Abstract
Activation of TLR signaling has been shown to induce autophagy in antigen-presenting cells (APCs). Using high-resolution microscopy approaches, we show that in LPS-stimulated dendritic cells (DCs), autophagosomes emerge from MHC class II compartments (MIICs) and harbor both the molecular machinery for antigen processing and the autophagosome markers LC3 and ATG16L1. This ENdosome-Mediated Autophagy (ENMA) appears to be the major type of autophagy in DCs, as similar structures were observed upon established autophagy-inducing conditions (nutrient deprivation, rapamycin) and under basal conditions in the presence of bafilomycin A1. Autophagosome formation was not significantly affected in DCs expressing ATG4BC74A mutant and atg4b−/− bone marrow DCs, but the degradation of the autophagy substrate SQSTM1/p62 was largely impaired. Furthermore, we demonstrate that the previously described DC aggresome-like LPS-induced structures (DALIS) contain vesicular membranes, and in addition to SQSTM1 and ubiquitin, they are positive for LC3. LC3 localization on DALIS is independent of its lipidation. MIIC-driven autophagosomes preferentially engulf the LPS-induced SQSTM1-positive DALIS, which become later degraded in autolysosomes. DALIS-associated membranes also contain ATG16L1, ATG9 and the Q-SNARE VTI1B, suggesting that they may represent (at least in part) a membrane reservoir for autophagosome expansion. We propose that ENMA constitutes an unconventional, APC-specific type of autophagy, which mediates the processing and presentation of cytosolic antigens by MHC class II machinery, and/or the selective clearance of toxic by-products of elevated ROS/RNS production in activated DCs, thereby promoting their survival.
Keywords: MHC class II, dendritic cell, autophagy, LC3, electron tomography
Introduction
An important step in the adaptive immune responses is the activation of T cells. Activation of CD4+ T cells is mediated by recognition of antigenic peptides presented by major histocompatibility complex (MHC) class II molecules on the cell surface of antigen presenting cells (APCs).1 Among APCs, dendritic cells (DCs) are considered the most efficient in antigen presentation.2 To perform this function, they constantly sample their extracellular environment in peripheral tissues. Endocytosed antigens are transported to MHC class II containing compartments (MIICs), which represent adapted endocytic organelles equivalent to the typical late endosomes/multivesicular bodies and lysosomes existing in other cell types.3 Intracellular cytosolic and nuclear proteins also get loaded and presented on MHC class II molecules.4 Examples of such proteins are the Epstein Barr virus nuclear antigen 1 (EBNA1), tumor antigen Mucin 1 (MUC1), and complement component.5 Delivery of cytoplasmic material to lysosomes for degradation can take place via three autophagy pathways (microautophagy, chaperone-mediated autophagy and macroautophagy).5,6 Macroautophagy is the best characterized and involves the bulk or selective sequestration of cytosolic material and/or entire organelles within double-membrane structures and subsequent delivery to lysosomes for degradation.7 Typically, autophagosome formation begins with a nucleation membrane, known as the phagophore, which expands by membrane addition, and upon completion, fuses with a late endosome/lysosome. The origin of the autophagosomal membrane has been subject of intense speculation until recently, when at least three different organelles, the endoplasmic reticulum (ER), the mitochondria and the plasma membrane have been shown to contribute to its formation.7-12
A growing body of evidence has implicated autophagy in immune cells as a surveillance mechanism of the intracellular environment for pathogens and changes in self proteins.5,13,14 One pathway inducing autophagy in APCs is through activation of toll-like receptors (TLRs), which belong to a family of pattern recognition receptors (PRRs) that play a key role in microbe detection.14 In contrast to the well-characterized mechanism of autophagy induction by nutrient deprivation,15 the way TLRs induce autophagy has just started to be defined. Many studies have been performed in macrophages treated with various TLR agonists including lipopolysaccharide (LPS), a potent TLR4 agonist.16-19 LPS stimulation in DCs has not led to a detectable increase in autophagy, which has been attributed to the high constitutive levels of autophagosome formation in professional APCs.5,17,20 Indeed, when this fast autophagosome turnover is blocked with chloroquine, the autophagosome marker LC3 extensively colocalizes with MIICs at the light microscopy level. These results have been considered as evidence for delivery of cytoplasmic material in MIICs for degradation through the classical autophagy pathway.20 However, a detailed characterization of this process at the ultrastuctural level is lacking. In addition, LPS stimulation in DCs has been shown to induce the transient formation of ubiquitinated protein aggregates, termed DALIS (DC aggresome-like induced structures).21,22 Similar ubiquitin-positive aggresome-like structures (ALIS) are also induced in other immune and nonimmune cells under various stress conditions, such as oxidative stress or starvation, and contain substrates for degradation by the proteasome or autophagosomes/lysosomes.23
In the present study, we used a combination of confocal microscopy, high-resolution immuno-electron microscopy (IEM) and EM tomography to characterize the process of autophagosome formation in the D1 mouse dendritic cell line and mouse bone marrow-derived dendritic cells (BMDCs). We showed that DCs form double-membrane structures reminiscent of autophagosomes on their MIIC limiting membrane. These structures, which become most evident in the presence of the autophagosome degradation inhibitor bafilomycin A1 (BAFA1), contain the molecular machinery involved in antigen processing, as well as the autophagosome markers LC3 and ATG16L1. Similar double membranes are formed in DCs upon nutrient deprivation or rapamycin treatment. Upon LPS stimulation, these endosome-mediated autophagosomes target ubiquitin- and sequestosome1 (SQSTM1)-positive DALIS for degradation. In addition to its presence on double membranes, LC3 localizes on DALIS. Using ATG4B-mutant or -deficient cells, we show that LC3 association to both locations is likely independent of its lipidation status. Finally, we describe that the LPS-induced DALIS constitute previously not described vesicular membrane assemblies. These vesicles could provide the membrane for the MIIC-associated autophagosome expansion, as they are positive for ATG9, ATG16L1 and VTI1B.
Results
LPS-induced formation of double-membrane structures in D1 cells
TLR-mediated induction of autophagy in APCs has been recently reported but the exact details of how this takes place at a morphological level have not been established. A morphological hallmark of autophagosomes is the formation of double membranes around cytoplasmic material or entire organelles. Therefore, the formation of autophagosomes from their precursors is best characterized by electron microscopy (EM).
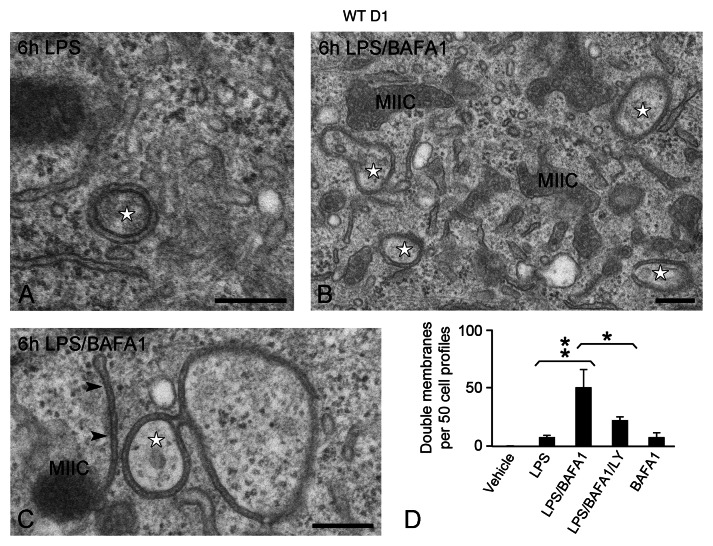
To address this question, we compared untreated and LPS-stimulated wild-type (WT) D1 cells grown in full medium by conventional EM. Whereas double-membrane structures were very rarely observed in unstimulated control cells (on average less than 1 per 50 cell sections), these were more frequently encountered in LPS-stimulated cells (on average 7 per 50 cell sections) (Fig. 1A and D). These structures exhibited often an electron-dense content and had morphological characteristics of the previously described phagophores and autophagosomes.24 Because “autophagic flux” is a dynamic and rapid process, inhibitors blocking their fusion to lysosomes and/or degradation are often used to facilitate their detection.25 In this regard, we stimulated D1 cells with LPS in the presence of BAFA1, an inhibitor of lysosomal acidification and autophagic degradation.26,27 This resulted in a profound accumulation of tubular phagophores and cup-shaped double membranes, reminiscent of those observed in different stages of autophagosome formation (Fig. 1B–D). Interestingly, these membranes appeared closely associated and sometimes connected to MIICs, the late endosomes of D1 cells (Fig. 1C).3 The number of double membranes observed upon LPS/BAFA1 treatment was significantly reduced in the presence of LY294002, a PtdIns3K inhibitor that blocks the initial steps of autophagosome formation.25 These results indicate that LPS stimulation leads to a small increase in the formation of double membranes in D1 cells, which is additional to the already high number that is formed at the basal state. These structures have the morphological characteristics of autophagosomes and respond to pharmacological treatments known to affect autophagic flux.
Figure 1. Formation of double-membrane structures in D1 cells. WT D1 cells were exposed to LPS for 6 h in the absence (A) or presence of BAFA1 (B and C). Double-membrane structures (stars) are detected in the cytosol after 6 h LPS stimulation and their number increases when autophagosome turnover is inhibited with BAFA1. Note in (C) the close connection of a double membrane (arrowheads) with a MIIC. (D) Quantitation of the number of double-membrane structures. No obvious change in D1 cell or nuclear size was observed upon the different treatments. Bars represent the mean of 3 independent experiments (n ≥ 50 cells per experiment) ± SD. **p < 0.01, *p < 0.04. Scale bars: 200 nm.
Electron tomography reveals cup-shaped membrane profiles connected to MIICs
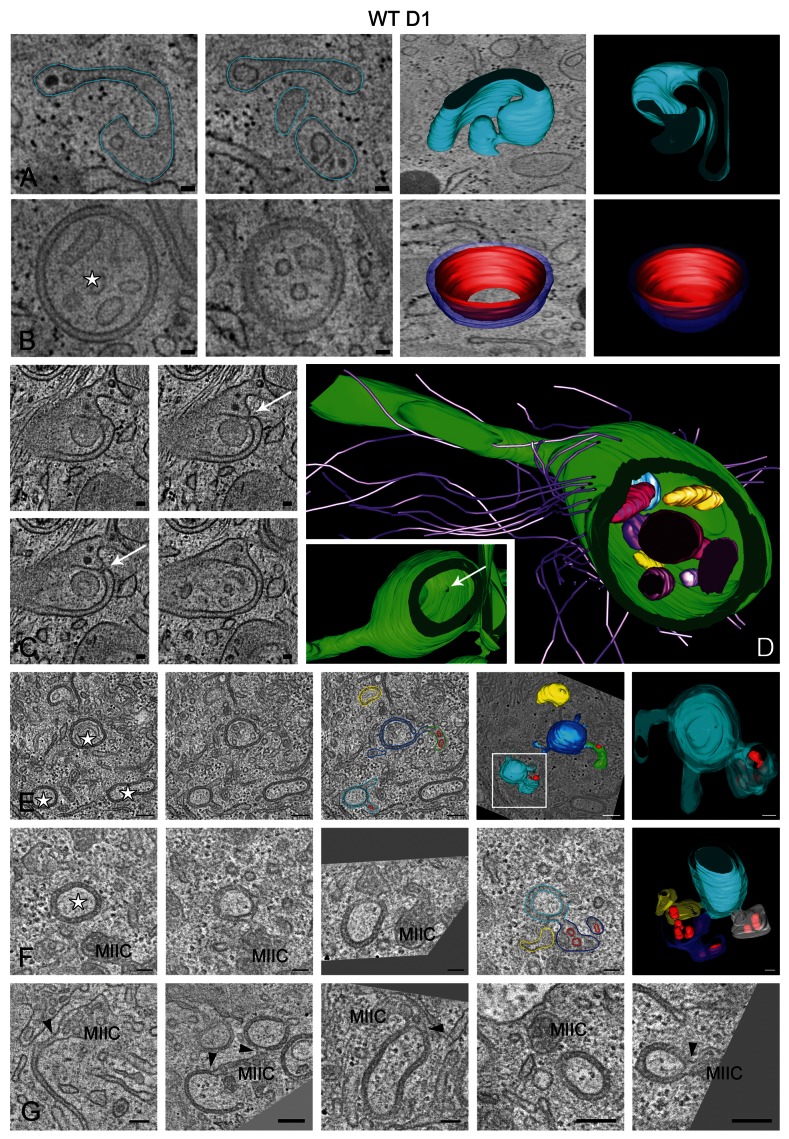
To further analyze the double-membrane structures and their association to MIICs, we performed EM tomography. The tomographic data confirmed the existence of typical sheet-like putative phagophores, which are continuous with the multivesicular MIIC compartments (Fig. 2A; Vid. S1). Regular-shaped concave double-membrane structures, reminiscent of autophagosomes, were also observed (Fig. 2B; Vid. S2), and appear to engulf cytoplasmic areas containing free-membrane vesicles (Fig. 2B–D). The lumenal distance between the two lipid bilayers measured 10.4 ± 1.3 nm. The MIIC-derived double membranes were frequently not completely closed and exhibited single small openings/pores connecting their inner cavity with the cytosol (Fig. 2C and D, arrows). Although its significance is not clear, this feature has been observed in most tomograms that were analyzed, and its high frequency must have certainly been underestimated by conventional EM analysis, underscoring the power of tomography. Additionally, the double membranes were commonly connected to the elaborate tubular/reticular network that is formed by MIICs upon LPS stimulation (Fig. 2C and D; Vid. S3),28,29 and this was especially apparent in the presence of BAFA1 (Fig. 2E–G; Vids. S4 and S5). Finally, the double-membrane structures were often surrounded by actin filaments (based on their cross-sectional diameter) that could provide mechanical support or drive their formation (Fig. 2D). Combined, our morphological analyses by EM and EM tomography suggested that, although the observed double membranes have characteristics of autophagosomes, they seem to be not conventional ones, because they appear to emerge from an endosomal compartment (see Fig. 2F and G for typical examples), rather than fusing with it after their completion forming the so-called amphisomes.27
Figure 2. EM tomography of double membranes formed upon treatment with LPS and LPS/BAFA1 for 6 h. Selected tomographic slices (~5 nm thick) and 3D model of (A) a putative phagophore connected to a multivesicular MIIC-like compartment containing internal vesicular and sheet-like membranes, (B) a regularly-shaped double-membrane structure surrounding cytoplasmic material and isolated vesicles (star), (C and D) a double-membrane structure exhibiting a ~20 nm pore connecting the engulfed material with the cytosol (arrows). Note in (C) the continuity between the double membrane and the multilaminar MIIC at the upper right corner. The 3D model in (D) illustrates the double-membrane interior, actin filaments associated to its outer membrane, and a long tubule emerging from it. Note that the model in (D) is rotated compared with the tomographic slices shown in (C). The inset shows the position of the pore in relation to the other features of the compartment. See also the corresponding Videos S1–S3. (E and F) Tomographic slices and 3D models of D1 cells treated with LPS/BAFA1 for 6 h. See also the corresponding Videos S4 and S5. (G) Tomographic slices showing connections (arrowheads) between MIICs and open and or seemingly closed double membranes. Scale bars: 50 nm.
The double-membrane structures contain MIIC but no other organelle markers
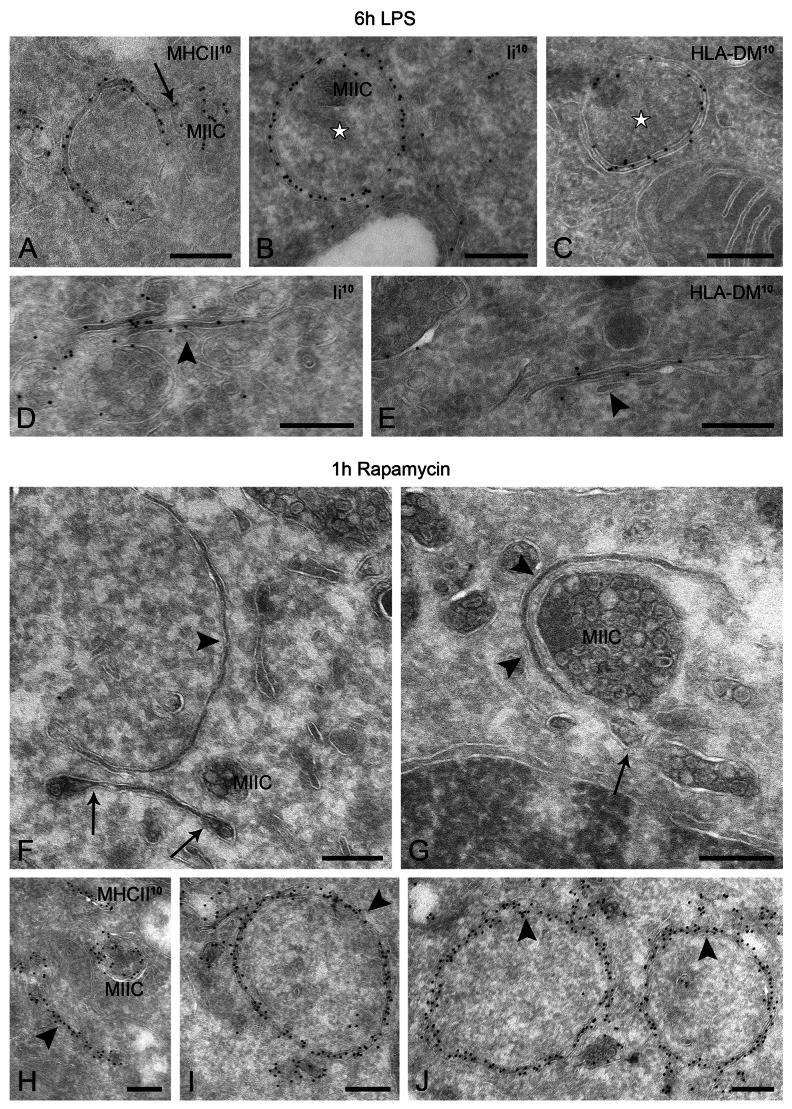
To characterize the molecular composition of the double membranes, we next performed a detailed IEM analysis on ultrathin cryosections. Abundant MHC class II labeling was found on these membranes (Fig. 3A), suggesting that they originate from the MIIC compartments. In line with the EM tomography, we found connections between the concave membrane structures and the multivesicular MIIC profiles (Fig. 3A, arrow). Other molecules known to be involved in MHC class II loading, such as HLA-DM and Ii, were also abundantly present on seemingly closed (Fig. 3B and C), as well as on nascent (open) double-membrane profiles (Fig. 3D and E).
Figure 3. LPS- and rapamycin-induced autophagosome-like structures contain MIIC markers. WT D1 cells were stimulated with LPS for 6 h (A–E) or treated with rapamycin for 1 h (F–J), processed for IEM, and labeled as indicated on the figures. (A) MHC class II-positive double-membrane structure connected to a MIIC (arrow) and engulfing cytoplasmic material. (B–E) Representative electron micrographs showing localization of invariant chain (Ii) and HLA-DM on autophagosomes (stars) and putative phagophores (arrowheads). (F–J) Phagophore and closed double membranes formed upon treatment with rapamycin around cytoplasmic material and organelles (arrowheads). Note the membranes extending from multivesicular compartments (arrows in F and G), likely MIICs. (H–J) Similar to the LPS-induced ones, these double membranes (phagophores and autophagosomes) are abundantly labeled for MHC class II. Scale bars: 200 nm.
To test whether the double-membrane structures share morphological and molecular features with autophagosomes, WT D1 cells were nutrient deprived (D-PBS) or treated with the autophagy-inducing drug rapamycin. Indeed, similar double-membrane structures were encountered under these conditions (Fig. 3F and G; Fig. S1A), which were labeled for MHC class II (Fig. 3H–J; Fig. S1B). To examine whether this is also true in primary cells, we cultured human monocyte-derived DCs (Fig. S1C and S1D) and mouse bone marrow-derived DCs (not shown) under autophagy-inducing conditions. Despite differences in their general MIIC architecture as compared with D1 cells, typical double-membrane structures reminiscent of autophagosomes were also detected in primary DCs. As in D1 cells, these structures often exhibited an electron-dense lumen (i.e., the space between the two limiting membranes) and were positive for MIIC markers, such as MHC class II (not shown) and the late-endosomal marker CD63 (Fig. S1C and S1D).
Considering recent reports that several organelles, including ER and mitochondria, can supply membrane for autophagosome formation in mammalian cells, we next examined whether markers for various membrane compartments could be detected on the LPS-induced phagophores and double membranes. These included EEA1 and transferrin receptor for early endosomes, TGOLN2/TGN38 and GOLGA2/GM130 for Golgi membranes, LMAN1/ERGIC53 for the ER-Golgi intermediate compartment, P4HB (protein disulfide isomerase, PDI) and KDEL for the ER and clathrin. None of these markers, however, was found specifically associated with the autophagic structures (Fig. S2A–S2D and not shown). Although ER cisternae were often in close proximity to the double membranes, no connections were observed between them in our tomograms. When autophagosome degradation was inhibited by BAFA1, P4HB-labeled ER membranes could be recovered in autolysosomes, indicating autophagic turnover of ER membranes (Fig. S2B). Additionally, we performed labeling for peroxisomes, which in D1 cells emerge from specialized ER subdomains through lamellar extensions30 that show striking structural similarity to autophagic membranes on EM micrographs. Although the peroxisome marker PEX13 was readily associated with the lamellar ER subdomains that were always next to peroxisomes, it was never found on MIIC-connected double membranes (Fig. S2E and S2F). The reverse has been shown for MHCII.31 Taken together, these results suggest that LPS stimulation drives the formation of autophagosomes from MIICs similar to these induced under conditions triggering the classical macroautophagy pathway.
Autophagosomal markers are associated with MIIC-connected double membranes
To test whether the double membranes that are connected to MIICs also contain the classical autophagy machinery, we transduced D1 cells to produce mGFP-MAP1LC3A/LC3A (for simplicity referred to as GFP-LC3 hereafter) using a retroviral expression system. LC3 is a cytosolic protein that, after cleavage and lipidation, generates the LC3-II form, which becomes associated with the autophagosome membranes soon after they form and remains attached until its degradation.25 GFP-LC3-expressing D1 cells behaved similar to WT D1 upon treatment with LPS, BAFA1 and LY294002 (Fig. S3A–S3D). Moreover, the molecular machinery involved in antigen processing, i.e., MHC class II (Fig S3E and S3F), HLA-DM and Ii (Fig. S3G–S3J), was equally present on phagophores and seemingly ‘closed’ double membranes.
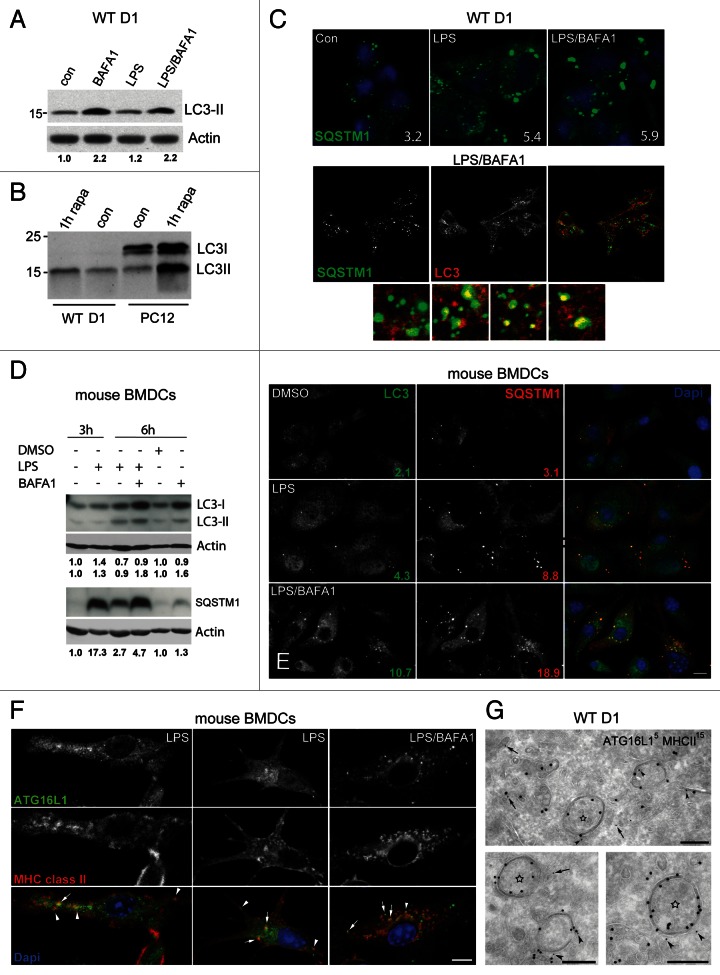
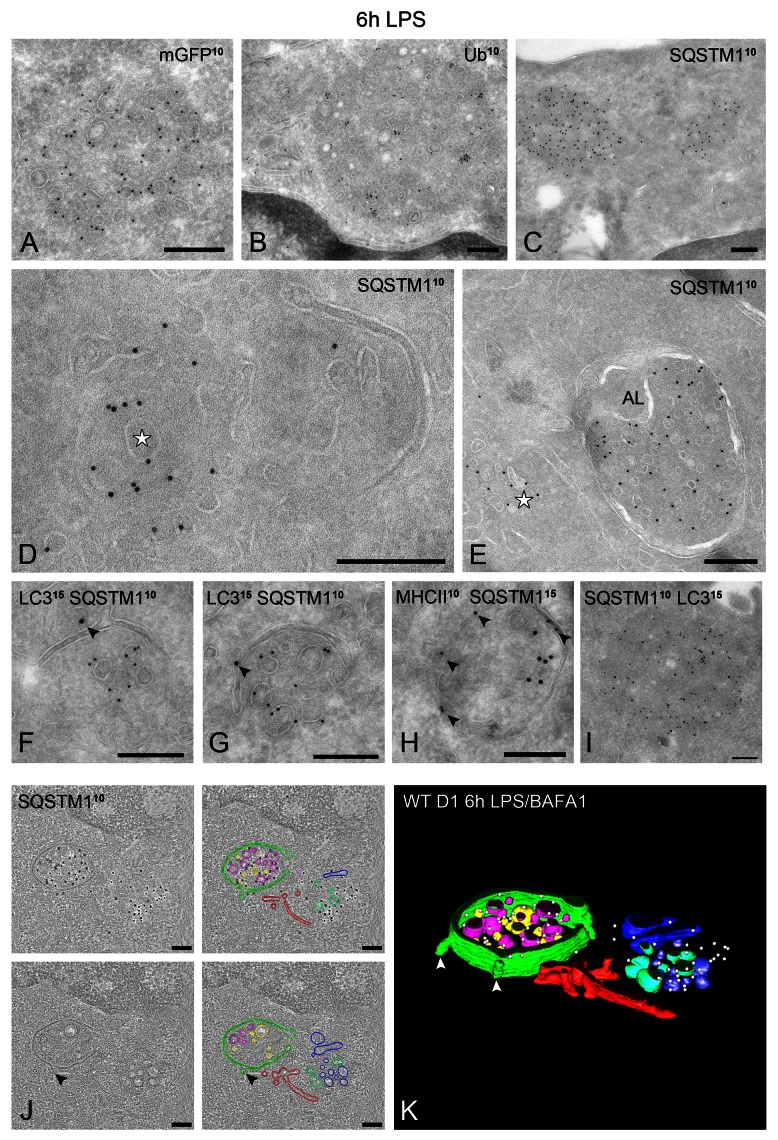
To monitor autophagy induction, we next analyzed by immunoblotting the levels of the cleaved and lipidated (LC3-II) and nonlipidated (LC3-I) form of LC3 in the presence or absence of BAFA1.32 As previously described for APCs,20 WT D1 cells exhibit a very active basal autophagy as indicated by the 2-fold increase in LC3-II upon BAFA1 treatment for 6 h (Fig. 4A), and the virtual absence of LC3-I as compared with PC12 cells that was used as control cell line (Fig. 4B). In agreement with the EM quantification, LPS alone induced only a small increase in the basal LC3-II levels, while this increase became pronounced upon cotreatment with BAFA1 (Fig. 4A). Similar observations were made in GFP-LC3 D1 cells using an anti-GFP or anti-LC3 antibody (Fig. S4A and S4B). To confirm these results in another DC type, we performed similar experiments in mouse primary bone marrow-derived DCs (BMDCs). In these cells, LC3-II appeared significantly increased upon incubation with BAFA1 alone and even higher when treated with LPS/BAFA1 (Fig. 4D). BAFA1 also led to an increase in LC3-I both in BMDCs (Fig. 4D) and GFP-LC3 D1 cells (Fig. S4A, lane 5). In addition, we quantified the number of SQSTM1 cytoplasmic aggregates, which are known substrates for autophagy33,34 (see below). SQSTM1 aggregates increased in number upon LPS stimulation both in D1 cells (Fig. 4C) and BMDCs (Fig. 4E), presumably due to its transcriptional upregulation induced by LPS.35 However, upon combined LPS/BAFA1 treatment, their number (Fig. 4C and E), as well as SQSTM1 total protein levels (Fig. D), increased further suggesting that their clearance takes place through autophagy.
Figure 4. Behavior of autophagosomal markers and SQSTM1 upon LPS and LPS/BAFA1 stimulation in WT D1 cells and BMDCs. (A and B) Representative immunoblot detection of endogenous LC3 in WT D1 cells treated for 6 h as indicated. Actin was used as loading control. Band intensities were quantified using ImageJ software, and the numbers represent fold increase compared with the ratio of the vehicle-treated sample that was set at 1.0. Note that LC3-II increases slightly upon LPS stimulation but is much stronger after incubation with BAFA1. (B) Endogenous LC3 in WT D1 and PC12 cells treated for 1 h as indicated. Note the absence of LC3-I in D1 cells. (C) Confocal images of WT D1 treated for 6 h as indicated, and quantification of SQSTM1 dots Lower panel, localization of LC3 and SQSTM1 upon LPS stimulation for 6 h in the presence of BAFA1. Colocalization of LC3 with SQSTM1 is shown in the small insets. (D) Immunoblot detection of LC3 and SQSTM1 in BMDCs treated as indicated. Quantification of LC3-I/actin, LC3-II/actin and SQSTM1/actin ratios was performed as in (A). (E) Representative confocal images and quantification of LC3 and SQSTM1 puncta in BMDCs. (F) BMDCs stimulated with LPS or LPS/BAFA1 for 3 h showing partial colocalization of ATG16L1 with MHC class II (arrows). Arrowheads mark puncta positive for the two proteins that are very closely positioned but not overlapping. (G) Representative electron micrographs showing low but specific labeling of ATG16L1 (5 nm gold) on MHC class II (15 nm gold) positive autophagosomes (arrowheads). A small fraction of ATG16L1 is localized on isolated vesicular structures (arrows) found in close proximity to the double membranes (star). Scale bars: 200 nm.
To study the distribution of LC3 in relation to MIICs and SQSTM1 aggregates upon LPS stimulation, we performed confocal microscopy. In D1 cells, GFP-LC3 puncta partially colocalized with MHC class II (~50%), and this became more prominent in the presence of BAFA1 (Fig. S4C). The same was true for endogenous LC3 and SQSTM1 (Fig. 4C). In BMDCs, the overlap between LC3 and SQSTM1 dots was higher, ranging from 72% to 95% in cells stimulated with LPS- and LPS/BAFA1, respectively. These data combined with the EM analyses suggest that the number of double-membrane structures observed upon incubation with LPS or LPS/BAFA1 correlate well with the increased level of LC3-II and the LC3 fluorescent puncta, indicating activation of autophagy.
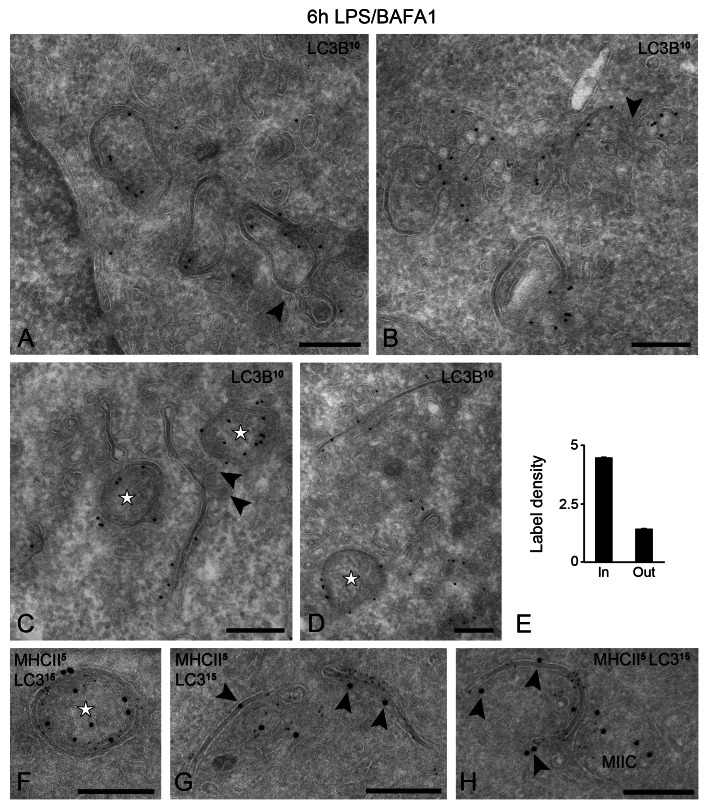
To better correlate these fluorescent distribution patterns to the double-membrane structures observed by EM, we performed a detailed IEM analysis both in WT and GFP-LC3-expressing cells. We have used an anti-LC3B antibody to detect both endogenous and GFP tagged LC3, and avoid potential artifacts, for instance from partial cleavage of the GFP tag from the GFP-LC3 chimeric protein. While a portion of LC3 was cytosolic, a significant fraction was specifically associated with both nascent and seemingly ‘closed’ double membranes that were connected to MIIC profiles (Fig. 5A–D). LC3 appeared to be preferentially associated with the convex membrane (Fig. 5E). In addition, LC3 was confined to cytoplasmic aggregate-like structures, which contained membrane vesicles (Fig. 5B) and accumulated when autophagic turnover was inhibited by BAFA1. Finally, LC3 was present in the membrane-bound autolysosomes, where it remained associated with these aggresome-like structures (not shown). Because GFP-LC3 expression has been associated with artifacts,36 we also detected endogenous LC3 in WT D1 cells, which exhibited a similar distribution pattern (i.e., associated with developing phagophores and MIIC limiting membrane), albeit at lower labeling density (Fig. S5). Since MIICs have been described to change their shape dramatically upon LPS stimulation,27,28 LC3-positive tubular profiles could be distinct from the MHC class II-positive ones. To this end, we performed immunogold double labeling and confirmed that both MHC class II and LC3 colocalize on the same phagophore membrane and double- membrane structures (Fig. 5F–H).
Figure 5. LC3 is enriched on the MIIC-derived autophagosomes. GFP-LC3 D1 cells were stimulated with LPS/BAFA1 for 6 h. (A–D) Thin cryosections were immuno-labeled with an anti-LC3B antibody. Representative electron micrographs are shown. Note the connections between double-membrane structures and MIIC profiles (arrowheads in A and B). (C and D) Putative phagophore membranes enriched in LC3. Vesicular continuities are frequently located at the concave site of the phagophore membrane (arrowheads in C). Stars indicate autophagosome profiles with ‘closed’ double membranes. (E) LC3 labeling density on convex (In) and concave (Out) side of the double membranes. (F–H) Immunogold double labeling of MHC class II (5 nm gold) and LC3 (15 nm gold) on autophagosome (F) and tubular (G) and concave (H) phagophore membranes. Scale bars: 200 nm.
Since LC3-II is present on mature autophagosomes and autolysosomes (late marker) rather than the phagophores,37 we also examined the localization of the more ‘upstream’ autophagic markers ATG16L1 and ATG12. These two proteins together with ATG5 form a complex that localizes predominantly to the phagophores,38 while ATG16L1 has recently been attributed an even earlier function.12 In line with their previously described role, LC3 exhibited partial colocalization with ATG16L1 and ATG12 in BMDCs (Fig. S4E). A fraction of ATG16L1 was also colocalizing with MHC class II by confocal microscopy (Fig. 4F). Similarly, IEM analysis showed that ATG16L1 was associated with the MHC class II-positive phagophore membranes, albeit at a low density (Fig. 4G). Interestingly, the majority of ATG16L1-positive fluorescent puncta, although very closely positioned, did not fully overlap with MHC class II puncta (Fig. 4F). As suggested by IEM, these large ATG16L1 puncta most likely represent the localization of the protein on cytoplasmic vesicular aggregates (Fig. S4F). Occasionally, small isolated vesicles in close proximity to double membranes were also labeled (Fig. 4G, arrow). Together, these data further suggest that the MHC class II-positive double membranes represent autophagosomes.
DALIS are cytosolic LC3 and SQSTM1-containing membrane assemblies that are cleared by the MIIC autophagosomes
To investigate whether the LC3-positive vesicular membrane aggregates are DALIS, we performed immuno-labeling for ubiquitin, whose LPS-induced aggregates in DCs define DALIS.21,22 Indeed, LPS-induced ubiquitin-positive dots were largely colocalizing with GFP-LC3, especially the larger ones, which were mostly distinct from the MHC class II-positive compartments (Fig. S4D). This suggests that the part of LC3 that does not colocalize with MHC class II is confined to DALIS. Similar to GFP-LC3 puncta, the ubiquitin-positive structures increased in number upon BAFA1 treatment (not shown). DALIS were equally detected in WT and GFP-LC3 D1 cells, and were nearly exclusively observed after stimulation with LPS, but not after starvation or treatment with rapamycin. IEM confirmed the concentration of LC3 and ubiquitin on the vesicle-containing cytoplasmic aggregates that are not enclosed by a limiting membrane (Fig. 6A and B). To our knowledge, the presence of membranes in DALIS has not been described before.
Figure 6. LPS-induced DALIS are cytosolic membrane assemblies that contain ubiquitinated proteins, LC3 and SQSTM1. (A–C) Immuno-localization of GFP-LC3, ubiquitin and SQSTM1 on DALIS. Note that the labeled structures consist of aggregated membranes embedded in a more electron-dense material compared with the surrounding cytoplasm. No limiting membrane is visible. (D) SQSTM1-positive DALIS (star) in close proximity to a putative phagophore. (E) SQSTM1 in cytosolic DALIS (star) and recovery in autolysosome (AL). (F–I) Immunogold double labeling as indicated on the figures. (J and K) EM tomography of SQSTM1-positive DALIS. (F). Four tomographic slices extracted from a 250 nm thick cryosection immuno-labeled for SQSTM1 (10 nm gold). The gold particles are confined to the top slices. (G) Reconstructed model showing the SQSTM1-positive membrane assemblies surrounded by a cup-shaped autophagosome (green). Arrowheads indicate vesicular membranes emerging from or fusing with the autophagosome. See also the corresponding Video S6. Scale bars: (A–I) 200 nm; (J) 165 nm.
A key component regulating the formation of ALIS (the DALIS equivalent in all other cell types) in macrophages upon LPS stimulation is SQSTM1.35 SQSTM1 is a multifunctional protein that undergoes self-oligomerization and acts as an autophagy adaptor by aggregating ubiquitinated proteins leading them to emerging autophagosomes through interaction with LC3. As previously mentioned, in this process, SQSTM1 itself becomes a substrate for autophagy.33,34 We performed IEM to test whether SQSTM1 was also associated with DALIS. As suggested by our IF data, SQSTM1 was exclusively confined to cytosolic DALIS (Fig. 6C, D and I) and the membrane-bound autolysosomes (Fig. 6E).
LC3 and SQSTM1-positive vesicular assemblies/DALIS were often situated in close proximity or surrounded by double membranes (Fig. 6D), which were positive for LC3 (Fig. 6F and G) and MHC class II (Fig. 6H), suggesting their selective turnover by autophagy. EM tomography combined with SQSTM1 immunolabeling confirmed that DALIS-associated membranes are vesicular in nature and become enwrapped by the cup-shaped autophagosomes (Fig. 6J and K; Vid. S6). Altogether, these results are consistent with the idea that cytosolic or membrane-associated ubiquitinated proteins formed upon LPS stimulation are aggregated by SQSTM1 (and perhaps other homologous proteins) leading to the formation of DALIS, which are selectively cleared by autophagy.39 Furthermore, as recently suggested for macrophages,35 our results indicate that LC3 could be recruited to DALIS independently of its association and role in autophagosome formation.
Because none of the proteins we had localized in the vesicle-containing DALIS (LC3, SQSTM1, ubiquitin and ATG16L1) contained a transmembrane domain, we were not certain whether they were bound to the observed membranes or the electron dense cytosolic material. This raised the question of the identity and role of the vesicular membranes. One possibility is that this membrane pool contributes to the expansion of MIIC-built autophagosomes. To this end, we examined the localization of ATG9, a transmembrane protein essential for autophagy that has been localized to vesicular-tubular clusters in yeast,40 and cycles between TGN and endosomes in mammalian cells.41 Accordingly, ATG9 in D1 cells was found in vesicles situated in close proximity to the TGN and to a lesser extent on DALIS membranes (Fig. S6A and S6B). Interestingly, ATG9-positive vesicles seem to fuse or bud off from the MIIC-associated autophagosomes (Fig. S6A, inset), a feature often observed also in our tomograms (Fig. 6J and K). In addition, we examined the localization of VTI1B (vesicle transport through interaction with t-SNAREs homolog 1B), a Q-SNARE implicated in early42 and late stages43 of autophagic fusion events. As shown previously, VTI1B was associated with small vesicular structures localizing at the TGN area. Strikingly, a significant portion of VTI1B was confined to DALIS-associated membranes (Fig. S6C and S6D) and vesicles in the vicinity of forming MIIC autophagosomes (Fig. S6E and S6F). These results indicate that at least part of the membrane vesicles localizing in DALIS may contribute to MIIC-based autophagosome expansion.
MIIC-driven autophagosome formation, but not DALIS-SQSTM1 turnover, is independent of ATG4B and LC3-II
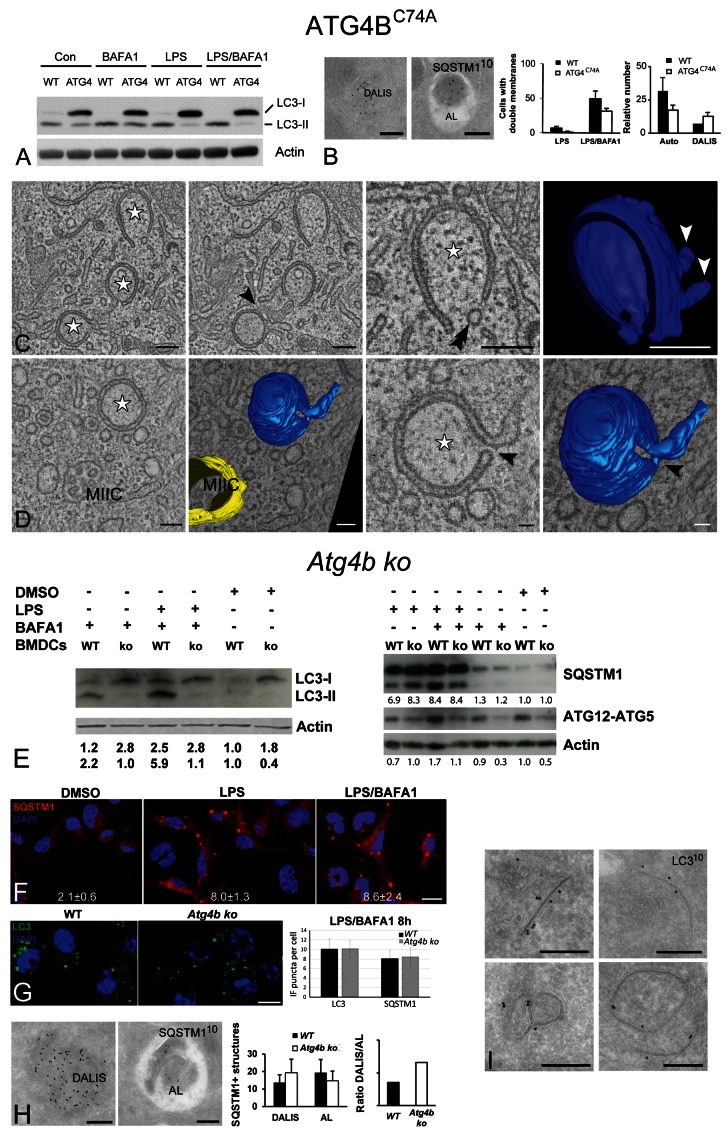
To test whether we can affect autophagy turnover by manipulating experimentally the MIIC-derived autophagosomes, we transduced D1 cells to express mStrawberry-ATG4BC74A. Overexpression of this dominant-negative ATG4B mutant has been shown to sequester and block the C-terminal cleavage of LC3, thereby inhibiting its lipidation and membrane association and leading to a defect in autophagosome closure and degradation.44,45 Indeed, ATG4BC74A D1 cells showed a clear defect in LC3 cleavage and lipidation, as indicated by the marked increase in LC3-I, both under basal conditions and upon LPS stimulation (Fig. 7A). Although upon EM quantification the number of cells containing double membranes, as well as their number per cell, were decreased in the ATG4BC74A mutant, this was not statistically significant. In addition, IEM analysis showed that SQSTM1-positive autolysosomes were decreased by 35%, and this was counteracted by a slight increase in the number of SQSTM1-positive autolysosomes (Fig. 7B). Moreover, no significant differences were encountered in the percentage of “closed” autophagosomes over total autophagic structures (37 ± 6% in WT and 33 ± 15% in the ATG4B mutant cells, n = 90 cell profiles). Upon EM tomography analysis, the LPS/BAFA1-treated ATG4BC74A D1 cells exhibited phagophore and double membranes that were connected to MIICs, essentially as in WT D1 cells (Fig. 7C and D; Vids. S7 and S8). Open vs. closed profiles were equally distributed in the tomograms (n = 8 for WT and n = 7 for mutant cells), and fusion/fission profiles and small openings on nascent double membranes (Fig. 7C and D, arrowheads) were as frequently observed in both cells.
Figure 7. Effect of ATG4BC74A overexpression and ATG4B deficiency upon LPS stimulation. (A) Immunoblot detection of endogenous LC3-I, LC3-II and actin in whole-cell lysates prepared from WT and ATG4BC74A D1 cells treated as indicated. (B) IEM localization of SQSTM1 in DALIS and autolysosomes (AL) in ATG4BC74A D1 cells. Quantification of the number of double membranes (left) and SQSTM1-positive structures in WT and ATG4BC74A-expressing cells. Data represent the mean of 3 independent experiments each representing 50 cells ± SD. The observed differences are not statistically significant. (C and D) Tomographic slices and 3D models of MIIC-associated double membranes in ATG4BC74A-expressing D1 cells, stimulated with LPS/BAFA1 for 6 h. (C) Cup-shaped phagophore membrane connected to a MIIC structure (black arrowheads). Double arrowhead indicates a vesicle closely located position to the expanding double membrane. White arrowheads in model indicate fusion/fission profiles extending from the outer phagophore membrane. (D) Tomographic slices and 3D models of a MIIC (yellow) and double-membrane structure (blue). Note the narrow opening close to the tubular membrane extension (arrowhead). See also corresponding Videos S7 and S8. (E) Immunoblot detection of LC3-I and II, SQSTM1 and ATG12–ATG5 complex in lysates from WT and Atg4b ko BMDCs treated as indicated. Band quantification was performed as in Figure 4. (F) IF localization of SQSTM1 in Atg4b ko BMDCs treated as indicated for 4 h. The average number of SQSTM1 fluorescent puncta per cell ± SD appears at the bottom of each figure (n = 3). (G) IF localization of LC3 and quantification of LC3 and SQSTM1 puncta in WT and Atg4b ko BMDCs treated with LPS/BAFA1 for 8 h. (H) IEM of SQSTM1 in DALIS and autolysosomes (AL) in Atg4b ko BMDCs. Left: Quantification of SQSTM1-positive structures in WT and Atg4b ko BMDCs. (I) LC3 detection on phagophores and double membranes of Atg4b ko BMDCs. Scale bars: (B, C, H and I) 200 nm; (D) 200 nm (left) and 55 nm (right); (F and G) 10 µm.
An explanation for this mild phenotype of ATG4BC74Aon MIIC-driven autophagosomes could be the variable expression levels of the mutant protein in different cells. To clarify this issue, we obtained bone marrow cells from Atg4b ko mice and littermate controls, differentiated them into BMDCs,46 and stimulated them with LPS in the presence or absence of BAFA1. As originally reported,46 ATG4B ablation led to a nearly complete inhibition in LC3-I to LC3-II conversion (Fig. 7E). In contrast, however, to the cytosolic redistribution observed in atg4b−/− MEFs, LC3 still exhibited a punctate localization pattern both in WT and ATG4B-deficient BMDCs (Fig. 7G). To further test whether autophagy is impaired, we examined the formation of the ATG12–ATG5 complex. As shown in Figure 7E, the formation of this complex was severely impaired in Atg4b ko BMDCs compared with the WT cells. Moreover, we assessed the turnover of SQSTM1 levels upon LPS stimulation in the presence or absence of BAFA1. Upon LPS stimulation for 8 h, SQSTM1 accumulated to a larger extent in Atg4b ko cells compared with WT cells, and the addition of BAFA1 did not increase it further, in contrast to the WT DCs (Fig. 7E). Similar results were obtained by IF quantification of the number of SQSTM1 aggregates, which did not increase further in Atg4b ko cells in the presence of BAFA1 (Fig. 7F). EM analysis also showed that, although there was no significant change in the total number of SQSTM1-positive structures (cytosolic DALIS and autolysosomes), there was a difference in their relative abundance leading to a nearly 2-fold increase in DALIS/autolysosome ratio (Fig. 7H). Taken together, these results suggest that autophagy is largely inhibited in ATG4B-deficient DCs, similar to what has been recently reported in atg4b−/− macrophages.47
Finally, as mentioned above, despite the fact LC3 was present almost exclusively in its non-lipidated LC3-I form, atg4b−/− BMDCs still exhibited LC3 fluorescent puncta colocalizing with SQSTM1, and their number was comparable to that observed in WT cells (Fig. 7G). To address the nature of LC3 fluorescent puncta, we performed IEM. Similar to ATG4BC74A D1 cells, ATG4B-deficient BMDCs still exhibited double-membrane profiles upon LPS/BAFA1 incubation. The low endogenous LC3 labeling was confined to MIICs and DALIS (not shown). Surprisingly, however, LC3 also appeared to be equally associated with the MIIC-driven double membranes in atg4b–/– BMDCs (Fig. 7I), as in WT DCs. These membranes were also positive for MHC class II (not shown). Although unconventional, these observations are in agreement with the study of Fujita et al.35 showing that in macrophages expressing a LC3 mutant that cannot be lipidated, LC3 fluorescent puncta were still colocalizing to SQSTM1-positive ALIS. Whether this membrane association of LC3-I is a peculiarity of DCs and other immune cells or reflects an additional autophagy-independent role of LC3 will require further investigation in the future.
Discussion
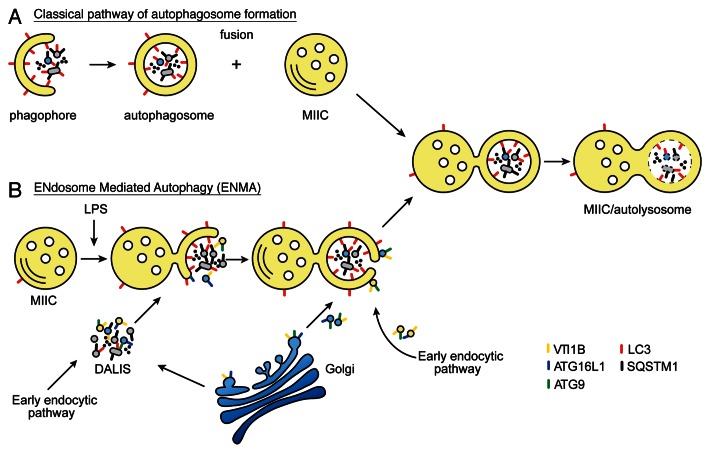
Activation of various PRRs, including TLR4, induces autophagy in antigen-presenting cells.14 Using a combination of high-resolution microscopy approaches (EM, IEM, IF and tomography), we have performed a detailed characterization of the autophagosome formation in DCs, showing for the first time that they emerge from the reorganization of the MIIC limiting membrane. Although we cannot formally exclude that some MIIC-connected double membranes represent amphisomes, the “hybrid organelles” deriving from the fusion of de novo formed autophagosomes with late endosomes (Fig. 8, model A),27,37 we believe that this is not the most common mechanism in these cells. Amphisomes are formed when completely closed autophagosomes fuse with late endosomes/lysosomes, so unless nascent open double membranes are able to fuse with endosomes in DCs, this means by definition that the structures we observed are not amphisomes. Engulfment of cytosolic material by a late endosomal compartment would be more consistent with microautophagy described extensively in yeast, and recently reviewed by Mijaljica et al.48 Because the etymology and original definition of microautophagy refers to small vesicles budding inwards from endosome limiting membranes, and due to the fact that we observed large portions of cytoplasmic material being sequestered (up to 1µm in diameter), we have introduced the generic term ENdosome- Mediated Autophagy (ENMA) (Fig. 8, model B).
Figure 8. Classical and proposed model of unconventional autophagosome formation in DCs. (A) During classical autophagosome formation, the phagophore membrane is built up from not yet clearly defined membrane sources in the cytoplasm. Several organelles have been proposed as potential membrane delivery vehicle. Cytoplasmic material and complete membrane-bound organelles become trapped in the cavity of the autophagosomes. Degradation of the engulfed cargo is achieved after fusion of the autophagosome with the MIIC. The molecular machinery present on the MIIC redistributes toward the autophagosomal membrane before degradation begins. (B) Proposed model of the unconventional, ENdosome Mediated Autophagy pathway. Newly formed sequestration membranes emerge from reorganization of the MIIC limiting membrane around SQSTM1- and LC3-positive, vesicle-containing aggregates (DALIS) that are formed upon LPS stimulation. Selective accumulation of VTI1B, ATG9 and ATG16L1 membrane carriers within these DALIS, possibly deriving either from TGN (blue vesicles) or the early endocytic pathway (yellow vesicles), could contribute to the delivery of membrane for the expansion of the MIIC-driven double membranes. Part of DALIS-associated membranes could also originate from damaged organelles (gray vesicles) that need to be eliminated. After being nearly or completely closed, these structures appear as amphisomes in ultrathin EM sections. However, they remain attached to the MIIC and become degraded in time. In addition to LPS stimulation, ENMA is likely to take place constitutively and upon starvation or rapamycin treatment in DCs without SQSTM1 being highly expressed.
Formation and molecular composition of MIIC-deriving autophagosomes in DCs
Several pieces of evidence suggest that the double-membrane structures represent various stages of autophagosome formation. First, the number of both early (phagophores) and late (closed double membranes) autophagosomal structures and autolysosomes was altered upon treatment with drugs known to affect autophagic flux. Second, nutrient starvation and rapamycin treatment, established conditions inducing autophagy,25 led to the formation of double-membrane structures with similar morphological and molecular characteristics. Third, both LC3 (a marker of mature autophagosomes/autolysosomes) and ATG16L1 (a more “upstream” marker of nascent autophagosomes) localized on these membranes. Fourth, upon LPS stimulation, these structures preferentially engulf cytoplasmic membrane assemblies positive for the autophagic substrate SQSTM1, which are also recovered in autolysosomes. Last, ATG4B-deficient BMDCs show impaired formation of LC3-II and the ATG12–ATG5 complex, increased SQSTM1 accumulation in the absence of BAFA1, and higher DALIS/autolysosome ratio, when compared with WT cells. Since no significant difference was observed by EM in the abundance of double membranes in these cells, these results indicate that the autophagy block is due to an impairment in late stages of double membrane closure or degradation, rather than in their formation/expansion. Similar results and conclusions have been reported in atg4b−/− macrophages,47 ATG4C74A-expressing cells45 and atg3−/− MEFs.49
Although the MIIC-associated autophagosomes share several characteristics with classical autophagosomes, they also exhibit unconventional ones. For instance, on EM micrographs, they display an electron-dense lumen (i.e., the space between the two limiting membranes), whereas classical autophagosomes are typically electron-lucent.9,25,50 This feature may be APC-, or even DC-specific, as very similar profiles of autophagosomes are illustrated in human DCs stimulated with a NOD2 agonist.51 The reason for this could be the different lipid/protein composition of the double membranes. The classical autophagosome membranes are known to be rich in lipids,52 which are not well preserved upon fixation and processing for EM. On the other hand, the high content of MIIC autophagosomes in transmembrane proteins (MHC class II, Ii) could make their structure more resistant to lipid extraction. Furthermore, another issue for future study is how the MIIC-driven autophagosome matures into an autolysosome to allow degradation of the engulfed material, considering that the MIIC limiting membrane is usually protected against proteolytic degradation through a thick glycocalyx.53
The origin of the membrane required for the nucleation and expansion of autophagosomes is currently under intense research. Several groups have shown autophagosomes emerging from ER membrane subdomains,8-10 while a high-resolution live cell imaging study demonstrated the involvement of outer mitochondrial membrane in supplying membrane for the autophagosome expansion through transient connections with the forming organelles.11 Plasma membrane has also been shown to contribute to this process.12 In addition, data from yeast and Dictiostelium have suggested that membrane transport from the trans-Golgi and late endosomes contribute to autophagosome biogenesis.7,54,55 These data imply that the origin of the autophagosomal membrane may vary depending on the functional adaptation of each cell type.
In DCs, both phagophores and closed double membranes were systematically connected to multivesicular MIICs, the late endosomal compartment in these cells. Furthermore, these membranes contain high levels of proteins involved in antigen processing, such as MHC class II, the peptide-loading chaperone HLA-DM and invariant chain (Ii).28,56 At least two possible models could explain how these membranes are formed. The first is an adaptation of the classical autophagy pathway and would predict that membranes of an unknown origin form de novo phagophores that are able to fuse with pre-existing MIICs, followed by rapid redistribution of MIIC molecular components into the autophagic structures (Fig. 8A). For reasons explained above we favor the alternative, not mutually exclusive model, where phagophore membranes emerge from the MIIC limiting membrane (Fig. 8B). Upon LPS stimulation, this could partly involve the structural reorganization of the previously described tubules formed from MIICs.28,29,57,58 Additional membrane could be supplied by membrane carriers trafficking from other organelles. For instance, ATG9 that was previously shown to traffic between the TGN and endosomes41 was found in close proximity and sometimes on vesicles connected to the double-membrane structures. This suggests that the membrane protrusions frequently observed in our tomograms could reflect fusion events of TGN-derived carriers. An alternative macroautophagy, which depends only on a subset of the ATG proteins implicated in the conventional pathway (BECN1/Beclin 1- and ULK1-dependent but LC3-II- and ATG5/ATG7-independent), has also been described in mouse embryonic tissues. This process depends on the late-endosomal marker RAB9,59 suggesting a role for endosomes in membrane delivery. Despite the abundant presence of LC3 and ATG16L1 on the MIIC-derived double membranes, the involvement of other essential ATG proteins, especially those that affect the formation/expansion of autophagosomes, such as BECN1, will have to be addressed in the future.
The use of MIIC membrane for autophagosome formation in DCs is logical, since the cytoplasm of these specialized cells is crowded with these organelles, therefore representing an abundant source of membrane. The involvement of MIICs could also be specifically linked to the main function of DCs in cytosolic protein degradation and presentation of antigenic peptides (see below).
LC3 localization on MIIC-derived autophagosomes and DALIS and its lipidation status
An unexpected observation in our study was that upon LPS or LPS/BAFA1 stimulation for 6 h, the autophagic marker LC3 was localized on the vesicle-containing cytoplasmic aggregates (DALIS), in addition to its distribution to the cytosol, the double membranes and the autolysosomes that has previously been described in other cell types.32 When we compared the number of these aggregates in WT- and GFP-LC3-expressing cells, we found no correlation between their number and the level of LC3, indicating that they are not triggered by LC3 overexpression.
LC3 localization in DALIS appears to be independent of its association with autophagosomes, as we could observe several LC3-positive DALIS without yet being in close proximity or engulfed by MIIC autophagosomes. In addition, both in Atg4b ko BMDCs and ATG4BC74A-expressing D1 cells, where LC3-I conversion to LC3-II is inhibited, the number of LC3 fluorescent puncta was unchanged, suggesting that LC3 can bind to DALIS in its nonlipidated LC3-I form. The same was recently proposed in macrophages, where GFP-LC3 dots were formed independently of the autophagic machinery and its lipidation status, but were cleared through classical macroautophagy.35 These observations are also in line with earlier studies challenging the notion that GFP-LC3 spots correspond only to autophagosomes.36,60
More surprising was our IEM observation that the accumulated LC3-I in Atg4b ko BMDCs was still associated with double-membrane structures as in WT cells, indicating an uncoupling between LC3-II presence and its binding to autophagosomal membranes. One possibility is that some residual LC3-II that is formed, perhaps by other ATG4 homologs, represents the double membrane-associated pool of LC3. However, since characterization of Atg4b ko MEFs has shown that LC3-I is completely redistributed to the cytosol,46 it seems more plausible that this membrane localization of LC3 is specific for immune cells. Indeed, accumulating evidence suggests that ATG proteins, including LC3, may have additional autophagy-independent roles, particularly in immunity. For instance, ATG12–ATG5-ATG16L1 complex was recently shown to play a role in IFNG/IFNγ-mediated host defense against mouse norovirus infection by blocking viral replication.47 Another alternative function of ATG proteins could be in the unconventional secretion of certain alarmins, including IL1B/IL-1β and HMGB1.61 Such alternative pathways involving ATG proteins can exist mutually with endosome-driven autophagy (ENMA) operating in DCs.
Formation and function of DALIS
DALIS formation was first described in DCs as a physiological response to LPS stimulation,21,22 and it has been proposed to function as cytosolic storage for unfolded/defective proteins destined for elimination22 or presentation by MHC class I62 after proteasome-mediated degradation. Equivalent structures (ALIS) have been described since in many cell types, including macrophages,63 under various stress conditions.23,64 Similar aggregates are also formed in vivo upon chronic pathological conditions, such as type II diabetes,65 and microbial infections.66 In these cases, the formation of reactive oxygen species (ROS) seems to drive their formation. Similarly, ALIS formation in macrophages was shown to be triggered by LPS-induced ROS through a MAPK11/12/13/14/p38-NFE2L2/Nrf2-dependent transcriptional upregulation of SQSTM1, which subsequently mediated the recruitment of LC3 to ALIS. This SQSTM1-interacting LC3 pool has been suggested to promote LC3-I conversion to LC3-II during the engulfment of ALIS by autophagosomes.35 A similar mechanism is likely underlying DALIS formation in DCs, since the majority of SQSTM1 is associated with membranous DALIS (83.7%, quantified by IEM), independent of the presence of an MIIC-driven double membrane around them.
Our EM analyses have revealed that DALIS, at least in D1 cells, contain abundant vesicular membranes. The identity of these membranes is currently unknown, as they were negative for all organelle-specific markers we have tested, and were only labeled for SQSTM1, LC3, ubiquitinated proteins, and to smaller extent for ATG16L1. One possibility is that these represent membranes of damaged intracellular compartments, resulting from DC activation, which need to be cleared. Alternatively, they could serve as a membrane pool for autophagosome expansion, thus representing the mammalian equivalent of the ATG9 membrane reservoirs observed in yeast.40 In line with this proposal, ATG9 was partially localized on these vesicles. Supporting this possibility, the size distribution of DALIS membranes (50 to 60 nm) is in the same range as described for the pre-autosomal ATG16L1 vesicles.42 Moreover, Q-SNARE VTI1B, which is upregulated in macrophages upon LPS stimulation,67 and has been implicated in early42 and late43 stages of autophagic fusion, was also confined to these membranes. As already noted, we found repeated membrane fission/fusion profiles particularly on the outer autophagosomal membrane. Whether VTI1B is implicated in the vesicular transfer of ATG16L1 to the expanding MIIC, either directly or via DALIS, remains to be determined. In this context it is noteworthy to mention that VTI1B has been implicated in the tubulation of late endosomes/lysosomes and simultaneous transport of TNF out of the Golgi in macrophages.67 We therefore suggest that VTI1B could also play a role in targeting membrane to the expanding MIIC.
An open issue is whether SQSTM1 could regulate autophagosome formation in addition to its role as adaptor for autophagic degradation of ubiquitinated substrates. SQSTM1 is targeted to early autophagic structures in MEFs. This is mediated through its Phox and Bem1 (PB1) domain in an LC3-II independent way,68 suggesting that it could play an organizational role in autophagosome expansion. In line with this study, overexpression of ATG4BC74A mutant in D1 cells or atg4b deficiency in BMDCs led to an increased ratio of SQSTM1-positive DALIS to autolysosomes, indicating a defective autophagic turnover presumably at a late stage,44,45 but also being compatible with an additional role of SQSTM1 early during the formation of MIIC autophagosomes. In neutrophils, the PB1 domain is important for the targeting of the Phox complex to the phagosomal membrane.69 Similarly, SQSTM1 could be involved in targeting membranes carrying ATG proteins (i.e., ATG16L1) from intracellular compartments, such as TGN, to the expanding MIIC autophagosome.
In summary, our study shows that DC set up a paradigm of a mammalian cell type exhibiting an unconventional, endosome-mediated autophagy (ENMA). The formation of MIIC-derived autophagosomes could mediate different but not mutually exclusive functions. First, upon LPS stimulation, they are likely to be involved in the specific clearance of the SQSTM1/ubiquitin-positive DALIS. In our EM analysis, we could identify all putative stages of this pathway: (1) nascent cytosolic membrane assemblies not surrounded by autophagic membranes, (2) structures located in the cavity of phagophores or closed double membranes, and (3) structures included in autolysosomes, especially in BAFA1-treated cells. These autophagosomes could be one of the cellular responses that protect APCs from cytotoxic by-products accumulated during their activation by pathogens, such as oxidized proteins and lipids caused by increased ROS. Additional survival mechanisms are also operational in LPS-stimulated DCs, as evidenced by a significant increase in mitochondrial SOD2/MnSOD and nitro-tyrosine (our unpublished data). Second, constitutive autophagy in DCs could be important for their function in conferring central and peripheral immune tolerance.70 Finally, they could play an important role in coordinating the process of antigen presentation by both MHC class I and MHC class II molecules and T cell activation. In this regard, a recent in vivo study in mice with DC-specific Atg5 deletion suggested that autophagy is required for MHC class II-mediated antigen presentation.71 Another study demonstrated that activation of intracellular PRR NOD2 also triggers ATG5-, ATG7- and ATG16L1-dependent autophagy, which is required for bacterial elimination and antigen presentation.51 Efficient T cell activation requires timely regulated antigen presentation. The presence of small pores that were observed in several of our tomograms of otherwise fully closed MIIC autophagosomes could represent such a regulatory mechanism. In this way, the autophagosomes may temporarily sequester from the cytosol the material that needs to be processed for presentation at the cell surface, but this process does not proceed until the DC has migrated close to its target T cell. Our prediction would be that only then these pores would close to allow degradation and antigen presentation to proceed.
Materials and Methods
Materials
All reagents were purchased from Sigma-Aldrich unless otherwise stated.
Cell culture
Mouse wild-type dendritic cell line D1 was cultured in Petri dishes essentially as described previously72 except that glutamine was substituted ultraglutamine1 (Lonza, BE17-605E/U1). mGFP-LC3A and mStrawberry-ATG4BC74A-transduced D1 cells were cultured under the same conditions as the WT D1 cells with the addition of 100 μg/ml G-418 (Invitrogen, 10131-027) in the medium.
Primary monocyte-derived human DCs were isolated from peripheral human blood and used immediately. Bone marrow-derived DCs, from WT and Atg4b ko mice (kind gift from Dr. Lopez-Otin) were generated according to Lutz et al.73 Briefly, bone marrow was harvested from femurs of 8- to 10-week-old BALB/c mice. Erythrocytes were lysed with ammonium-chloride-potassium buffer and the remaining cells were cultured in plastic bacteriologic dishes in RPMI 1640 with 10% fetal bovine serum, 2 mM L-glutamine, 50 μM 2-Mercaptoethanol and 20 ng/ml murine granulocyte/macrophage-colony stimulating factor (GM-CSF). The cultures were refreshed with 10 ml of medium containing 20 ng/mL GM-CSF on days 3, 6 and 8. On day 8, the nonadherent DCs were collected and replated, while the experiments were performed on day 10 after washing with PBS.
DNA-constructs and retroviral transduction of DCs
cDNAs encoding green fluorescent protein-LC3A (GFP-LC3) and the dominant negative construct mStrawberry-ATG4BC74A were a kind gift from Dr. Yoshimori. (Osaka University; Suita-Osaka, Japan). The rat Map1lc3a/LC3A cDNA was first subcloned to exchange the dimerizing GFP tag with the monomeric GFP tag (mGFP-LC3A). The tagged cDNAs were amplified by PCR and cloned into pQCXIN (Clontech, 631514), and the sequences were verified. Retroviral particles were produced by 293T Phoenix-Eco cells after cotransfection with the pQ and pCl-Eco vectors (provided by H. Rozemuller, Utrecht Medical Center, Utrecht). Harvested viruses were filtered, mixed with 4 μg/ml polybrene (Sigma, H-9268) and used to infect D1 by spin occulation 7 d after isolation. Transduced D1 cells were analyzed 2 d later by immunofluorescence or FACS, and maintained as stable cell cultures.
LPS stimulation and drug treatment
For DC stimulation, LPS was added to the culture medium at a final concentration of 10 μg/ml. The cells were stimulated with LPS for the time periods indicated, in the presence or absence of 100 nM BAFA1. DC maturation was verified by the cell surface expression of MHC class II as analyzed by FACS or IF. For induction of autophagy, 100 nM rapamycin (final concentration) was added to the culture medium for the time periods indicated. Nutrient deprivation-induced autophagy was achieved by growing the cells in D-PBS (Gibco, 14040-091) for 30 or 60 min. The phosphatidylinositol 3-kinase (PtdIns3K) inhibitor 2-(4-morpholinyl)-8-phenyl-chromone (LY294002) was used at a final concentration of 10 μg/ml.
Antibodies
The following primary antibodies were used in our study: Monoclonal rat anti-mouse MHC class II antibody (M5/114)73 (BD-PharMingen, 556999), rat anti-mouse H2DM (2E5A) directed to the HLA-DM β-chain,28 and rat anti-mouse invariant chain antibody directed to the cytosolic tail (In-1) were gifts from Dr. L. Karlsson (Scripps Research Institute, La Jolla, CA), rat anti-mouse invariant chain antibody, directed to the lumenal part (S23) was a kind gift from Dr P. Cresswell (Dept. of Immune Biology, Yale School of Medicine, New Haven, CT), monoclonal anti-P4HB antibody (protein disulfide isomerase, PDI) was from Stressgen (P4HB, clone ID3), mouse monoclonal anti-KDEL (clone 10C3) (Calbiochem, 420400), mouse monoclonal anti-LC3 (clone 5F10) (Nanotools, 0231-100), rabbit anti-LC3B (Abgent, AP1805a and AP1806a), rabbit anti-GFP (Clontech, 8362-2), mouse monoclonal against mono- and poly-ubiquitinated conjugates (clone FK2) (Enzo Life Sciences, PW8810), goat anti-Actin (clone I-19) (Santa Cruz, sc-1616), mouse monoclonal anti-α-Tubulin (Sigma-Aldrich, T6074), anti-PEX13 was raised against part of the SH3 domain (amino acids 275 to 454) of human PEX13 fused to the N terminus of dihydrofolatereductase, guinea pig anti-SQSTM1/p62 (Progen, GP62-C), mouse anti-ATG9 and anti-VTI1B were kind gifts from Dr. S. Tooze and Dr. Fischer von Mollard, respectively, rabbit anti-ATG16L (Abcam, ab47946), anti-ATG5 (Cell Signaling, 2630) and anti-ATG12 (Cell Signaling, 2011S). Secondary goat anti-guinea pig antibodies conjugated to HRP and donkey anti-guinea-pig Alexa-488 or 594 were from Santa Cruz and Invitrogen, respectively. Goat anti-mouse FITC was from Abcam. The bridging antibodies rabbit anti-mouse IgG and rabbit anti-rat IgG were from Dako.
Immunoblotting
WT, mGFP-LC3A and mStrawberry ATG4BC74A D1 cells were either left untreated (0.1% DMSO or buffer), or treated as indicated. The cells were lysed on ice in 1% Triton-X100, 1mM ethylenediamine tetraacetic acid (EDTA), 20 mM Tris/HCl (pH 7.5), 150 mm NaCl, 10mM N-ethylmaleimide and complete protease inhibitor mix (Roche, 11836170001). Twenty-five microliter aliquots were loaded and fractionated by SDS-PAGE on 10 or 15% gels under reducing conditions and immunoblotted on PVDF membranes (Millipore, IPVH00010). Bands were detected after incubation with primary antibodies (LC3, SQSTM1 or actin) and HRP (Amersham, NA9310V and NA934V) or fluorophore-conjugated secondary antibodies using ECL Plus detection reagent (Amersham, RPN2132) or an Odyssey scanning system (LI-COR Biosciences). Densitometry for LC3-I, LC3-II, SQSTM1 or ATG12–ATG5 was performed using imageJ analysis software (NIH) and the ratio of each protein over actin calculated. The ratio in the control-treated sample for each time-point was set at 1.
Immunofluorescence
WT and transfected D1 cells were fixed with 4% paraformaldehyde in PBS. The cells were washed with PBS and quenched with 50 mM NH4Cl, followed by permeabilization in 0.1% Triton-X-100 and blockage with 0.2% fish-skin gelatin. The cells were stained with primary antibodies, followed by secondary IgGs coupled to Alexa488, 568 or 647 (Molecular Probes). The coverslips were mounted with Vectashield containing Dapi (Vector Laboratories, H-1200) on microscopic slides and images were acquired using a Zeiss LSM-510 confocal microscope (Carl Zeiss). The acquisition settings were kept equal for the different conditions within each experiment.
Immunoelectron microscopy
For IEM analysis, WT and mGFP-LC3A D1 cells were cultured as described above. After drug treatment the cells were fixed in a mixture of 2% paraformaldehyde and 0.2% glutaraldehyde in 0.1 M phosphate buffer. The cells were washed in PBS-lysine to quench free aldehydes, then embedded in gelatin,50 and infiltrated in 2.3 M sucrose, followed by rapid freezing in liquid N2. 50-nm thick cryosections were cut at -120°C using an Ultracut-S ultramicrotome (Leica Microsystems). The sections were collected on formvar-coated grids using a mixture of 1.8% methylcellulose and 2.3 M sucrose,74 and incubated with primary antibodies and proteinA-gold.75 Rabbit anti-mouse- and anti-rat IgG were used as bridging antibodies. Immunogold single and double labeling were performed using 5-nm, 10-nm and 15-nm proteinA-gold.76 After labeling, the sections were fixed with 1% glutaraldehyde, counterstained with uranyl-acetate, and embedded in methyl cellulose-uranyl-acetate. The sections were viewed in a JEOL 1200CX electron microscope (Jeol Ltd.).
For quantification, 50 randomly selected cells were evaluated from three independent labeling experiments and the total number of double-membrane structures and phagophores was determined. The relative distribution patterns of endogenous LC3, SQSTM1, ATG16L1, VTI1B and GFP-LC3 were determined on randomly selected thin frozen sections by counting the number of gold particles associated with the different intracellular compartments. To evaluate the specificity of immunogold labeling of LC3, the density of gold particles over autophagosomes was compared with that of randomly dispersed points (point-hit method).
Electron tomography
For EM tomography, cells were cultured in formvar- and gelatin-coated golden grids.77 After LPS treatment28 the cells adhering to the grids were sandwiched between 2 aluminum cups (flat side toward the grid) and immediately cryo-immobilized using a high-pressure freezer (Leica HPF, Leica Microsystems). The samples were transferred to the freeze substitution apparatus (Leica AFS, Leica Microsystems), containing precooled anhydrous acetone as substitution fluid, and fixed with 0.5% osmium tetroxide and 0.25% glutaraldehyde in anhydrous acetone at a temperature of −90°C for 48 h. The temperature was increased by 1°C/h to −30°C and this temperature was further maintained for 8 h. The substitution media was washed away by anhydrous acetone and the cells and grids were embedded in Epon.
Sections of 200- to 400-nm thickness were collected on 1 hole formvar-coated copper grids, and post stained with uranyl-acetate and lead citrate. Fiducial markers (10- or 15-nm gold particles) were applied to both sides of the section and 1 extra layer of formvar was applied on top of the section for stability. Dual axis tilt series of the autophagic structures were recorded using a Tecnai 20 transmission electron microscope (FEI Company) equipped with a slow-scan CCD camera (Temcam F214, TVIPS GmbH).78,79 Tilt series were acquired using a single axis high tilt tomography holder (model 670, Gatan GmbH) or a Fischione tomography holder (model 2020, Fischione Instruments Inc., K-Vision BV). Double-membrane structures in close proximity to MIICs were selected on thin sections. Next, 250- to 300-nm thick serial sections were prepared and single and serial section tomograms were reconstructed of the double-membrane profiles using the IMOD program package, (http://bio3d.colorado.edu/imod),80,81 essentially as previously described.29 Membranes were traced manually, and videos from the models were made with adobe premiere software.
Statistical analysis
Statistical significance was assessed by two-tailed unpaired Student’s t-tests and is indicated by asterisks in the histograms and figure legends.
Supplementary Material
Acknowledgments
This research was funded by grant ALW 813.08.001 (H.E.vNtP.) from The Netherlands Organization of Scientific Research (NWO). We thank Dr. P. Ricciardi-Castagnoli for kindly providing the D1 cell line, Drs. S. Tooze and G. Fischer von Mollard for antibody gifts, Dr. T. Yoshimori for the GFP-LC3 and mStrawberry-ATG4BC74A constructs, Dr. C.Lopez-Otin for sharing bone marrow from Atg4b ko and WT litter mates. We would like to thank Drs Catherine Rabouille, Fulvio Reggiori and Attila Kovacs for the many fruitful discussions, Dr. E. O’Toole (University of Colorado, Boulder, CO USA) for her advice on 3D tomographic modeling, Marc van Peski and Rene Scriwanek for their help with the preparation of the videos and figures.
H.E.vNtP. was supported by The Netherlands Organization of Scientific Research, grant ALW 813.08.001. V.K. was supported by a Horizon Programme from National Regie-Orgaan Genomics (050-71-029).
Glossary
Abbreviations:
- APC
antigen-presenting cell
- DC
dendritic cell, BMDC, bone marrow-derived DC
- MHC class II
major histocompatibility complex class II
- MIIC
MHC class II containing compartment
- ER
endoplasmic reticulum
- TLR
toll-like receptor
- PRR
pattern recognition receptor
- LPS
lipopolysaccharide
- BAFA1
bafilomycin A1
- PtdIns3K
phosphatidylinositol 3-kinase
- LC3
microtubule-associated protein 1 light chain 3
- ATG16L1
autophagy-related gene 16-like 1 (S. cerevisiae)
- ATG9
autophagy-related 9
- ATG4B
autophagy related 4B, cysteine peptidase
- VTI1B
vesicle transport through interaction with t-SNAREs 1B
- DALIS
dendritic cell aggresome-like induced structures
- EM
electron microscopy
- IEM
Immuno-EM
- SQSTM1
sequestosome 1
- IF
immunofluorescence
Disclosure of Potential Conflicts of Interest
The authors declare to have no competing financial interests.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/24111
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24111
References
- 1.Bryant P, Ploegh H. Class II MHC peptide loading by the professionals. Curr Opin Immunol. 2004;16:96–102. doi: 10.1016/j.coi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–8. doi: 10.1016/S0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 3.Geuze HJ. The role of endosomes and lysosomes in MHC class II functioning. Immunol Today. 1998;19:282–7. doi: 10.1016/S0167-5699(98)01269-9. [DOI] [PubMed] [Google Scholar]
- 4.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Müller M, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005;102:7922–7. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–49. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–5. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 8.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 10.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–5. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 11.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotzer VL, Blum JS. Autophagy and adaptive immunity. Immunology. 2010;131:9–17. doi: 10.1111/j.1365-2567.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–82. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi C-S, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–82. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–21. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 20.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lelouard H, Gatti E, Cappello F, Gresser O, Camosseto V, Pierre P. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature. 2002;417:177–82. doi: 10.1038/417177a. [DOI] [PubMed] [Google Scholar]
- 22.Pierre P. Dendritic cells, DRiPs, and DALIS in the control of antigen processing. Immunol Rev. 2005;207:184–90. doi: 10.1111/j.0105-2896.2005.00300.x. [DOI] [PubMed] [Google Scholar]
- 23.Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–99. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 24.Fengsrud M, Roos N, Berg T, Liou W, Slot JW, Seglen PO. Ultrastructural and immunocytochemical characterization of autophagic vacuoles in isolated hepatocytes: effects of vinblastine and asparagine on vacuole distributions. Exp Cell Res. 1995;221:504–19. doi: 10.1006/excr.1995.1402. [DOI] [PubMed] [Google Scholar]
- 25.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–12. [PubMed] [Google Scholar]
- 27.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–50. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 28.Kleijmeer M, Ramm G, Schuurhuis D, Griffith J, Rescigno M, Ricciardi-Castagnoli P, et al. Reorganization of multivesicular bodies regulates MHC class II antigen presentation by dendritic cells. J Cell Biol. 2001;155:53–63. doi: 10.1083/jcb.200103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Nispen tot Pannerden HE, Geerts WJ, Kleijmeer MJ, Heijnen HF. Spatial organization of the transforming MHC class II compartment. Biol Cell. 2010;102:581–91. doi: 10.1042/BC20100046. [DOI] [PubMed] [Google Scholar]
- 30.Geuze HJ, Murk JL, Stroobants AK, Griffith JM, Kleijmeer MJ, Koster AJ, et al. Involvement of the endoplasmic reticulum in peroxisome formation. Mol Biol Cell. 2003;14:2900–7. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabak HF, Murk JL, Braakman I, Geuze HJ. Peroxisomes start their life in the endoplasmic reticulum. Traffic. 2003;4:512–8. doi: 10.1034/j.1600-0854.2003.00110.x. [DOI] [PubMed] [Google Scholar]
- 32.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–4. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–8. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A. 2011;108:1427–32. doi: 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–8. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 37.Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep. 2011;3:25. doi: 10.3410/B3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 39.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–69. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 40.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–22. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 42.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–17. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–10. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–76. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 45.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–9. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mariño G, Fernández AF, Cabrera S, Lundberg YW, Cabanillas R, Rodríguez F, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang SS, Kim K, Lee W, Lee GR. Aberrant expression of IFN-γ in Th2 cells from Th2 LCR-deficient mice. Biochem Biophys Res Commun. 2012;424:512–8. doi: 10.1016/j.bbrc.2012.06.146. [DOI] [PubMed] [Google Scholar]
- 48.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–82. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 49.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liou W, Geuze HJ, Geelen MJ, Slot JW. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol. 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 52.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4:286–9. doi: 10.4161/auto.5377. [DOI] [PubMed] [Google Scholar]
- 53.Peters C, von Figura K. Biogenesis of lysosomal membranes. FEBS Lett. 1994;346:108–14. doi: 10.1016/0014-5793(94)00499-4. [DOI] [PubMed] [Google Scholar]
- 54.van der Vaart A, Reggiori F. The Golgi complex as a source for yeast autophagosomal membranes. Autophagy. 2010;6:800–1. doi: 10.4161/auto.6.6.12575. [DOI] [PubMed] [Google Scholar]
- 55.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol. 2010;188:527–36. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleijmeer MJ, Raposo G, Geuze HJ. Characterization of MHC class II compartments by immunoelectron microscopy. Methods. 1996;10:191–207. doi: 10.1006/meth.1996.0095. [DOI] [PubMed] [Google Scholar]
- 57.Barois N, de Saint-Vis B, Lebecque S, Geuze HJ, Kleijmeer MJ. MHC class II compartments in human dendritic cells undergo profound structural changes upon activation. Traffic. 2002;3:894–905. doi: 10.1034/j.1600-0854.2002.31205.x. [DOI] [PubMed] [Google Scholar]
- 58.Boes M, Cuvillier A, Ploegh H. Membrane specializations and endosome maturation in dendritic cells and B cells. Trends Cell Biol. 2004;14:175–83. doi: 10.1016/j.tcb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 60.Ciechomska IA, Tolkovsky AM. Non-autophagic GFP-LC3 puncta induced by saponin and other detergents. Autophagy. 2007;3:586–90. doi: 10.4161/auto.4843. [DOI] [PubMed] [Google Scholar]
- 61.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30:4701–11. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herter S, Osterloh P, Hilf N, Rechtsteiner G, Höhfeld J, Rammensee H-G, et al. Dendritic cell aggresome-like-induced structure formation and delayed antigen presentation coincide in influenza virus-infected dendritic cells. J Immunol. 2005;175:891–8. doi: 10.4049/jimmunol.175.2.891. [DOI] [PubMed] [Google Scholar]
- 63.Canadien V, Tan T, Zilber R, Szeto J, Perrin AJ, Brumell JH. Cutting edge: microbial products elicit formation of dendritic cell aggresome-like induced structures in macrophages. J Immunol. 2005;174:2471–5. doi: 10.4049/jimmunol.174.5.2471. [DOI] [PubMed] [Google Scholar]
- 64.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–51. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 65.Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH. Ubiquitinated-protein aggregates form in pancreatic beta-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes. 2007;56:930–9. doi: 10.2337/db06-1160. [DOI] [PubMed] [Google Scholar]
- 66.Kaniuk NA, Lam GY, Ma C, Galindo-Mata E, Jones N, Vallance BA, et al. Citrobacter rodentium infection induces MyD88-dependent formation of ubiquitinated protein aggregates in the intestinal epithelium. J Innate Immun. 2011;3:83–98. doi: 10.1159/000320644. [DOI] [PubMed] [Google Scholar]
- 67.Murray RZ, Wylie FG, Khromykh T, Hume DA, Stow JL. Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis Factor-alpha. J Biol Chem. 2005;280:10478–83. doi: 10.1074/jbc.M414420200. [DOI] [PubMed] [Google Scholar]
- 68.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, et al. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–8. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 70.Amodio G, Gregori S. Dendritic cells a double-edge sword in autoimmune responses. Front Immunol. 2012;3:233. doi: 10.3389/fimmu.2012.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–39. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winzler C, Rovere P, Rescigno M, Granucci F, Penna G, Adorini L, et al. Maturation stages of mouse dendritic cells in growth factor-dependent long-term cultures. J Exp Med. 1997;185:317–28. doi: 10.1084/jem.185.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lutz MB, Kukutsch N, Ogilvie AL, Rössner S, Koch F, Romani N, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/S0022-1759(98)00204-X. [DOI] [PubMed] [Google Scholar]
- 74.Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- 75.Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–35. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slot JW, Geuze HJ. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol. 1985;38:87–93. [PubMed] [Google Scholar]
- 77.Murk JL, Posthuma G, Koster AJ, Geuze HJ, Verkleij AJ, Kleijmeer MJ, et al. Influence of aldehyde fixation on the morphology of endosomes and lysosomes: quantitative analysis and electron tomography. J Microsc. 2003;212:81–90. doi: 10.1046/j.1365-2818.2003.01238.x. [DOI] [PubMed] [Google Scholar]
- 78.Murk JL, Humbel BM, Ziese U, Griffith JM, Posthuma G, Slot JW, et al. Endosomal compartmentalization in three dimensions: implications for membrane fusion. Proc Natl Acad Sci U S A. 2003;100:13332–7. doi: 10.1073/pnas.2232379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murk JL, Lebbink MN, Humbel BM, Geerts WJ, Griffith JM, Langenberg DM, et al. 3-D Structure of multilaminar lysosomes in antigen presenting cells reveals trapping of MHC II on the internal membranes. Traffic. 2004;5:936–45. doi: 10.1111/j.1600-0854.2004.00235.x. [DOI] [PubMed] [Google Scholar]
- 80.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 81.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–52. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.