagy in various pathological conditions.1 Chemical or genetic inhibition of histone deacetylases (HDACs) leads to autophagy induction. HDAC regulates the specification of P19 embryonic carcinoma cells into cardiac lineage.118 In the future, more detailed studies on the regulation of autophagy would be essential to understand the specification and differentiation of cardiac stem cells.
Third, does autophagy play a role in cardiac stem cell proliferation? The process of self-renewal requires the coordination of cell-cycle progression and cell-fate determination. Indeed, increased cardiac differentiation is associated with decreased proliferation.114 It would be of great interest to investigate whether autophagy directly regulates cardiac stem cell proliferation.
It should be noted that the recent work discussed above analyzed the function of autophagy in embryonic cardiac stem cells. Since FGF signaling also regulates the differentiation of adult cardiac stem cells, although in an opposite way,119 it would be interesting to investigate whether autophagy performs the same function in adult cardiac stem cells. Likewise, as cardiac stem cells are also capable of differentiating into smooth muscle cells and endothelial cells besides cardiomyocytes, it would also be interesting to examine whether autophagy is required for the differentiation of cardiac stem cells into the other two cell types.
Autophagy and Mesenchymal Stem Cells
Mesenchymal stem cells (MSCs) are a diverse group of multipotent precursors capable of differentiating to mesenchymal lineages, including adipose, bone, cartilage and muscle.120 Although originally isolated from human bone marrow, MSCs have also been identified in many other adult tissues including muscles, adipose tissues, kidney, pancreas, brain and liver.120 It is important to note that little is known about the MSCs in vivo regarding their identity and function. In addition, MSCs cultured by current methods are a heterogeneous population.121 Therefore, it is disputable whether it is correct to use the term “mesenchymal stem cells.” Instead, MSCs are often called multipotent mesenchymal stromal cells.120,122 Currently, the role of autophagy in MSCs is largely unknown. The very limited knowledge about autophagy and MSCs is entirely derived from a very few studies using bone marrow MSCs. Recently it was shown that primary human bone marrow MSCs exhibit a high level of constitutive autophagy, whereas this basal autophagy decreases after these cells have differentiated into osteoblasts.123 In agreement with this observation, we found that primary mouse bone marrow plastic-adherent stromal cells isolated from GFP-LC3 transgenic mice124 have high levels of GFP-LC3 puncta. However, these GFP-LC3 puncta disappeared after these cells differentiated into osteoblast-like cells (Liu et al., unpublished). To date, the role of constitutive autophagy in bone marrow MSCs is unknown. Another recent report demonstrated that autophagy protects primary rat bone marrow MSCs from apoptosis under hypoxia/serum deprivation.125 It was shown that compared with the hypoxia/serum deprivation control group, 3-MA-treated rat bone marrow MSCs had higher rate of apoptosis.125 In agreement with this, another study showed that autophagy is essential for human bone marrow MSCs (cell line) survival during serum starvation, and these MSCs can release anti-apoptotic or pro-survival factors during serum deprivation to facilitate solid tumor survival and growth.126
Besides this sporadic knowledge in MSCs, there is emerging evidence for the important role of autophagy in MSCs-derived cells including adipocytes, chondrocytes and osteoblasts/osteocytes. It has been shown that adipose-specific deletion of Atg7 leads to decreased adipose mass and enhanced insulin sensitivity, and autophagy is important in normal adipogenesis.82,111 Massive autophagy is activated when wild-type primary MEFs are induced for adipocyte differentiation. Importantly, the autophagy-deficient primary atg5−/− MEFs exhibit dramatically reduced efficiency in adipogenesis.112 On the one hand, silencing of Becn1 results in enhanced chondrocyte death.127 On the other hand, treatment with 3-MA renders the chondrocytes refractory to killing, suggesting that sustained autophagy promotes cell death. Thus, autophagy may play both a cytoprotective and a death-promoting role in chondrocytes.127 Some autophagy proteins including ULK1, BECN1 and MAP1LC3A are constitutively expressed in normal human articular cartilage, whereas the expression is reduced in osteoarthritis chondrocytes and cartilage, suggesting a possible role of autophagy in the development of osteoarthritis.128 It has been shown that osteocytes also utilize autophagy for their homeostasis. Glucocorticoid treatment can induce the development of autophagy, preserving osteocyte viability depending on glucocorticoid dose.129,130 Osteoblasts are the precursor of osteocytes and are responsible for bone formation. As osteoblasts actively produce mineralized matrix during bone formation, it is conceivable that autophagy is activated to meet the high-energy demands of active osteoblasts. Indeed, we can now demonstrate that suppression of autophagy by deleting RB1CC1/FIP200 in osteoblasts leads to severely compromised bone development in mice. In vitro studies show that the consequence of RB1CC1/FIP200 deletion is compromised osteoblast terminal differentiation by limiting osteoblast condensational growth. Furthermore, autophagy inhibitors 3-MA and chloroquine mimic the effects of Rb1cc1/Fip200 deletion on osteoblast condensation and terminal differentiation (Liu et al., unpublished).
Taken together, current data demonstrate the activation of autophagy in cultured bone marrow MSCs. However, the detailed functions of autophagy in MSC biology such as stemness maintenance, self-renewal and differentiation are largely unknown. Recent studies reveal the role of autophagy in the terminal differentiation of MSC-derived cell lineages including osteoblast, osteocyte, chondrocyte and adipocyte. The detailed cellular and molecular mechanisms for the regulation of these cells during differentiation demand future investigation.
Autophagy in Cancer Stem Cells
CSCs (also termed tumor-initiating cells or tumor-propagating cells) are a subpopulation of cancer cells that have some characteristics of stem cells, being capable of self-renewal and differentiation that is responsible for the heterogeneity in bulk tumors.131-134 Recent studies suggest that autophagy also plays a crucial role in the origin, maintenance and systemic distribution of CSCs besides its many functions in normal embryonic and tissue stem cells.
Hypoxia- and starvation-related autophagy: A cytoprotective adaptive mechanism of cancer stem cells against microenvironmental stresses
The physiology of the microenvironment in solid epithelial tumors is characterized by lower oxygen levels (hypoxia), higher lactate levels, extracellular acidosis and depletion of nutrients (e.g., glucose and glutamine) compared with normal tissues.135-137 These changes are usually called tumor microenvironment stresses, and some of these stresses, particularly hypoxia, play critical roles during the evolution of the tumor stromal microenvironment and formation of the putative CSC niches.138,139 The ability of an undifferentiated hypoxic microenvironment to provide essential cellular interactions and environmental signals for the preferential maintenance of CSCs may therefore result in the specific expansion of CSC pools vs. non-CSC tumor cells. Recent studies have explored the hypothesis that hypoxia could induce tumor cell autophagy through hypoxia-inducible factors as a cytoprotective adaptive response. Because hypoxia-mediated autophagy promotes tumor cell survival in response to antiangiogenic therapies, and antiangiogenic agents have been recently shown to increase the population of CSCs via HIF1A,140,141 it might be tempting to suggest that autophagy is instrumental in hypoxia-driven CSC stimulation. Unfortunately, few studies have illustrated how autophagy could causally allow a co-promotion of enhanced tolerance of CSCs to biophysical stressful microenvironmental conditions, maintenance of CSC functionality and the expansion of CSC populations.
Beyond evading biophysical constraints, starving CSCs must use alternative sources of energy via activation of catabolic processes that permit them to recycle intracellular components to maintain metabolic homeostasis, quiescence and cell viability during periods of metabolic stress. Furthermore, CSC functioning and viability depend on their ability to ensure complementary bioenergetic sources at times other than during the development and growth of premalignant, pre-invasive lesions. Migrating CSCs experience hypoxia immediately after extravasation at secondary organs, and metastatic CSCs eventually become oxygen- and nutrient-starved, once the secondary tumors outstrip the blood supply.142-144 These considerations support the notion of protein catabolism functions of autophagy in protecting CSCs under starvation conditions. A causal relationship between activation of autophagy and enhanced cell survival of CSCs was initially suggested by the specific spatio-temporal upregulation pattern observed for BECN1 in certain pre-malignant lesions of the breast. In human comedo-ductal carcinoma in situ (DCIS), BECN1 is notably upregulated at the viable rim of intraductal cells within the hypoxic ductal niche.143,144 The expression levels of the autophagic effector MAP1LC3A are also increased in DCIS lesions vs. normal breast tissues.145 Remarkably, a BECN1-related enhancement of the autophagic flux appears to occur in DCIS tumor-founding progenitor cells because pre-malignant, cytogenetically abnormal DCIS spheroids-forming cells directly isolated from DCIS lesions exhibit an increased expression of autophagy-associated proteins that persists in culture as well as in tumors generated by malignant precursor cells in immunosuppressed mice. Treatment with chloroquine is sufficient to completely suppress the generation of DCIS spheroids/3D structures and ex vivo invasion of autologous breast stroma, induce apoptosis and eliminate cytogenetically abnormal spheroid-forming cells in the organ culture, and abrogate xenograft tumor formation via reduced expression of autophagy-associated proteins,144 confirming that autophagy is necessary for the survival of CSC-like precursor cells pre-existing in pre-malignant lesions.
Starvation-independent activation of autophagy: From adaptive to intrinsic metabolic feature in cancer stem cells
Homeostatic autophagy could operate in a starvation-independent manner to constitute an intrinsic metabolic feature of CSC cellular states. Comparing mammospheres from patients and well-characterized human breast cancer cell lines, autophagic flux is significantly higher in the mammospheres than the adherent cells, both at basal levels and under starvation conditions.146,147 Breast cancer cells bearing CSC-like phenotypes appear to constitutively display higher autophagic fluxes than non-CSC cells because both basal and starvation-induced autophagy are higher in aldehyde dehydrogenase 1 (ALDH1)-positive subpopulations derived from mammospheres than in the bulk tumor cell population.146,147 The specific depletion of BECN1 notably reduces both the size and formation efficiency of mammospheres, thus confirming and expanding the suggestion that the expression status of BECN1 may be pivotal for the tumorigenicity of breast CSCs.146,147 Given that depletion of BECN1 in CSCs also reduces tumor development in nude mice and that decreased survival in autophagy-deficient cells during anoikis does not contribute to any deficiency in mammosphere formation, it might be reasonable to conclude that autophagy is necessarily required for the maintenance and expansion of breast CSC populations.
Autophagy and the glycolytic metabotype in CSCs: Friends or enemies?
An interesting question that remains to be answered relates to the possible active contribution of autophagy to the metabolic shift of cancer cells to enhanced glycolysis (the Warburg effect)148-150 during malignant transformation and acquisition of stemness by CSC-like cell populations. If enhanced glycolysis indeed plays a causal role in the gain of stem-like properties by protecting tumor-initiating cells from the pro-senescent effects of mitochondrial respiration-induced oxidative stress, the ability of autophagy to functionally engage the glycolytic metabotype may generate a cellular state that is metabolically endowed to immortalization.
Friends: Autophagy as a facilitator of the Warburg effect
A Warburg-like glycolytic metabolism, and its facilitation by autophagy may accompany the acquisition of stemness and perhaps, both the occurrence and maintenance of CSC cellular states. Some malignant cells, particularly those driven by the Kras oncogene, appear to depend on elevated levels of autophagy for survival, even in the absence of external stressors.151 Kras [Kras(V12)]-induced malignant transformation is accompanied by upregulation of Atg5 and Atg7, and autophagic vacuole formation;152 pharmacological inhibition of autophagic activity and genetic ablation of Atg5 or Atg7 completely block Kras-induced anchorage-independent cell growth in soft agar and tumor formation in nude mice, partially establishing a causal role of autophagy induction in malignant transformation.152 In autophagy-deficient MEFs expressing oncogenic Kras, there is a decrease in both glucose uptake and in the glycolytic flux, thus suggesting dependency on autophagy for the glycolytic capacity of oncogenically transformed cells.153 Remarkably, it has been recently confirmed that a significant impairment of glucose uptake and intracellular lactate production and decreased sensitivity to reduced glucose availability occur in Rb1cc1/fip200-null primary MEFs transformed by Kras.10 Loss of autophagy, therefore, leads to deficient glycolysis and appears to intrinsically contribute to the decreased proliferation of mammary tumor cells. Further studies are required to clarify whether the inhibition of mammary tumor initiation, progression and metastasis imposed by an increased accumulation of both total and healthy mitochondria mass in autophagy-null tumors can be exclusively explained in terms of the necessary and/or sufficient “autophagy/glycolysis addiction” of CSCs.
Another body of evidence is related to the ability of autophagy to actively refuel the high-consuming, glycolysis-driven, anabolic phenotype. If the Warburg-like glycolytic metabotype can be correctly redefined in terms of the obligatory dependence of cancer cells to rapidly produce other metabolic end products in an unrestricted manner (i.e., independently of the generally starved microenvironmental conditions), the ability of autophagy to generate an internal nutrient pool that is recycled back and reused to synthesize essential cellular components can be expected to pleiotropically synergize with the anabolic side of the Warburg effect. Importantly, glycolysis can produce ATP at a higher rate without the undesirable presence of endogenous ROS and, by promoting flux through the pentose phosphate pathway, glycolysis can also support antioxidant defenses by inducing NADPH production, which is required for generating the key antioxidant enzyme, reduced glutathione.154,155 It should be noted that systemic dissemination of CSCs occurs at earlier stages of tumor development than previously thought, and fewer chromosomal aberrations are found in these disseminated CSCs compared with the matched primary tumors.156 Because failure to maintain autophagy-driven generation of metabolites of low molecular weight for continued use in the energetic and biosynthetic metabolism of the cells associates with increased DNA damage, gene amplification and aneuploidy, the ability of autophagy to alleviate cellular energy and anabolic deficits may significantly limit genomic instability during early tumorigenesis,157,158 thus functioning as a driving force of more stable CSC-like cellular states.
Enemies: Autophagy as an antagonist of the Warburg effect
Phenotypically diverse cancer cells can be efficiently reversed back to a less-tumorigenic iPSC-like stage of common ancestry that appears to be fully capable of originating nontumorigenic ontogenies.159-162 If mitochondrial restructuring and bioenergetics intimately reflect the molecular dynamics fundamental for the resetting and redirection of cell fate (i.e., the glycolytic metabotype supports the anabolic and catabolic requirements of pluripotent cell homeostasis, whereas redirection of pluripotency into defined lineages requires mitochondrial biogenesis and maturation of efficient oxidative energy generation and distribution networks), the generation of terminally differentiated phenotypes following (re)-expression of pluripotency-specific genes might imply that activation of a glycolytic Warburg effect in malignant cells apparently impedes the activation of CSC-like properties, including tumorigenesis. In this alternate scenario, does autophagy promote mitochondria-driven oxidative phosphorylation (OXPHOS)? Autophagy is constitutively activated in most Kras-mutated pancreatic intraepithelial neoplasms and high-grade pancreatic ductal carcinomas, but not in normal pancreatic ductal epithelium or low-grade pancreatic intraepithelial neoplasms.163 Genetic and pharmacological inhibition of autophagy attenuates growth and tumorigenicity of pancreatic cancer cells through a mechanism involving a significant decrease in OXPHOS and tricarboxylic acid intermediates. In agreement with a putative pro-mitochondrial OXPHOS activity of autophagy, oncogenic upregulation of autophagy can support cell survival and transformation primarily through the maintenance of mitochondrial metabolic function and energy levels.164
Regardless of the pro- or anti-glycolytic activity of oncogene-driven autophagy, both the activation of some oncogenes and/or the loss of some tumor suppressor genes might impose a metabolic insult on CSCs that depletes energy sources, thereby making them indirectly dependent on autophagy either to empower the Warburg-like glycolytic metabotype or preserve mitochondrial function and to directly provide catabolically-derived metabolic substrates.
Mitophagy: A direct way to connect autophagy with the metabolic reprogramming of CSCs
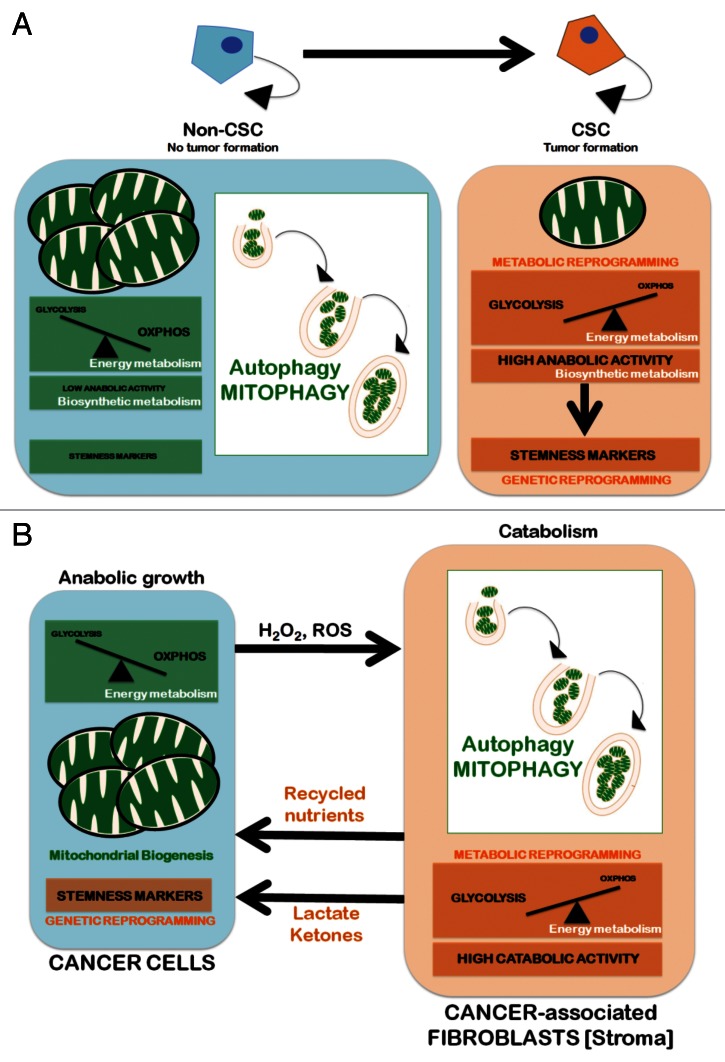
The cellular energy deficit triggered by cellular states of glucose deficiency imposed by oncogenic transformation (e.g., Kras) can induce a decline in mitochondrial respiration that is not related to changes in mitochondrial biogenesis; instead, it is inversely associated with the increased formation of acidic vesicles enclosing mitochondria, a process accompanied by the induction of autophagy-related proteins, such as BECN1, ATG5, LC3-II and vacuolar ATPases (i.e., mitophagy).164 Conversely, genetic ablation and pharmacological inhibition of autophagy promote the efficient recovery of respiratory protein expression and respiratory activity. Mitophagy might also have an impact on mitochondrial dynamics to segregate the mitochondria destined for clearance through autophagy, which may result in the loss of mitochondrial function and the accelerated onset of the glycolytic metabolism.165 Our group has explored if somatic cells reprogrammed to iPSCs bio-energetically utilize mitophagy. Interestingly, pharmacological promotion of mitochondrial fusion using mdivi-1 (for mitochondrial division inhibitor) at concentrations that rapidly induce the formation of mitophagy-refractory mitochondrial net-like or collapsed perinuclear mitochondrial structures largely impede somatic cell reprogramming to pluripotency.166 Although these findings strongly suggest that mitophagy is functionally integrated into the transcriptional network that specified the unique pluripotency of stem cells, future studies should elucidate whether the ability to directly shift the oxidative:glycolytic production ratios closer to those of pluripotent cells can molecularly explain the impact of autophagy-regulated mitochondria fusion-fission dynamics on the acquisition and maintenance of CSC cellular states (Fig. 3A).
Figure 3. Autophagy/mitophagy and the CSC cellular state: Two models. Mitochondria can be eliminated through nonspecific autophagic processes or selectively removed by mitophagy. The former is generally related to metabolic responses that are imposed by a lack of nutrients. Mitophagy, however, permits a tight adjustment in the number of cellular mitochondria by regulating the selective degradation of damaged, dysfunctional and superfluous mitochondria and adjust to changing physiological demands. Mitophagy, therefore, can play a crucial role in adapting the number and quality of mitochondria to new microenvironmental conditions. (A) Cell-autonomous model. Upregulation of mitophagy leading to significant reductions in both the number and the size of mitochondria (i.e., the “mitochondrial phenotype” that is commonly associated with stem cells) concurrently enhances a bioenergetic shift from somatic oxidative mitochondria toward an alternative glycolytic phenotype (“direct Warburg effect”), an energetic infrastructure associated with the transcriptional networks responsible for stemness and pluripotency. (B) Non-cell-autonomous model (“autophagic tumor stroma model of cancer metabolism”). Cancer cells’ mitochondrial oxidative phosphorylation (OXPHOS) can induce oxidative stress in adjacent fibroblasts (cancer-associated fibroblasts [CAFs] via H2O2 and reactive oxygen species (ROS), resulting in the onset of a myofibroblastic autophagic phenotype in CAFs. The CAFs autophagic phenotype leads to a loss of mitochondria via mitophagy, forcing CAFs to undergo aerobic glycolysis (“reverse Warburg effect”). The products of aerobic glycolysis (such as L-lactate and ketones) are then reused by cancer cells for OXPHOS, resulting in increased mitochondrial mass in cancer cells. The utilization of the high-energy metabolites L-lactate and ketones in cancer cells can promote the transcriptional activation of CSC-like phenotypes.
Mitophagy and the “reverse Warburg effect”: Connecting autophagic stroma with cancer stem cells
Both autophagy inhibition and promotion appear to similarly impair the occurrence of cancer cells with tumor-initiating capacities. In normal tissues, autophagy-mediated damage mitigation may efficiently suppress tumorigenesis; conversely, macromolecular recycling may support CSC survival by buffering bioenergetic demands under stressful metabolic and microenvironmental conditions.167-169 Therefore, activation of autophagy in normal tissues operates as a bona fide tumor suppressive mechanism, whereas autophagy inhibition may be extremely beneficial for anti-CSC therapy in established tumors. However, both autophagy inhibitors (e.g., chloroquine) and autophagy promoters (e.g., MTOR inhibitors) block tumorigenesis and cancer progression by eliminating CSCs.143,144,170,171 Although the simplest solution to this “autophagy paradox” could be just to claim that different types of tumors undergo different bioenergetic adjustments, the answer may also lie with the interaction between tumor cells and adjacent, autophagic, stromal cells. First, it has been observed that the stromal fibroblasts associated with breast cancer epithelial cells are glycolytic.172,173 The conversion of cancer-associated fibroblasts to glycolytic metabolism is induced by hydrogen peroxide secreted from the adjacent cancer cells, which results in loss of CAV1 from the fibroblasts.174 Second, a loss of stromal CAV1 causes the induction of mitophagy, which actively reduces the number of mitochondria in the stromal fibroblasts, thus reducing mitochondrial function and switching the tumor stroma metabolism to glycolysis. As a consequence, the autophagic/mitophagic tumor stroma generates recycled nutrients that can be used as chemical building blocks by anabolic epithelial cancer cells.175 Indeed, the increased glycolysis due to enhanced mitochondrial turnover generates excessive stromal cell lactate and ketones, which are secreted into the intracellular space.176 The cancer cells then take up the high-energy metabolites (lactate and ketones) and use them to feed cancer cell mitochondrial energy production and generate mitochondrial precursors for the biogenesis of new cancer cells. Lactate and ketones from the autophagic tumor stroma increase the transcriptional expression of gene profiles normally associated with stemness, including genes commonly upregulated in embryonic stem cells, and may therefore promote the dynamic appearance of CSC cellular states, resulting in significant decreases in patient survival.177 This host-parasite autophagic/mitophagic relationship from the tumor stroma to the epithelial cancer cells, which has been designated “the autophagic tumor stromal model of cancer cell metabolism,” “battery-operated tumor growth,” “stromal-epithelial metabolic coupling” and the “reverse Warburg effect”177-181 (Fig. 3B), functionally resolves the “autophagy paradox” because the systemic induction of autophagy prevents epithelial cancer cells from using recycled nutrients, whereas the systemic inhibition of autophagy prevents stromal cells from producing recycled nutrients, both effectively “starving” cancer cells.
The autophagy-regulated migratory/invasive phenotype in cancer stem cells
CSCs can be generated by the microenvironment through induction of CSC molecular features in more differentiated tumor cells. In addition to a role in CSC maintenance, the microenvironment is hypothesized to be involved in metastasis by the induction of the epithelial-to-mesenchymal transition (EMT) phenomenon, leading to the dissemination and invasion of tumor cells. Not surprisingly, EMT and CSC-like features have been linked to one another.182-186 In breast cancer, the undifferentiated CD44+CD24-/low antigenic state commonly attributed to breast cancer subpopulations with CSC properties is highly enriched with EMT transcriptional factors and mesenchymal markers.187,188 Our group recently explored whether autophagy might be important not only for survival but also for the maintenance of a migratory, invasive, treatment-resistant CD44+CD24-/low mesenchymal cellular state.189,190 Impairment of the autophagic flux drastically decreases the number of breast cancer cells bearing CD44+CD24-/low cell surface antigens and appears to impede the ontogeny of generating the invasive CD44+CD24-/low mesenchymal phenotype by preventing the full acquisition of a post-EMT status. The essential autophagy gene Atg12 might be pivotal for the autophagy-regulated migratory phenotype because its genetic ablation activates the transcription of the epithelial marker CD24 while concomitantly repressing the gene coding for the mesenchymal filament vimentin.189 The deficiency of ATG12 in glioma cancer cells, while failing to significantly have an impact on many tumor cell functions in 2D cultures (e.g., proliferation or cell viability), notably reduces their capacity to invade an organotypic matrix consisting of human fibroblasts embedded in polymerized collagen.191,192 Further supporting a previously unrecognized role of autophagy in the metastatic potential of migratory CSCs, the autophagy-associated genes DRAM1 and SQSTM1 regulate the motility and invasion of glioblastoma stem cells.193 The fact that a number of autophagy regulators are highly expressed in tumor cells carrying a mesenchymal signature, which intrinsically defines aggressiveness and metastatic invasion, might suggest that autophagy positively acts in concert with EMT-related processes to reduce proliferation in favor of enhanced cell motility, thereby providing a selective advantage during early stages of cancer metastasis, while supporting the survival of disseminated cells with CSC-like properties.
Targeting autophagy in cancer stem cells: A therapeutic corollary
Much work remains before developing an approach for pharmacologically altering CSC-related autophagy in a clinical setting.194-196 First, our current ability to accurately measure basal autophagic flux or determine whether autophagy is activated or inhibited in patient samples is very limited and of unproven utility in human patients. Similarly, we are awaiting more tests to identify and isolate CSCs in epithelial tumors; circulating CSCs directly obtained from the portal vein and peripheral blood of cancer patients could represent a powerful biomarker for patient monitoring during autophagy-based, CSC-targeted therapies. Second, all human clinical trials currently exploring autophagy inhibition as a therapeutic strategy use chloroquine or the derivative hydroxychloroquine due to its long track record of safety in human patients.197 However, whether chloroquine and its derivatives represent the most efficacious drugs for inhibiting autophagy is debatable. Hydroxychloroquine when combined with autophagy inducers, such as chemotherapy (i.e., temozolamide), proteasome inhibitors (i.e., bortezomib), MTOR inhibitors (i.e., temsirolimus) and/or radiation, has low response rates in preliminary clinical trials, indicating that hydroxychloroquine is not a potent autophagy inhibitor at clinically tolerable doses. The fact that an encouraging achievement of stable disease in 76% of advanced melanoma patients has been observed when combining hydroxychloroquine with temsirolimus clearly motivates the development of new, more potent, autophagy inhibitors (e.g., Lys05, Spautin-1).195 Ultimately, we may be able to develop very effective anticancer treatment modalities by combining a certain class of autophagy inhibitors with molecular-targeted drugs directed against specific cellular targets in CSCs. In this scenario, autophagy-based, “customized” combinatorial approaches will likely provide new powerful methods to better manage CSC-driven cancer origin and evolution in the near future.
Concluding Remarks
The past decade has witnessed significant growth in interest regarding stem cells and autophagy, but research investigating the intersection of these two exciting areas is very much in its infancy. Thus, we can expect tremendous growth in our understanding of the role of autophagy in various stem cells and more importantly of the mechanisms by which the autophagy pathway and different autophagy proteins control and balance the quiescence, self-renewal and differentiation of stem cells. Given the nature of these stem cell-specific characteristics, much of the future research will necessarily be performed using in vivo model systems. So far, conditional knockout of specific autophagy genes in mouse models have provided the strongest direct evidence for autophagy in different stem cells.67,75 It is anticipated that studies using other genetic models such as the fly and worm will also likely be fruitful based on the power of genetics to look for extensive gene interactions as well as the availability of well-defined stem cells in these systems. Likewise, future investigations into autophagy in stem cell functions will be facilitated by development of assays in cultured cells (especially for human stem cells) that allow monitoring of selective autophagy targets such as the mitochondrion in live cells.
Besides increasing our understanding of basic stem cell biology, studies of autophagy in stem cells also have significant therapeutic implications. Embryonic stem cells hold promise as therapeutic tools. However, if the potential is to be realized it is important that ESCs are maintained in a viable, pluripotent state and free of genetic damage. Although the lack of autophagy does not appear to affect the ability of ES cells to form full-term embryos, the lack of autophagic activity appears to have an effect on cells cultured in vitro under particular circumstances and stresses (e.g., inability to cavitate). It is not clear what the individual stressors are under these circumstances, but they may relate to the absence of particular growth factors and/or correct growth conditions. Autophagy may play an important role in protecting ESCs against unwanted outcomes either during renewal or differentiation in vitro either through the provision of an energy source or the elimination of cellular damage including DNA and defective mitochondria. It remains to be determined as to whether interference with autophagy during the early stages of ES cell expansion or differentiation has long-term consequences for the mature organism. If the hypothesis is correct that mitophagy plays a role in eliminating substandard mitochondria from ES and early progenitor cells, one would predict that tissues originating from the autophagy-deficient ES cells would have increased loads of mtDNA mutations and/or display premature aging phenotypes. With the advent of iPSC technology in which pluripotent stem cells destined for regenerative medicine applications are generated from individuals of advanced age (autophagic/mitophagic) quality control processes would need to be carefully examined and controlled to ensure long-term safety of any cellular grafts. Similarly, further studies of autophagy in tissue stem cells such as HSCs may shed light on the pathogenesis of hematopoietic disorders, serve as a basis for the development of new therapeutic strategies for such disorders, prove useful for the expansion of HSCs in vitro as a possible replacement for blood transfusion, and provide vital insight into general stem cell biology.
Recent studies suggested radiation therapy-induced autophagy contributes to the radio-resistance of glioma stem cells.198 Interestingly, the autophagy inhibitor bafilomycin A1 and silencing of Atg5 and Becn1 sensitizes the CD133+ glioma stem cells to γ-radiation and significantly decreases the viability of the irradiated cells and their ability to form neurospheres. Therefore, modulation of autophagy could be used in future therapies for cancer. However, we should acknowledge that a close affinity of normal tissue stem cells and CSCs in terms of autophagic fluxes might underlie a cautionary note for off-target effects of autophagy-based anti-CSC therapies. Discerning the specific autophagy requirements for normal tissue stem cells and CSCs may be an essential requisite when assessing autophagy targets for discriminatory targeting of CSCs with minimal sequelae. Only through a rigorous scrutiny of the nuances of the autophagy machinery in normal tissue stem cells and CSCs can we clarify the pathogenic mechanisms of autophagy and reveal the autophagic processes exploited by cancer cells to co-opt stem cell traits. Other interesting questions for future studies on the role of autophagy in CSCs include: Do CSCs necessarily require the activation of the autophagy machinery to ensure survival and/or functioning? Does autophagy endow tumor cells with self-renewal capabilities? Does autophagy constitute a shared metabolic feature of the stemness signatures among all of the intra-individual and inter-individual heterogeneity of CSCs cellular states? It is time for the autophagy community199 to make the responses as unambiguous as possible.
Acknowledgments
J.L.G. is supported by NIH grants (primarily GM052890 for work related to this review). A.K.S. is funded by the Oxford Biomedical Research Center, NIHR. Work at Dr. Menendez’s Laboratory is financially supported through funding from the Instituto de Salud Carlos III [Ministerio de Sanidad y Consumo, Fondo de Investigación Sanitaria (FIS), Spain, Grants CP05-00090 and PI06-0778 and RD06-0020-0028], the Fundación Científica de la Asociación Española Contra el Cáncer (AECC, Spain) and the Ministerio de Ciencia e Innovación (SAF2009-11579, Plan Nacional de I+D+ I, MICINN, Spain). Alejandro Vazquez-Martin received the Sara Borrell post-doctoral contract [CD08/00283, Ministerio de Sanidad y Consumo, Fondo de Investigación Sanitaria (FIS), Spain]. F.L is supported by NIAMS of the NIH under Award Number R01AR062030.
Glossary
Abbreviations:
- 3-MA
3-methyladenine
- ATG
autophagy-related
- CSCs
cancer stem cells
- DCIS
ductal carcinoma in situ
- EBs
embryoid bodies
- EMT
epithelial-to-mesenchymal transition
- ESCs
embryonic stem cells
- hESCs
human ESCs
- HDACs
histone deacetylases
- HSCs
hematopoietic stem cells
- iPSC
induced pluripotent stem cell
- LIF
leukemia inhibitory factor
- MEFs
mouse embryonic fibroblasts
- mESC
mouse ESC
- MSCs
mesenchymal stem cells
- NSCs
neuronal stem cells
- ROS
reactive oxygen species
- SVZ
subventricular zone
- ULK
Unc51-like kinase
- UPS
ubiquitin-proteasome system
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24132
References
- 1.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–95. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Hematology Am Soc Hematol Educ Program. 2006;2006:1–12, 505-6. doi: 10.1182/asheducation-2006.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 9.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–7. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–27. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phadwal K, Watson AS, Simon AK. Tightrope act: autophagy in stem cell renewal, differentiation, proliferation, and aging. Cell Mol Life Sci. 2013;70:89–103. doi: 10.1007/s00018-012-1032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vessoni AT, Muotri AR, Okamoto OK. Autophagy in stem cell maintenance and differentiation. Stem Cells Dev. 2012;21:513–20. doi: 10.1089/scd.2011.0526. [DOI] [PubMed] [Google Scholar]
- 14.Zeng M, Zhou JN. Roles of autophagy and mTOR signaling in neuronal differentiation of mouse neuroblastoma cells. Cell Signal. 2008;20:659–65. doi: 10.1016/j.cellsig.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel LB, Wen J, Lees M, Høyer-Hansen M, Farkas T, Krogh A, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628–41. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tooze J, Davies HG. Cytolysomes in amphibian erythrocytes. J Cell Biol. 1965;24:146–50. doi: 10.1083/jcb.24.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent G, Minick OT, Volini FI, Orfei E. Autophagic vacuoles in human red cells. Am J Pathol. 1966;48:831–57. [PMC free article] [PubMed] [Google Scholar]
- 19.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–6. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 27.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima NA. A(β) generation in autophagic vacuoles. J Cell Biol. 2005;171:15–7. doi: 10.1083/jcb.200508097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–20. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 30.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 31.Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–8. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernebring M, Brolén G, Aguilaniu H, Semb H, Nyström T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:7700–5. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–62. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denton D, Chang TK, Nicolson S, Shravage B, Simin R, Baehrecke EH, et al. Relationship between growth arrest and autophagy in midgut programmed cell death in Drosophila. Cell Death Differ. 2012;19:1299–307. doi: 10.1038/cdd.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tra T, Gong L, Kao LP, Li XL, Grandela C, Devenish RJ, et al. Autophagy in human embryonic stem cells. PLoS One. 2011;6:e27485. doi: 10.1371/journal.pone.0027485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Su P, Wang L, Chen J, Zimmermann M, Genbacev O, et al. mTOR supports long-term self-renewal and suppresses mesoderm and endoderm activities of human embryonic stem cells. Proc Natl Acad Sci U S A. 2009;106:7840–5. doi: 10.1073/pnas.0901854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–8. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fathi A, Hatami M, Hajihosseini V, Fattahi F, Kiani S, Baharvand H, et al. Comprehensive gene expression analysis of human embryonic stem cells during differentiation into neural cells. PLoS One. 2011;6:e22856. doi: 10.1371/journal.pone.0022856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mijaljica D, Nazarko TY, Brumell JH, Huang WP, Komatsu M, Prescott M, et al. Receptor protein complexes are in control of autophagy. Autophagy. 2012;8:1701–5. doi: 10.4161/auto.21332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todd LR, Damin MN, Gomathinayagam R, Horn SR, Means AR, Sankar U. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell. 2010;21:1225–36. doi: 10.1091/mbc.E09-11-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Todd LR, Gomathinayagam R, Sankar U. A novel Gfer-Drp1 link in preserving mitochondrial dynamics and function in pluripotent stem cells. Autophagy. 2010;6:821–2. doi: 10.4161/auto.6.6.12625. [DOI] [PubMed] [Google Scholar]
- 43.Kuo TC, Chen CT, Baron D, Onder TT, Loewer S, Almeida S, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–23. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lonergan T, Bavister B, Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7:289–96. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, et al. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–9. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 46.Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–16. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fritz V, Fajas L. Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene. 2010;29:4369–77. doi: 10.1038/onc.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–53. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- 50.Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J Mol Cell Cardiol. 2010;48:725–34. doi: 10.1016/j.yjmcc.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prigione A, Adjaye J. Modulation of mitochondrial biogenesis and bioenergetic metabolism upon in vitro and in vivo differentiation of human ES and iPS cells. Int J Dev Biol. 2010;54:1729–41. doi: 10.1387/ijdb.103198ap. [DOI] [PubMed] [Google Scholar]
- 52.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varum S, Momcilović O, Castro C, Ben-Yehudah A, Ramalho-Santos J, Navara CS. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res. 2009;3:142–56. doi: 10.1016/j.scr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park J, et al. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348:1472–8. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 55.Sauer H, Wartenberg M. Reactive oxygen species as signaling molecules in cardiovascular differentiation of embryonic stem cells and tumor-induced angiogenesis. Antioxid Redox Signal. 2005;7:1423–34. doi: 10.1089/ars.2005.7.1423. [DOI] [PubMed] [Google Scholar]
- 56.Eliasson P, Jönsson JI. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J Cell Physiol. 2010;222:17–22. doi: 10.1002/jcp.21908. [DOI] [PubMed] [Google Scholar]
- 57.Salemi S, Yousefi S, Constantinescu MA, Fey MF, Simon HU. Autophagy is required for self-renewal and differentiation of adult human stem cells. Cell Res. 2012;22:432–5. doi: 10.1038/cr.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–7. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lemischka IR, Raulet DH, Mulligan RC. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell. 1986;45:917–27. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- 60.Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, Coller HA. Staying alive: metabolic adaptations to quiescence. Cell Cycle. 2012;11:1680–96. doi: 10.4161/cc.19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haneline LS, White H, Yang FC, Chen S, Orschell C, Kapur R, et al. Genetic reduction of class IA PI-3 kinase activity alters fetal hematopoiesis and competitive repopulating ability of hematopoietic stem cells in vivo. Blood. 2006;107:1375–82. doi: 10.1182/blood-2005-05-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–82. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–22. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 64.Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, et al. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci U S A. 2008;105:19384–9. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moran-Crusio K, Reavie LB, Aifantis I. Regulation of hematopoietic stem cell fate by the ubiquitin proteasome system. Trends Immunol. 2012;33:357–63. doi: 10.1016/j.it.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–90. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–67. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson LA, Fleming WH, Galbraith TA. Intrapleural doxycycline control of malignant pleural effusions. Ann Thorac Surg. 1993;55:1115–21, discussion 1121-2. doi: 10.1016/0003-4975(93)90017-C. [DOI] [PubMed] [Google Scholar]
- 69.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 70.Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–8. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–66. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–20. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- 73.Phadwal K, Alegre-Abarrategui J, Watson AS, Pike L, Anbalagan S, Hammond EM, et al. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8:677–89. doi: 10.4161/auto.18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dykstra B, Olthof S, Schreuder J, Ritsema M, de Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med. 2011;208:2691–703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–14. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu F, Guan JL. FIP200, an essential component of mammalian autophagy is indispensible for fetal hematopoiesis. Autophagy. 2011;7:229–30. doi: 10.4161/auto.7.2.14125. [DOI] [PubMed] [Google Scholar]
- 77.Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 79.Yahata T, Takanashi T, Muguruma Y, Ibrahim AA, Matsuzawa H, Uno T, et al. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood. 2011;118:2941–50. doi: 10.1182/blood-2011-01-330050. [DOI] [PubMed] [Google Scholar]
- 80.Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–14. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 81.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–28. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–5. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A. 2010;107:832–7. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kang YA, Sanalkumar R, O’Geen H, Linnemann AK, Chang CJ, Bouhassira EE, et al. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012;32:226–39. doi: 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–63. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim M, Cooper DD, Hayes SF, Spangrude GJ. Rhodamine-123 staining in hematopoietic stem cells of young mice indicates mitochondrial activation rather than dye efflux. Blood. 1998;91:4106–17. [PubMed] [Google Scholar]
- 88.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–41. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 90.Kharas MG, Okabe R, Ganis JJ, Gozo M, Khandan T, Paktinat M, et al. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 2010;115:1406–15. doi: 10.1182/blood-2009-06-229443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoshii T, Tadokoro Y, Naka K, Ooshio T, Muraguchi T, Sugiyama N, et al. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J Clin Invest. 2012;122:2114–29. doi: 10.1172/JCI62279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–92. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 93.Dixon JS. “Phagocytic” lysosomes in chromatolytic neurones. Nature. 1967;215:657–8. doi: 10.1038/215657a0. [DOI] [PubMed] [Google Scholar]
- 94.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 95.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 96.Liang CC, Wang C, Peng X, Gan B, Guan J-L. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–16. doi: 10.1016/S0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 98.Palmer TD, Takahashi J, Gage FH. The adult rat hippocampus contains primordial neural stem cells. Mol Cell Neurosci. 1997;8:389–404. doi: 10.1006/mcne.1996.0595. [DOI] [PubMed] [Google Scholar]
- 99.Mazumdar J, O’Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, et al. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12:1007–13. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol. 2010;12:999–1006. doi: 10.1038/ncb2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 102.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–53. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–39. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 106.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 107.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 108.Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, et al. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vázquez P, Arroba AI, Cecconi F, de la Rosa EJ, Boya P, de Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy. 2012;8:187–99. doi: 10.4161/auto.8.2.18535. [DOI] [PubMed] [Google Scholar]
- 110.Chin TY, Kao CH, Wang HY, Huang WP, Ma KH, Chueh SH. Inhibition of the mammalian target of rapamycin promotes cyclic AMP-induced differentiation of NG108-15 cells. Autophagy. 2010;6:1139–56. doi: 10.4161/auto.6.8.13564. [DOI] [PubMed] [Google Scholar]
- 111.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–39. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–30. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takano-Ohmuro H, Mukaida M, Kominami E, Morioka K. Autophagy in embryonic erythroid cells: its role in maturation. Eur J Cell Biol. 2000;79:759–64. doi: 10.1078/0171-9335-00096. [DOI] [PubMed] [Google Scholar]
- 114.Zhang J, Liu J, Huang Y, Chang JY, Liu L, McKeehan WL, et al. FRS2α-mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ Res. 2012;110:e29–39. doi: 10.1161/CIRCRESAHA.111.255950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–79. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 116.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11:951–7. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–81. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 118.Karamboulas C, Swedani A, Ward C, Al-Madhoun AS, Wilton S, Boisvenue S, et al. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J Cell Sci. 2006;119:4305–14. doi: 10.1242/jcs.03185. [DOI] [PubMed] [Google Scholar]
- 119.Rosenblatt-Velin N, Lepore MG, Cartoni C, Beermann F, Pedrazzini T. FGF-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. J Clin Invest. 2005;115:1724–33. doi: 10.1172/JCI23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nombela-Arrieta C, Ritz J, Silberstein LE. The elusive nature and function of mesenchymal stem cells. Nat Rev Mol Cell Biol. 2011;12:126–31. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pevsner-Fischer M, Levin S, Zipori D. The origins of mesenchymal stromal cell heterogeneity. Stem Cell Rev. 2011;7:560–8. doi: 10.1007/s12015-011-9229-7. [DOI] [PubMed] [Google Scholar]
- 122.Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10:709–16. doi: 10.1016/j.stem.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 123.Oliver L, Hue E, Priault M, Vallette FM. Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells Dev. 2012;21:2779–88. doi: 10.1089/scd.2012.0124. [DOI] [PubMed] [Google Scholar]
- 124.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Q, Yang YJ, Wang H, Dong QT, Wang TJ, Qian HY, et al. Autophagy activation: a novel mechanism of atorvastatin to protect mesenchymal stem cells from hypoxia and serum deprivation via AMP-activated protein kinase/mammalian target of rapamycin pathway. Stem Cells Dev. 2012;21:1321–32. doi: 10.1089/scd.2011.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sanchez CG, Penfornis P, Oskowitz AZ, Boonjindasup AG, Cai DZ, Dhule SS, et al. Activation of autophagy in mesenchymal stem cells provides tumor stromal support. Carcinogenesis. 2011;32:964–72. doi: 10.1093/carcin/bgr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bohensky J, Shapiro IM, Leshinsky S, Terkhorn SP, Adams CS, Srinivas V. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207–14. doi: 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- 128.Caramés B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xia X, Kar R, Gluhak-Heinrich J, Yao W, Lane NE, Bonewald LF, et al. Glucocorticoid-induced autophagy in osteocytes. J Bone Miner Res. 2010;25:2479–88. doi: 10.1002/jbmr.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jia J, Yao W, Guan M, Dai W, Shahnazari M, Kar R, et al. Glucocorticoid dose determines osteocyte cell fate. FASEB J. 2011;25:3366–76. doi: 10.1096/fj.11-182519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–9. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 132.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 134.Vazquez-Martin A, Oliveras-Ferraros C, Del Barco S, Martin-Castillo B, Menendez JA. The anti-diabetic drug metformin suppresses self-renewal and proliferation of trastuzumab-resistant tumor-initiating breast cancer stem cells. Breast Cancer Res Treat. 2011;126:355–64. doi: 10.1007/s10549-010-0924-x. [DOI] [PubMed] [Google Scholar]
- 135.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 136.Gillies RJ, Gatenby RA. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis? J Bioenerg Biomembr. 2007;39:251–7. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- 137.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 138.Lin Q, Yun Z. Impact of the hypoxic tumor microenvironment on the regulation of cancer stem cell characteristics. Cancer Biol Ther. 2010;9:949–56. doi: 10.4161/cbt.9.12.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Quail DF, Taylor MJ, Postovit LM. Microenvironmental regulation of cancer stem cell phenotypes. Curr Stem Cell Res Ther. 2012;7:197–216. doi: 10.2174/157488812799859838. [DOI] [PubMed] [Google Scholar]
- 140.Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN, et al. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci U S A. 2012;109:2784–9. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Piao Y, Liang J, Holmes L, Zurita AJ, Henry V, Heymach JV, et al. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro Oncol. 2012;14:1379–92. doi: 10.1093/neuonc/nos158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–72. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Espina V, Mariani BD, Gallagher RI, Tran K, Banks S, Wiedemann J, et al. Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PLoS One. 2010;5:e10240. doi: 10.1371/journal.pone.0010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Espina V, Liotta LA. What is the malignant nature of human ductal carcinoma in situ? Nat Rev Cancer. 2011;11:68–75. doi: 10.1038/nrc2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mitchener JS, Shelburne JD, Bradford WD, Hawkins HK. Cellular autophagocytosis induced by deprivation of serum and amino acids in HeLa cells. Am J Pathol. 1976;83:485–92. [PMC free article] [PubMed] [Google Scholar]
- 146.Gong C, Song E, Codogno P, Mehrpour M. The roles of BECN1 and autophagy in cancer are context dependent. Autophagy. 2012;8:1853–5. doi: 10.4161/auto.21996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 148.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–70. [PubMed] [Google Scholar]
- 149.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bayley JP, Devilee P. The Warburg effect in 2012. Curr Opin Oncol. 2012;24:62–7. doi: 10.1097/CCO.0b013e32834deb9e. [DOI] [PubMed] [Google Scholar]
- 151.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pani G, Galeotti T, Chiarugi P. Metastasis: cancer cell’s escape from oxidative stress. Cancer Metastasis Rev. 2010;29:351–78. doi: 10.1007/s10555-010-9225-4. [DOI] [PubMed] [Google Scholar]
- 153.Pani G, Giannoni E, Galeotti T, Chiarugi P. Redox-based escape mechanism from death: the cancer lesson. Antioxid Redox Signal. 2009;11:2791–806. doi: 10.1089/ars.2009.2739. [DOI] [PubMed] [Google Scholar]
- 154.Hüsemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 155.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mahalingam D, Kong CM, Lai J, Tay LL, Yang H, Wang X. Reversal of aberrant cancer methylome and transcriptome upon direct reprogramming of lung cancer cells. Sci Rep. 2012;2:592. doi: 10.1038/srep00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ramos-Mejia V, Fraga MF, Menendez P. iPSCs from cancer cells: challenges and opportunities. Trends Mol Med. 2012;18:245–7. doi: 10.1016/j.molmed.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 161.Zhang X, Cruz FD, Terry M, Remotti F, Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2012 doi: 10.1038/onc.2012.237. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Folmes CD, Nelson TJ, Dzeja PP, Terzic A. Energy metabolism plasticity enables stemness programs. Ann N Y Acad Sci. 2012;1254:82–9. doi: 10.1111/j.1749-6632.2012.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–62. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, et al. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–11. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- 166.Abollo-Jiménez F, Jiménez R, Cobaleda C. Physiological cellular reprogramming and cancer. Semin Cancer Biol. 2010;20:98–106. doi: 10.1016/j.semcancer.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 167.Chen HY, White E. Role of autophagy in cancer prevention. Cancer Prev Res (Phila) 2011;4:973–83. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–23. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Calabretta B, Salomoni P. Inhibition of autophagy: a new strategy to enhance sensitivity of chronic myeloid leukemia stem cells to tyrosine kinase inhibitors. Leuk Lymphoma. 2011;52(Suppl 1):54–9. doi: 10.3109/10428194.2010.546913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Martelli AM, Evangelisti C, Follo MY, Ramazzotti G, Fini M, Giardino R, et al. Targeting the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin signaling network in cancer stem cells. Curr Med Chem. 2011;18:2715–26. doi: 10.2174/092986711796011201. [DOI] [PubMed] [Google Scholar]
- 171.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Martinez-Outschoorn UE, Whitaker-Menezes D, Pavlides S, Chiavarina B, Bonuccelli G, Casey T, et al. The autophagic tumor stroma model of cancer or “battery-operated tumor growth”: A simple solution to the autophagy paradox. Cell Cycle. 2010;9:4297–306. doi: 10.4161/cc.9.21.13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pavlides S, Tsirigos A, Migneco G, Whitaker-Menezes D, Chiavarina B, Flomenberg N, et al. The autophagic tumor stroma model of cancer: Role of oxidative stress and ketone production in fueling tumor cell metabolism. Cell Cycle. 2010;9:3485–505. doi: 10.4161/cc.9.17.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Martinez-Outschoorn UE, Prisco M, Ertel A, Tsirigos A, Lin Z, Pavlides S, et al. Ketones and lactate increase cancer cell “stemness,” driving recurrence, metastasis and poor clinical outcome in breast cancer: achieving personalized medicine via Metabolo-Genomics. Cell Cycle. 2011;10:1271–86. doi: 10.4161/cc.10.8.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Capparelli C, Guido C, Whitaker-Menezes D, Bonuccelli G, Balliet R, Pestell TG, et al. Autophagy and senescence in cancer-associated fibroblasts metabolically supports tumor growth and metastasis via glycolysis and ketone production. Cell Cycle. 2012;11:2285–302. doi: 10.4161/cc.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Castello-Cros R, Bonuccelli G, Molchansky A, Capozza F, Witkiewicz AK, Birbe RC, et al. Matrix remodeling stimulates stromal autophagy, “fueling” cancer cell mitochondrial metabolism and metastasis. Cell Cycle. 2011;10:2021–34. doi: 10.4161/cc.10.12.16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, et al. Transcriptional evidence for the “Reverse Warburg Effect” in human breast cancer tumor stroma and metastasis: similarities with oxidative stress, inflammation, Alzheimer’s disease, and “Neuron-Glia Metabolic Coupling”. Aging (Albany NY) 2010;2:185–99. doi: 10.18632/aging.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pavlides S, Vera I, Gandara R, Sneddon S, Pestell RG, Mercier I, et al. Warburg meets autophagy: cancer-associated fibroblasts accelerate tumor growth and metastasis via oxidative stress, mitophagy, and aerobic glycolysis. Antioxid Redox Signal. 2012;16:1264–84. doi: 10.1089/ars.2011.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–9. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 180.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–2. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 181.Hollier BG, Evans K, Mani SA. The epithelial-to-mesenchymal transition and cancer stem cells: a coalition against cancer therapies. J Mammary Gland Biol Neoplasia. 2009;14:29–43. doi: 10.1007/s10911-009-9110-3. [DOI] [PubMed] [Google Scholar]
- 182.Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 185.Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA. Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle. 2011;10:3871–85. doi: 10.4161/cc.10.22.17976. [DOI] [PubMed] [Google Scholar]
- 186.Amaravadi RK. Autophagy and tumor cell invasion. Cell Cycle. 2012;11:3718–9. doi: 10.4161/cc.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Macintosh RL, Timpson P, Thorburn J, Anderson KI, Thorburn A, Ryan KM. Inhibition of autophagy impairs tumor cell invasion in an organotypic model. Cell Cycle. 2012;11:2022–9. doi: 10.4161/cc.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Galavotti S, Bartesaghi S, Faccenda D, Shaked-Rabi M, Sanzone S, McEvoy A, et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 189.Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, et al. Autophagy is activated by TGF-β and potentiates TGF-β-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res. 2009;69:8844–52. doi: 10.1158/0008-5472.CAN-08-4401. [DOI] [PubMed] [Google Scholar]
- 190.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 191.Suzuki HI, Kiyono K, Miyazono K. Regulation of autophagy by transforming growth factor-β (TGF-β) signaling. Autophagy. 2010;6:645–7. doi: 10.4161/auto.6.5.12046. [DOI] [PubMed] [Google Scholar]
- 192.Tu YF, Kaipparettu BA, Ma Y, Wong LJ. Mitochondria of highly metastatic breast cancer cell line MDA-MB-231 exhibits increased autophagic properties. Biochim Biophys Acta. 2011;1807:1125–32. doi: 10.1016/j.bbabio.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 193.Caja L, Bertran E, Campbell J, Fausto N, Fabregat I. The transforming growth factor-beta (TGF-β) mediates acquisition of a mesenchymal stem cell-like phenotype in human liver cells. J Cell Physiol. 2011;226:1214–23. doi: 10.1002/jcp.22439. [DOI] [PubMed] [Google Scholar]
- 194.Mancias JD, Kimmelman AC. Targeting autophagy addiction in cancer. Oncotarget. 2011;2:1302–6. doi: 10.18632/oncotarget.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109:8253–8. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Amaravadi R. Autophagy can contribute to cell death when combining targeted therapy. Cancer Biol Ther. 2009;8:130–3. doi: 10.4161/cbt.8.21.10416. [DOI] [PubMed] [Google Scholar]
- 198.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, et al. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717–22. doi: 10.1002/ijc.24402. [DOI] [PubMed] [Google Scholar]
- 199.Klionsky DJ. The autophagy community. Autophagy. 2012;8:1003. doi: 10.4161/auto.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]