Abstract
Wholesale depletion of membrane organelles and extrusion of the nucleus are hallmarks of mammalian erythropoiesis. Using quantitative EM and fluorescence imaging we have investigated how autophagy contributes to organelle removal in an ex vivo model of human erythroid differentiation. We found that autophagy is induced at the polychromatic erythroid stage, and that autophagosomes remain abundant until enucleation. This stimulation of autophagy was concomitant with the transcriptional upregulation of many autophagy genes: of note, expression of all ATG8 mammalian paralog family members was stimulated, and increased expression of a subset of ATG4 family members (ATG4A and ATG4D) was also observed. Stable expression of dominant-negative ATG4 cysteine mutants (ATG4BC74A; ATG4DC144A) did not markedly delay or accelerate differentiation of human erythroid cells; however, quantitative EM demonstrated that autophagosomes are assembled less efficiently in ATG4BC74A-expressing progenitor cells, and that cells expressing either mutant accumulate enlarged amphisomes that cannot be degraded. The appearance of these hybrid autophagosome/endosome structures correlated with the contraction of the lysosomal compartment, suggesting that the actions of ATG4 family members (particularly ATG4B) are required for the control of autophagosome fusion with late, degradative compartments in differentiating human erythroblasts.
Keywords: ATG4B, ATG4D, erythropoiesis, electron microscopy, autophagosome, amphisome, mitochondrion
Introduction
Macroautophagy (henceforth simply “autophagy”) is an important catabolic process in which cytoplasm is delivered to lysosomes for degradation and recycling. It contributes to cellular homeostasis, protects cells from a variety of environmental stresses, and plays roles during development and cellular differentiation.1,2 Autophagy has also been implicated in human diseases including cancer, neurodegeneration and infection, and is thought to contribute to life- span extension.3,4 Coordinated by the ATG gene family, autophagy recycles toxic or redundant components by sequestering regions of cytoplasm (including whole organelles) within double- membrane autophagosomes that are trafficked to, and subsequently fuse with lysosomes. Substrates derived from hydrolysis of cytoplasmic material are then retro-translocated into the cytoplasm to meet the cell’s ongoing anabolic needs in times of nutrient and/or environmental stress.
Autophagy can either be nonselective or highly substrate specific. For example, pathways exist for the isolation and degradation of damaged mitochondria (a process known as mitophagy) (see ref. 5). A vital step during autophagosome biogenesis in both selective and nonselective autophagy is the covalent linkage of phosphatidylethanolamine (PE) to the exposed C-terminal glycine of the autophagosome marker Atg8 (the lipidation step). The Atg4 endopeptidase is a crucial regulatory component of this pathway: it primes newly synthesized pro-Atg8 by cleaving at the critical glycine residue; then later deconjugates (delipidates) lipid-bound Atg8 to recycle Atg8 and possibly also to facilitate autophagosomal maturation.6-8 Four paralogs of yeast Atg4 (ATG4A-D6) are expressed in mammals, and these act on several mammalian protein paralogs of yeast Atg8 (e.g., LC3A/B/C (MAP1LC3A/B/C); GABARAP; GABARAPL1/ATG8L; GABARAPL2/GATE-16). Possible redundancy, and/or specialist or tissue-specific roles for ATG4 and ATG8 paralog partners makes interpretation of their individual functions difficult in mammalian systems. Clear examples of redundancy can be seen in the phenotypes of mice genetically deficient in either atg4b or atg4c.7,8 Both have mild phenotypes,7,8 despite quite robust suppression of the autophagic response in cell lysates from the atg4b knockout model.7
Dramatic changes in cellular architecture symbolize the formation of the functional red cell, and these are coordinated by altered patterns of gene expression, controlled by erythroid-specific transcription factors.9 During erythropoiesis the entire organellar content of the nascent erythrocyte is eliminated. Although there is ample morphological evidence for an upregulation of autophagy during erythropoiesis,10-15 mouse genetics has provided the strongest evidence for a functional role for autophagy during erythropoiesis.16-19 This is perhaps best demonstrated by the persistence of mitochondria in mouse knockouts of the key autophagy genes, ulk1 and atg7,16,17,19 and in knockouts of the BH3 (BCL2/Bcl-2 homology domain 3)-only protein, bnip3l/nix.20,21 BNIP3L is a mitochondrial resident thought to trigger mitochondrial depolarization leading to mitophagy.20 bnip3l−/− mice are anemic and display compensatory expansions in erythrocyte precursor numbers, allied to elevated levels of erythropoietin (EPO).20 Surviving erythrocytes in the peripheral circulation retain their mitochondria, but these have reduced life spans due to elevated ROS.20 BNIP3L interacts with ATG8 paralogs including GABARAPL1,22 suggesting a direct link with the autophagosome biogenesis pathway. Importantly, a conditional knockout of ATG7 in the mouse hematopoietic system phenocopies the bnip3l−/− mouse in that erythocytes retain mitochondria, show elevated ROS, and are short-lived causing severe anemia.16,20 Controversy still remains concerning the relevance of autophagy for erythroid organelle clearance, because a RAB9A-dependent organelle clearance pathway has been described in atg5−/− and atg7−/− mice.18
In this study, we have investigated the influence of autophagy during human in vitro erythropoiesis. Our motivation has been to determine whether autophagy contributes to, and can therefore be manipulated to improve, human ex vivo erythroid differentiation. We find that autophagy is indeed functional during human erythropoiesis, and that reducing its efficiency using dominant-negative ATG4 constructs profoundly alters the organellar clearance pathway in late human erythroid cells. Despite this, only minor changes in differentiation profiles are observed. Our approach has also revealed a possible role for ATG4 proteins in mammalian autophagosome maturation, because erythroid cells expressing dominant-negative ATG4 accumulate enlarged autophagosomes and amphisomes that contain undegraded organelle remnants.
Results
Characterizing the declining endomembrane system in human erythroid cells
Refinement of in vitro erythroblast culture protocols has greatly improved the efficiency and reproducibility of production of reticulocyte cell populations from circulating, peripheral adult human CD34+ cells.23,24 Despite this, there are still many questions concerning how closely these in vitro differentiated erythroid cells compare with their in vivo counterparts, particularly in relation to cellular remodeling and the establishment of correct cytoplasmic macromolecular architecture. One of the defining features of mammalian erythrocytes is the absence of organelles. These are dismantled and eliminated progressively during normal differentiation. Autophagy has long been suspected to contribute to this, although specific aspects of the regulation and significance of autophagy during erythropoiesis remain undetermined, particularly in humans.
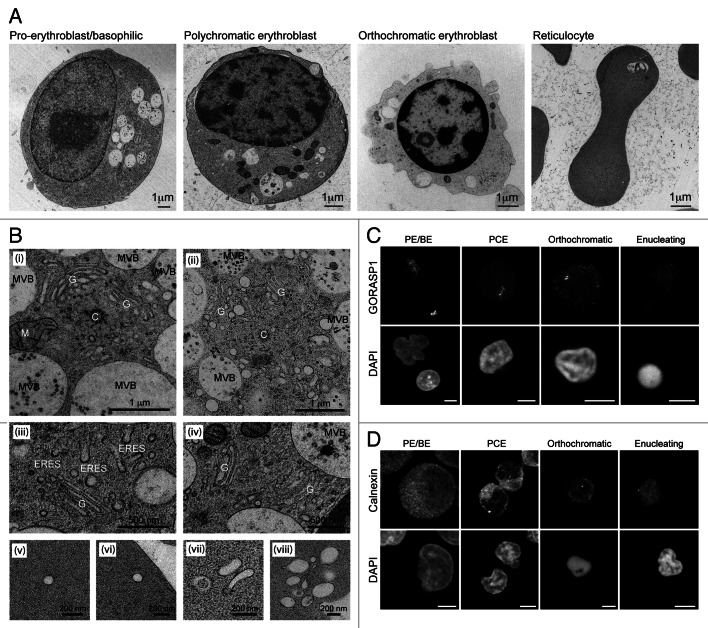
To differentiate human erythroid precursors in vitro, we used a two-stage protocol.25,26 Examples of cellular differentiation profiles are shown in Figure S1. We routinely achieved ~30% enucleated reticulocytes after 8 d in culture (post-shift to differentiation medium). For ultrastructural analysis, we used high-pressure freezing/freeze substitution (HPF), a method that preserves cellular and organellar shape better than standard aldehyde fixation/dehydration.27 In particular, autophagic and endocytic organelles retained their circular profiles unlike the misshapen/collapsed structures typically encountered in aldehyde-fixed samples (data not shown). Examples of cells at different differentiation stages processed by HPF are shown in Figure 1A. Chromatin condensation and progressive organelle depletion is apparent, preceding the appearance of enucleated reticulocytes that contain one or few large vacuoles (Fig. 1A).24
Figure 1. Progressive changes in endomembrane compartments in differentiating human erythroid cells. (A) Electron micrographs of staged human erythroid cells differentiated in vitro and processed by high-pressure freezing. Vacuoles and mitochondria are obvious features in PE/BE and PCE cells. Clear evidence of chromatin condensation can be seen in orthochromatic erythroblasts, which, by this stage, have a depleted organelle content. Reticulocytes are characterized by absence of a nucleus, the presence of one or few large vacuolar compartments in an otherwise depleted cytoplasm. (B) High magnification images of membrane compartments in early erythroid cells. (i and ii) Centrosomal areas in PE/BE cells showing centrosomes (C), extensive Golgi apparatus (G), mitochondria (M) and multivesicular bodies (MVBs). (iii and iv) Examples of small dispersed Golgi stacks in PCE cells. The appearance of abundant ER exit sites provides evidence for continued anterograde membrane trafficking. (v–viii) In reticuolocytes, small membrane compartments probably of endocytic origin are encountered at the cell periphery. (C and D) Confocal maximum projections of Golgi (GORASP1 labeling) and ER (calnexin labeling) in staged human erythoid cells. Evidence for Golgi fragmentation/dispersal can be seen at the PCE stage. Scale bars: (C and D) 5 µm.
In PE/BE cells, organelles of the secretory and endocytic compartments were observed clustered around the centrosome (Fig. 1B, i and ii). Here, extensive Golgi ribbons, and multivesicular bodies (MVBs) were often seen, suggesting active membrane traffic. At the PCE stage, Golgi profiles were still encountered, but these were smaller, representing individual (fragmented) stacks of Golgi membranes (Fig. 1B, iii and iv). Nevertheless, the presence of abundant ER exit sites indicated sustained secretory transport at this stage (Fig. 1B, iii). In reticulocytes, peripheral endocytic profiles were recorded (Fig. 1B, v–viii). To examine the fate of the Golgi and the ER during differentiation, we used confocal microscopy with anti-GORASP1/GRASP65 and anti-calnexin antibodies, respectively (Fig. 1C and D). These images agreed with the ultrastructural observations in that the Golgi apparatus is fragmented/scattered at the PCE stage (Fig. 1C). At later differentiation stages, any remaining Golgi signal was dispersed (Fig. 1C). The ER remained peripheral and reticular in appearance throughout in vitro erythropoiesis, and while a gradual reduction in calnexin-positive membrane signal was recorded, most partitioned away from the nucleus during enucleation (Fig. 1D).
Autophagy is upregulated during early erythroid differentiation
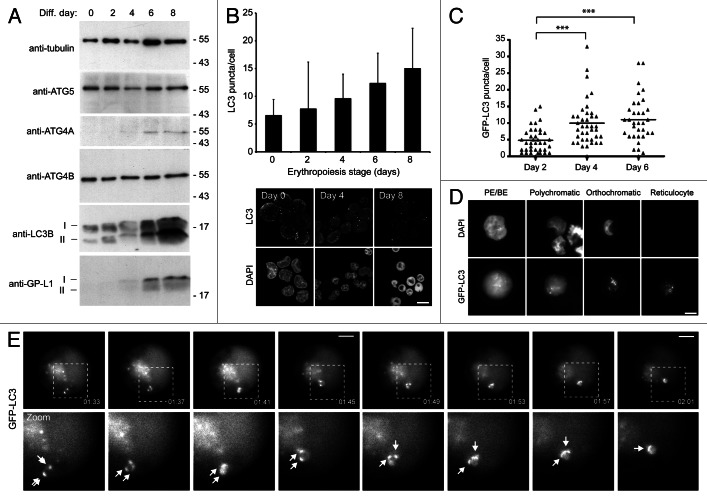
Erythropoiesis is tightly controlled at the transcriptional level, and a recent study has revealed GATA1/GATA-1 dependent upregulation of the expression of several autophagy genes during differentiation.9 Unpublished microarray data from our laboratory confirm upregulation of several autophagy genes in 1-stage differentiation protocols using adult human and cord CD34+ cells (data not shown; see ref. 28); however, to verify that transcriptional regulation was also taking place in the two-stage system, we used Q-RT-PCR (Fig. S2A). Transcripts with previously demonstrated erythroid expression patterns (e.g., TFRC/CD71, BNIP3) followed the expected expression profiles. With respect to autophagy genes, we saw upregulation of ULK1 (the mammalian ortholog of yeast ATG1) and BNIP3L; both are important for organelle clearance in mice.19,21 ATG5, an essential gene for autophagosome biogenesis, did not show transcriptional upregulation, meanwhile expression of BECN1/Beclin 1, a component of the autophagic PIK3C3/VPS34 (catalytic subunit type 3 of PtdIns 3-kinase) complex, was only marginally elevated at very late stages. Focusing on the ATG4 and ATG8 classes, we observed dramatic upregulation of expression of LC3B, GABARAP, GABARAPL2 and, particularly, GABARAPL1 (Fig. S2A). The products of these genes are degraded in the lysosome as a consequence of their association with the inner autophagosomal membrane, implying that enhanced expression might compensate for increased turnover. Interestingly, GABARAPL1 binds strongly to the mitophagy adaptor, BNIP3L, suggesting that it might be selectively upregulated to facilitate mitochondrial clearance. Of the mammalian ATG4 paralogs, only ATG4C showed reduced expression, while expression of both ATG4A and ATG4D increased markedly (Fig. S2A). Expression of ATG4B, which has the highest apparent activity and widest substrate specificity, did not change during erythropoiesis. Importantly, transcriptional upregulation was only seen in differentiating cells (Fig. S2E).
Immunoblotting indicated that expression patterns of autophagy proteins mirrored the changes in transcript levels [Fig. 2A; note: ATG4D antibodies (e.g., see ref. 29) did not give a reliable signal in erythroid cells]. Immunoblots for LC3B also provided evidence of increased autophagy, because most of the additional LC3B protein was in the cleaved and lipidated (LC3B-II) form in late erythroid stages (Fig. 2A). To test whether the upregulation of expression of autophagy genes and their products translated into increased autophagosome numbers, we first used immunofluorescence confocal imaging of erythroid cells fixed and stained with anti-LC3 antibodies to count puncta numbers in CD34+ cells differentiated using standard protocols (Fig. 2B). We recorded a gradual increase in steady-state LC3 puncta numbers using MetaMorph29 during 8 d of differentiation (Fig. 2B). We confirmed this pattern using lentivirus to express GFP-LC3B in differentiating human erythroid cells. These cells were visualized by wide-field fluorescence microscopy (Fig. 2C and D). In PE/BE cells, the majority of the GFP signal was cytosolic, indicative of low basal levels of autophagy; however, as cells progressed through subsequent differentiation stages, GFP-LC3B became progressively punctate (Fig. 2D). The GFP-LC3B puncta were highly dynamic, and examples of autophagosome movement and coalescence were documented during live-cell imaging experiments (Fig. 2E).
Figure 2. Induction of autophagy in differentiating human erythroid cells. (A) Immunoblots of selected autophagy proteins in staged human erythroid cultures. LC3B and GABARAPL1 (GP-LI) exist in 2 forms: soluble form I and lipid-bound form II which migrates slightly faster. TUBA/α-tubulin is included as a loading control. (B) LC3 puncta in differentiating human erythroid cells labeled with anti-LC3 antibodies. At the top, LC3 puncta counts in erythroid cells (data shown are means and s.d. of 3 independent cultures for which > 200 cells were scored). At the bottom, example images of fields of erythroid cells labeled with anti-LC3 antibodies and DAPI. Scale bar: 15 µm. (C) Quantification of LC3B puncta numbers in differentiating erythroblasts transduced with GFP-LC3B lentivirus. Cells were assessed at days 2, 4 and 6 following transfer to differentiation medium. Tukey’s multiple-comparison test: ***p ≤ 0.001. (D) Erythroid cells transduced with GFP-LC3B lentivirus, imaged by wide-field fluorescence microscopy. GFP-LC3B signal becomes progressively punctate during differentiation. Scale bar: 7.5 µm. (E) Live-cell imaging of autophagosome dynamics in a proerythroblast expressing GFP-LC3B. Arrows in zoomed images show individual autophagosomes. Scale bars: 5 µm.
Ultrastructural characterization of organelles in differentiating human erythroid cells
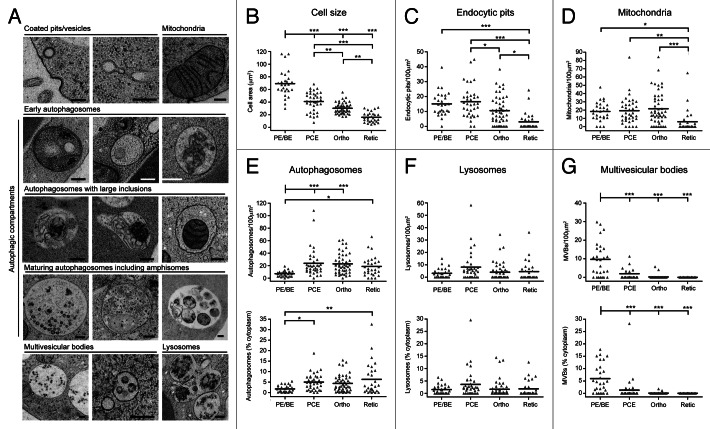
To characterize the endomembrane system during differentiation of human erythroid cells, we performed quantitative EM. Whole sections of cells were classified by differentiation stage (see Materials and Methods), and MetaMorph was used to calculate the abundance of various classes of organelles as a function of cytoplasmic area or length of plasma membrane. Figure 3A shows high magnification examples of the types of organelles that we scored. We considered autophagic compartments to be those structures having double-limiting membranes and/or content of obvious cytoplasmic origin, and these included amphisomes (autophagosome/endosome hybrid compartments en route to the lysosome for degradation).30 MVBs were electron translucent vacuoles containing 50- to 80-nm vesicles (Fig. 3A).30
Figure 3. Quantitative ultrastructural analysis of erythroid organelle content. (A) Example electron micrographs of the various categories of organelles that have been measured. Scale bars: 200 nm. (B) Maximal cellular cross-sectional area in micrographs of sections of whole erythroid cells. Erythroid differentiation stage was categorized morphologically. (C) Number of endocytic pits encountered per 100 µm of plasma membrane. (D) Number of mitochondria recorded in 100 µm2 cytoplasm. (E–G) Numbers/100 µm2 cytoplasm and % cytoplasmic occupancy of autophagosomes (E), lysosomes (F) and MVBs (G) in staged human erythroid cells. In (B–G), black bars represent the means. Numbers of cells analyzed: 28 for PE/BE; 36 for PCE; 49 for orthochromatics; 28 for reticulocytes. Tukey’s multiple comparison test: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
As expected, maximal cell area decreased progressively during differentiation (Fig. 3B). Endocytic pits were abundant features at early differentiation stages, but were less frequent in orthochromatic and reticulocyte cells (Fig. 3C). Mitochondrial density was stable until the transition to reticulocyte stage, at which point there occurred a rapid and significant depletion of the numbers of mitochondria encountered per unit area of cytoplasm (Fig. 3D). Consistent with the increase in GFP-LC3B puncta at early stages of differentiation (Fig. 2C and D), we recorded a significant increase in autophagic compartments at the PCE stage by quantitative EM (Fig. 3E); this upregulation occurred as a function of number and cytoplasmic occupancy, suggesting that there were no marked changes in the sizes of autophagic compartments during differentiation. Lysosomal number did not change significantly during differentiation (Fig. 3F). Finally, we recorded a significant decline in number and cytoplasmic volume occupancy of MVBs at the transition to PCEs (Fig. 3G). This general pattern of organelle restructuring was confirmed in two further ex vivo human erythroid cultures, and these data are summarized in Figure S3.
Expression of dominant-negative ATG4 mutants does not hamper human erythroid differentiation
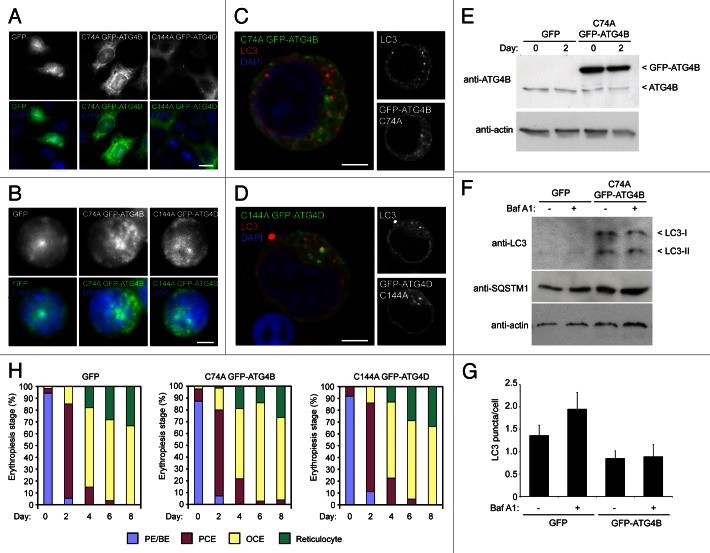
To examine its roles during human erythroid differentiation, we attempted to block autophagy at the autophagosome assembly stage by overexpressing dominant-negative cysteine mutants of ATG4B (C74A)31 or ATG4D (C144A)29 which sequester ATG8 family members at the priming step.31 These ATG4s were selected based on their recognized regulatory dominance (for ATG4B) and their expression profiles during human erythopoiesis (ATG4D) (we have so far been unable to generate mutant ATG4AC77A lentivirus). This approach was used in preference to siRNA/shRNA-mediated silencing, because suppressing gene expression human ex vivo erythroid cells to the levels needed to block autophagy is highly unlikely using current reagents/protocols. ATG4BC74A is a potent inhibitor of autophagy,31 whereas ATG4DC144A prevents the formation of GABARAPL1, but not LC3-positive autophagosomes.29 Lentiviral expression of GFP-tagged mutants was tested in HeLa cells (Fig. 4A), before human CD34+ erythroid progenitors were transduced (Fig. 4B). GFP-ATG4BC74A was primarily cytosolic, while GFP-ATG4DC144A showed the characteristic cytosolic and punctate staining that we have previously reported (Fig. 4A).29 GFP-expressing erythroid precursors were then selected by FACs (Fig. S4), and allowed to expand in culture before differentiation. Example confocal optical sections of erythroid precursors expressing ATG4BC74A or ATG4DC144A, and labeled with anti-LC3 antibodies, are shown in Figure 4C and D. Immunoblotting of lysates from GFP and GFP-ATG4BC74A-expressing erythroid cells using anti-ATG4B suggested that we had achieved an overexpression level greater than 10-fold for GFP-ATG4BC74A (Fig. 4E).
Figure 4. Expression of cysteine mutants of ATG4 in human erythroid precursors. (A) Wide-field fluorescence images of HeLa cells transduced with lentiviruses expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A. Scale bars: 10 µm. (B) Wide-field fluorescence images of human erythroid precursors cells transduced with lentiviruses expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A. Scale bars: 5 µm. (C and D) Confocal single sections of human pro-erythroblasts stably transduced with GFP-ATG4BC74A (C) or GFP-ATG4DC144A (D), labeled with anti-LC3 antibodies (red) and DAPI (blue). Scale bar: 3 µm. (E) Immunoblot of protein samples from human erythroid cells transduced with lentivirus expressing GFP-ATG4BC74A. Samples were collected at day 0 and day 2 of differentiation, and blots were probed with anti-ATG4B and anti-actin (loading control). The 2 bands detected on the anti-ATG4B blot correspond to endogenous ATG4B and GFP-ATG4BC74A. (F) Immunoblot of human erythroid cells expressing GFP or GFP-ATG4BC74A incubated in the absence or presence of BafA1 (6 h) at 2 d post-switch to differentiation medium. Samples were blotted using anti-LC3, anti-SQSTM1 and anti-actin (loading control) antibodies. LC3 is barely detectable in the GFP-expressing cells; however, in cells expressing GFP-ATG4B prominent LC3-I and LC3-II bands are visible, with no apparent change in LC3-II levels in the BafA1 treated sample. (G) LC3 puncta quantitation in human erythroid cells lentivirally-transduced with GFP or GFP-ATG4B BafA1 treatment increases LC3 puncta numbers in GFP expressing erythroid cells, but not in GFP-ATG4B expressing erythroid cells. Bars show means and standard error of > 40 individual cells at day 2 of differentiation. (H) Differentiation profiles of human erythroid cells expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A, assessed by light microscopy (10 to 15 random fields, 77 to 301 cells per time point for each treatment).
To begin to determine the impact of overexpression of dominant-negative ATG4 on erythroid autophagy and differentiation, we first immunoblotted lysates of GFP and GFP-ATG4BC74A cells cultured in the absence or presence of BafA1 for endogenous LC3 (Fig. 4F). In GFP-expressing erythroid cells analyzed at day 2 after a switch to differentiation medium, very little LC3 could be detected with the material available for analysis; however, there was a marked increase in the detection of both LC3-I and LC3-II in the GFP-ATG4BC74A-expressing cells (Fig. 4F). Interestingly, the ratio of LC3-II to LC3-I did not change during incubation in BafA1, suggesting that autophagosomal flux through the lysosomal system might be suppressed (Fig. 4F). Blotting for the autophagy cargo adaptor SQSTM1 showed a small increase in basal levels in ATG4BC74A-expressing cells compared with the GFP expressing population, but marginal increases in both cell populations following BafA1 treatment (Fig. 4F). Finally, and despite the clear differences in the relative amounts of lipidated LC3 in the GFP and GFP-ATG4BC74A-expressing erythroid cells, there were fewer LC3 positive puncta recorded in the latter (Fig. 4G). Addition of BafA1 caused an increase in LC3 puncta numbers in the GFP expressors, but no change in LC3 puncta in the GFP-ATG4BC74A-expressing cells (Fig. 4G). Although possibly pointing to a block in autophagic flux in cells expressing GFP-ATG4BC74A, these data should be viewed with some caution, not least because the apparent amounts of LC3-II as judged on immunoblots do not appear to correlate with puncta numbers assessed by imaging. Also, the number of autophagosomes recorded in these experiments was substantially lower than in earlier experiments performed in the same way (see Fig. 2B), and we believe this inconsistency to be due to the use of different batches of a commercial antibody. Nevertheless, the differentiation profiles of control (GFP) and mutant ATG4 overexpressors were then compared by light microscopy (Fig. 4H). This analysis suggested that overexpression of these ATG4 cysteine mutants has minimal impact on the kinetics of human erythroid differentiation in vitro (Fig. 4H), and cell size was also unaffected (Fig. S5A).
Organelle clearance in human erythroid cells with compromised autophagy
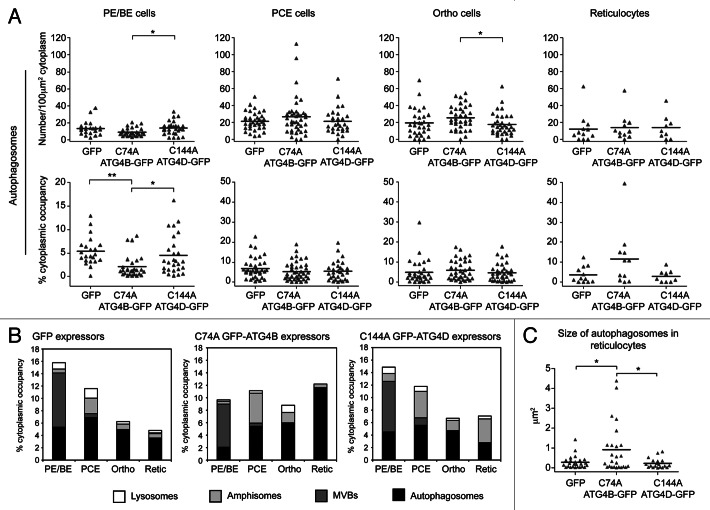
To measure the impact of ATG4 cysteine mutants on erythroid organelle clearance, we used quantitative EM. Prompted by the possible impact on autophagic flux in GFP-ATG4BC74A-expressing cells (Fig. 4F and G), we chose to treat autophagosomes and amphisomes as separate compartments to enable assessment of changes in autophagosome maturation events. In PE/BE cells expressing GFP-ATG4BC74A, we recorded a significant reduction in the cytoplasmic occupancy of autophagosomes, suggesting that this mutant abrogates autophagosome assembly at early differentiation stages (Fig. 5A). At the PCE stage autophagosome numbers increased under all conditions, suggesting that autophagosome assembly is supported in differentiating human erythroblasts expressing ATG4BC74A or ATG4DC144A (Fig. 5A), although from these observations it is not possible to assess the influence of altered autophagosomal flux upon steady-state autophagosome numbers. Interestingly, although similar numbers of autophagosomes were observed in reticulocytes in each condition, the cytoplasmic occupancy of autophagosomes in ATG4BC74A -expressing reticulocytes was increased suggesting differences in autophagosome size at this late differentiation stage (Fig. 5A; see below).
Figure 5. Quantitative ultrastructural assessment of organelles in human erythroid cells expressing cysteine mutant ATG4. (A) Abundance of autophagosomes in differentiating human erythroid cells expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A, assessed as a function of number/100 µm2 (top) or % cytoplasmic occupancy (bottom). Each data point represents a single cell, and the bar represents the mean. (B) Stacked columns of vacuolar compartments in differentiating human erythroblasts expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A expressed as % cytoplasmic occupancy. Data for autophagosome occupancy in individual cells are depicted in (A), while similar data sets for amphisomes and lysosomes are shown in Figures S6 and S7 respectively. (C) Analysis of the sizes of individual autophagosomes in reticulocytes expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A. Numbers of cells analyzed: (A and B) GFP: 22, 36, 31, 12; GFP-ATG4BC74A: 28, 40, 35, 11; GFP-ATG4DC144A: 27, 29, 35, 9 for PE/BE, PCE, orthochromatics and reticulocytes respectively. Tukey’s multiple comparison test: (A and C) *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
To examine in more detail the membrane compartments in differentiating human erythroid cells expressing ATG4 cysteine mutants, we also compared the numbers and cytoplasmic occupancy of endocytic pits, MVBs, amphisomes and lysosomes. Data for individual cells can be seen in Figure S5B (endocytic pits), Figure S6 (amphisomes) and Figure S7 (lysosomes), and mean values for MVBs, amphisomes and lysosomes, combined with the autophagosome data (depicted as individual data points in Fig. 5A), are presented as stacked columns in Figure 5B. We saw no differences in the decline in numbers of endocytic pits (Fig. S5B), but recorded a significant reduction in lysosomes in GFP-ATG4BC74A expressors at the PE/BE and PCE stages (Fig. S7). An expanded amphisomal compartment was also apparent in both GFP-ATG4BC74A and GFP-ATG4DC144A-expressing cells (Fig. 5B), although these data were not statistically significant (Fig. S6). Interestingly, when we measured the cross-sectional surface area of individual autophagosomes in reticulocytes, we saw a significant increase in autophagosome size in ATG4BC74A -expressing cells (Fig. 5C). These data suggest that human erythroid cells overexpressing GFP-ATG4BC74A (and to a lesser extent, ATG4DC144A) show altered maturation and clearance of autophagosomes (Fig. 5B), culminating in the retention of engorged autophagosomes at the reticulocyte stage.
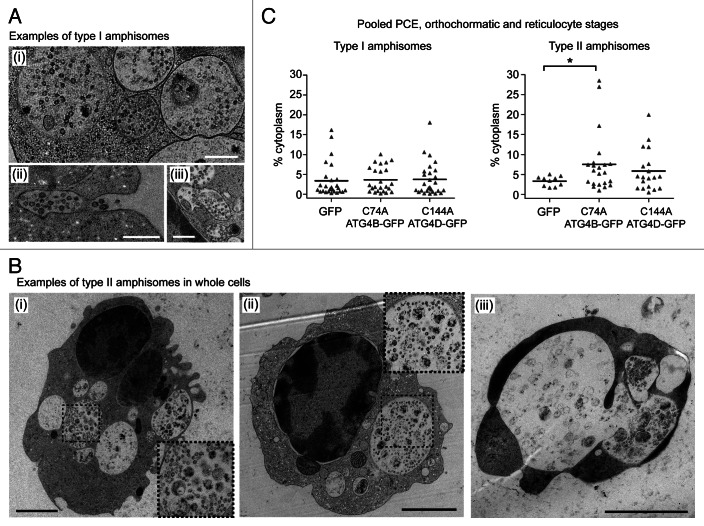
At late erythroid differentiation stages, much of the remaining organellar material was encountered within amphisomes. On close inspection, we were able to identify two distinct types of amphisomes in differentiating human erythroid cells: type I amphisomes (~1µm diameter) contained internal vesicles interspersed with small structures of cytoplasmic origin (Fig. 6A), and these were the dominant type in untreated cultures (data not shown); type II amphisomes were larger (~2µm diameter), and were characterized by the presence of large and/or intact, undegraded organelle (Fig. 6B). In pooled populations of PCE, orthochromatic and reticulocyte cells, type I amphisomes were recorded in 32.9% of GFP expressors (n = 79 cells), in 27.9% of GFP-ATG4BC74A expressors (n = 86 cells) and in 37.0% of GFP-ATG4DC144A expressors (n = 73 cells). By contrast, type II amphisomes were recorded in just 12.7% of GFP expressors, but were encountered in 25.6% GFP-ATG4BC74A and 26.0% GFP-ATG4DC144A expressors. Accordingly, expression of GFP-ATG4BC74A caused a significant increase in cytoplasmic occupancy of type II amphisomes compared with the GFP control (Fig. 6C). Interestingly, some cells containing extremely large type 2 amphisome compartments were clearly unviable (Fig. 6B, iii), and erythroid corpses were often encountered having abundant type II amphisomes (data not shown).
Figure 6. Two types of amphisomes in erythroid cultures: increase in large, type II amphisomes in cells expressing mutant ATG4. (A) Example electron micrographs of classical, type I amphisomes, containing internal vesicles and cytoplasmic material. An amphisome undergoing exocytosis can be seen in (ii). Scale bars: 500 nm. (B) Type II amphisomes in GFP-ATG4BC74A (i and iii) and GFP-ATG4DC144A (ii) expressors. The cell in (iii) contains several very large vacuoles and may not have been viable. Scale bars: 2 µm. (C) Quantitative analysis of type I and type II amphisomes in pooled PCE, orthochromatic and reticulocyte cells expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A. Tukey’s multiple comparison test: *p ≤ 0.05.
Mitochondria are cleared normally in mutant ATG4B and ATG4D expressing erythroid cells
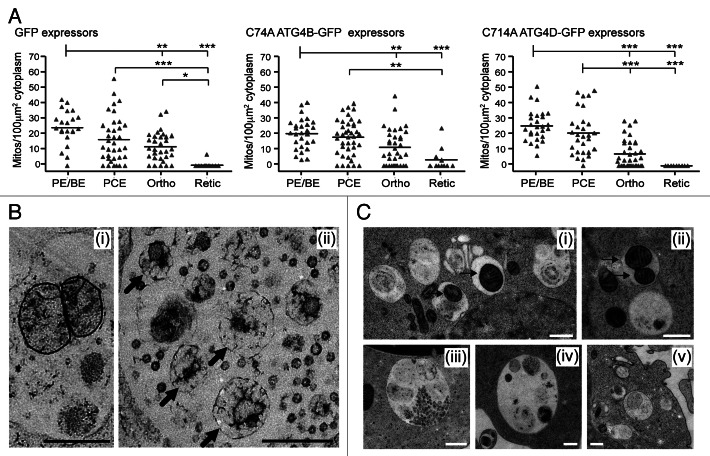
Evidence from knockout mice suggests that clearance of erythroid mitochondria is crucial for erythrocyte viability. We therefore checked whether mitochondria were removed normally in the mutant ATG4-expressing cells. Analysis of the numbers of mitochondria present in the cytoplasm of control, ATG4BC74A and ATG4DC144A expressing cells suggested little variability, although we did encounter some reticulocytes retaining mitochondria in the ATG4BC74A -expressing populations (Fig. 7A). On close inspection, it was apparent that type II amphisomes in mutant ATG4 expressing cells contained mitochondria in the process of degradation (Fig. 7B). In many cases, these were swollen and distorted, although membrane cristae were often apparent (Fig. 7B, i). Delayed degradation points to a role for ATG4 in autophagosome maturation and/or fusion with lysosomes. We therefore predicted that we would encounter similar late autophagic structures accumulating in erythroid cells treated with BafA1. Indeed control erythroid cultures treated for 6 h with BafA1 possessed enlarged autophagic structures containing undegraded organelles and cytoplasmic material, including mitochondria (Fig. 7C).
Figure 7. Mitochondrial clearance in human erythroid cells expressing cysteine mutants of ATG4. (A) Morphometric analysis of mitochondrial number in the cytoplasm of differentiating erythroblasts expressing GFP, GFP-ATG4BC74A or GFP-ATG4DC144A. Bars represent means. Tukey’s multiple comparison test: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. (B) Examples electron micrographs of type II amphisomes, showing intact mitochondria (i) and distorted, partially degraded mitochondria (ii, arrows). Scale bars: 500 nm. (C) Examples of vacuolar compartments in control erythroid cells treated with bafilomycin A1 (6 h). Mitochondria can be seen within autophagosomes and amphisomes (i and ii, arrows), while enlarged structures similar to the type II amphisomes that we have described are also prominent features (iii–v). Scale bars: 500 nm.
Discussion
We have characterized the progressive clearance of organelles during differentiation of human erythroid cells in vitro, and have studied the influence of autophagy on this process. To our knowledge, this is the first comprehensive morphometric analysis of cytoplasmic remodeling in differentiating human erythroid cells. In addition, we have established a system for the stable expression of dominant-negative trans-genes in primary human erythroid progenitor stem cells. This system could quite easily be modified to allow stable integration of shRNA constructs for targeted gene silencing, and provides an amenable system for manipulation of gene function in the context of ex vivo red cell production.
We have used quantitative RT-PCR and EM to study the expression of autophagy genes and the changes in organelle content respectively during erythropoiesis. Our RT-PCR data revealed differences in the expression patterns of ATG8 and ATG4 mammalian paralog family members, with marked increases in expression of LC3, GABARAP, GABARAPL2 and particularly GABARAPL1, and selective enhancement of ATG4A and ATG4D expression (Fig. S2). These data suggest that there may be erythroid-specific roles for some family members; a topic that will require further analysis. Changes in gene expression correlate with a dramatic increase in autophagic activity in differentiating human erythroid cells. Our ultrastructural analyses suggest that the numbers of autophagosomes increase statistically at the transition from PE/BE to PCE stages (Fig. 3E). At the same point, MVBs decline, and erythroid cells begin to accumulate amphisomes, suggesting convergence of endocytic and autophagic pathways (Fig. 3G). Mitochondria—which require autophagy for their systematic depletion17,19,21—remain abundant and maintain their proportional cytoplasmic volume occupancy until the transition to reticulocytes (Fig. 3D). The induction of mitophagy is therefore probably decoupled from the upregulation of general autophagy, perhaps instead linked with the increased expression of BNIP3L (Fig. S2).
To determine the impact of autophagy suppression on human erythroid differentiation, we overexpressed ATG4BC74A and ATG4DC144A. ATG4 cysteine mutants impair autophagy by forming stable complexes with ATG8 mammalian paralog family members at the priming step.31 In GFP-ATG4BC74A expressing PE/BE cells, autophagosome numbers were significantly reduced; however, because autophagy was subsequently stimulated to control levels, we concluded that only limited suppression of autophagosome biogenesis had been achieved (Fig. 5A). This could be because: (1) the levels of ATG4BC74A expression may have been insufficient to compete with the upregulated ATG8 paralog expression (Fig. S2); (2) endogenous ATG4 paralogs are present, allowing priming of sufficient ATG8 paralogs in the background of mutant ATG4; (3) autophagosomes may be assembled through LC3/ATG8-independent pathways. Despite the absence of a canonical autophagosome assembly pathway, reticulocytes from atg7−/− mice clear most of their organelles and contain identifiable autophagosome-like membrane compartments.16,17 In fact, an ATG5/ATG7-independent, RAB9-dependent autophagosome assembly pathway has been described in mouse embryonic fibroblasts,18 although this pathway has not yet been reported in other systems. Importantly, the differentiation profiles of control and mutant ATG4-expressing cells were very similar (Fig. 4H), suggesting that erythroid cells can compensate for impaired or absent autophagosome assembly pathways. It should be noted that in the GFP-ATG4BC74A-expressing populations, there was a greater number of highly vacuolated, nonviable cells (at the PE/BE stage, 5.4% GFP expressors were annexin V positive whereas 17.9% GFP-ATG4BC74A expressors were annexin V positive in one representative culture), suggesting that at high expression levels, this construct may not have been so well tolerated.
On careful inspection it was apparent that the organelle contents of erythoid cells expressing mutant ATG4B or ATG4D differed from GFP-expressing controls (Fig. 5B). In particular, large autophagosomes were encountered in reticulocytes in GFP-ATG4BC74A expressors, and a general enlargement of the amphisome compartments was recorded in cells expressing either mutant. The appearance of enlarged amphisomes containing undegraded cytoplasmic material implicates these proteins in autophagosome maturation and/or lysosomal fusion. ATG4 family members primarily function during autophagosome assembly, through the priming and regulated deconjugation of membrane-bound ATG8 (LC3);32 however, there is evidence that ATG4 may also act during late stages of autophagosome maturation in yeast.33 Many poorly understood fusion steps occur during autophagosome maturation,34 and degradation of mature autophagosomes has been shown to rely heavily on the acquisition of factors from late endosomes/lysosomes (including RAB7, LAMP1 and LAMP2). Interestingly, in the absence of LAMPs, or upon overexpression of dominant-negative RAB7T22N, cells accumulate abnormally large autophagic vacuoles similar to the type II amphisomes described in this study.35-38 RAB7 is a regulator of late endosomes, which also facilitates the recruitment of LAMP1 and lysosomal enzymes to maturing autophagosomes.37,39 Interestingly, LAMP1 recruitment and autophagosome acidification occur independently of lysosomal enzyme delivery.34,35,38 This is significant because the enlarged amphisomes that we have observed in mutant ATG4-expressing cells contained undegraded material, suggesting that the delivery of lysosomal enzymes and/or fusion with extant lysosomes may have been perturbed.
Two hypotheses could explain the impairment of autophagosome maturation by the expression of mutant ATG4 in erythroid cells. First, the ATG8 substrates of ATG4B and ATG4D may not be correctly deconjugated from the outer membrane of the autophagosome. In yeast expressing pre-primed ATG8, delivery of ATG8 to the vacuole is impaired, supporting a role for ATG4-mediated delipidation in late maturation.33 Second, considering that ATG4BC74A sequesters LC3 during priming,31 a similar stable interaction may also occur on the surface of autophagosomes when ATG4 cysteine mutants attempt to delipidate ATG8 paralogs on maturing autophagosomes. ATG4DC144A colocalizes with GABARAPL1 puncta in HeLa cells,29 and with LAMP1-positive structures in reticulocytes (data not shown). Binding of ATG4 cysteine mutants to ATG8 mammalian paralogs on the surface of late autophagosomes could block important ATG8-interactions or may directly interfere with ATG8-mediated fusion events. ATG8 regulates the elongation of the phagophore,33,40 possibly through its ability to trigger membrane tethering and fusion.41,42 Sequestration by ATG4 cysteine mutants may also interfere with an indirect role of ATG8 paralogs in fusion with late endosomes or lysosomes. For instance, LC3B binds to TBC1D25/OATL1, a RABGAP that regulates fusion events between autophagosomes and lysosomes.43 Meanwhile, GABARAP family members bind NSF (AAA ATPase N-ethylmaleimide sensitive factor), an essential factor in SNARE-mediated membrane fusion.44,45 Determining how the ATG4 cysteine mutants perturb autophagosome maturation during human erythropoiesis will require further investigation.
In conclusion, we have presented a comprehensive ultrastructural assessment of organelle restructuring during human erythropoiesis, and have began to assess the impact of autophagy during this process. We confirmed that autophagy is upregulated in human erythroid cells differentiating in vitro, culminating in reticulocytes containing one or few autophagic vacuoles.24 Using dominant-negative mutants we also showed that ATG4 endopeptidase activity is needed for late stages of autophagosome maturation, illustrated by the retention of expanded amphisome compartments in late erythroid cells. Despite this, in vitro differentiation of human erythroid cells proceeds with normal kinetics, highlighting how erythroid cells are able to adapt to diminished autophagic activity during terminal differentiation.
Materials and Methods
Reagents and antibodies
Unless stated, reagents were obtained from Sigma. Anti-GABARAPL129 and anti-GORASP1/GRASP6546 antibodies were raised in sheep. Monoclonal anti-TUBA/α-tubulin (B5-1-2/T5168), polyclonal anti-LC3B, polyclonal anti-ATG5 (Cell Signaling, 2360), polyclonal anti-LC3B (L7543), monoclonal anti-LC3B (Enzo Life Sciences, 5F10/ALX-803-080; MBL, PML036), polyclonal anti-ATG4A (from Dr I. Tanida), polyclonal anti-ATG4B (from Dr M. Koike), anti-SQSTM1 (Abnova, H00008878-M01), anti-calnexin (Enzo, SPA860), anti-actin (Santa Cruz, I-19/sc1616) were stored at −20°C. Alexa tagged secondary antibodies for immunofluorescence were from Molecular Probes. HRP-tagged secondary antibodies for immunoblotting were from Jackson Immunochemicals. Bafilomycin A1 (BafA1; Enzo Lifesciences, BML-CM110-0100, 200 µM stock) was stored at −20°C.
Erythroid cell culture
A > 95% pure population of CD34+, hemopoietic, progenitor cells (HPCs) was isolated from human blood donor mononuclear cells by magnetic bead separation according to the manufacturer’s protocol (Miltenyi Biotech Ltd.). Blood donor mononuclear cells, a waste fraction from a donation of platelets by apheresis, were donated with informed consent (reviewed by the National Health Service National Research Ethics Service, Review of Ethics Committee reference number 08/H0102/26).
Samples were diluted 1:1 with Hanks balanced salt solution (HBSS, H6648) and layered on Histopaque-1077 (H8889). PBMCs were harvested, washed with HBSS and red blood cells were lysed (150 mM Ammonium chloride, 1 mM K2EDTA.2H2O, 10 mM potassium bicarbonate, pH 7.5). CD34+ cells were isolated using the Direct CD34+ progenitor cell isolation kit (Miltenyi Biotech Ltd., 130-046-072) according to the manufacturer’s instructions. Isolated CD34+ cells were cultured using a two-stage system adapted from refs. 25 and 26. They were cultured for 5 d in serum-free Stemspan medium (Stem Cell Technologies, 09650) supplemented with stem cell factor (SCF, 10 ng/ml, R&D Systems, 255-SC), IL3 (1 ng/ml, R&D Systems, 203-IL), EPO (3 Ul/ml, Roche, NeoRecormon), bovine lipoprotein (1 ml/ml, L4646) and Prograf (0.1 ng/ml, Fujisawa) or up to 10 d when supplemented with dexamethasone (Dex, 1 µM, D4902) in the absence of IL3 from day 4 (expansion media). Cells were then transferred to a Stemspan-based differentiation medium supplemented with 1 mg/ml Holotransferrin (T0665), 3% AB human serum (H4522), 10 UI/ml EPO, 10 ng/ml insulin (I9278) and 1 µM 3,5,3′-triiodo-L-thyronine sodium salt (T6397).
Production and use of lentiviruses
Lentiviral vectors containing the sequences for ATG4BC74A and ATG4DC144A with N-terminal GFP tags were obtained by cloning mutated cDNAs29 in pxlg3-gfp (a modified pSEW sin vector kindly provided by Dr G. Cory). Lentiviruses were produced by cotransfection with the envelope plasmid pMD.G and the packaging plasmid pCMVR8.91 in HEK 293T cells. Viruses were harvested and stored at −80°C. Erythroid progenitors were transduced overnight, at 3 d post-CD34+ isolation, at a concentration of 1 × 105/ml in the presence of Polybrene (Hexadimethrine bromide, 8 µg/ml). Lentiviruses were then washed away with HBSS and cells were cultured in expansion medium containing dexamethasone (1 µM) without IL3, as recommended for long-term expansion of human erythroblasts.26 GFP-expressing erythroblasts were sorted by flow cytometry at day 7 of culture and expanded a further 3 d before being transferred to differentiation medium.47 The differentiation stage of each cell was determined using a combination of cell size and nuclear condensation (the main criteria), negative stain density (by EM) and DAPI staining by fluorescence wide-field imaging.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted with TRIZOL (Invitrogen, 15596-018) according to manufacturer’s instructions from erythroid cells at 2-d intervals from the start of differentiation (day 0). RNA was quantified using a NanoDrop 1000 spectrophotometer (Labtech International). First strand cDNA was synthesized from 300 ng of total RNA using AffinityScript multiple temperature cDNA synthesis kit (Stratagene, 200436) at 42°C according to manufacturer’s instructions.
qPCR gene-specific primers were designed using web-based Primer 3 software (Table S1). Efficiency of all primer sets were tested on serial dilutions of cDNA produced from 1 µg Universal human reference RNA (Stratagene, 750500), using the Pfaffl method.48 As recommended, only primer sets with 95 to 110% efficiency were used. qPCR reactions were set up with 1× SYBRgreen (Finnzymes, F-400L) and 1 µl of template (25 ng of cDNA) in 96-well plates (Biorad) and run in a DNA Engine Opticon 2 thermocycler (MJ Research, GRI). Each sample was tested in triplicate and gene expression was normalized to a reference gene, PABPC1 whose expression remains constant during erythroid differentiation.28 The normalized results were expressed as a fold increase compared with the day 0 sample.
Immunoblotting
Cultured erythroblasts were lysed in SDS-PAGE sample buffer (62.5 mM Tris/HCl, pH 6.8, 10% v/v glycerol, 3% w/v SDS, 5% v/v β-mercaptoethanol, 0.0025% w/v bromophenol blue) and resolved using a Mini-Protean 3 electrophoresis system (BioRad). Proteins were transferred to nitrocellulose membrane using a Genie electrophoretic transfer system (Idea Scientific). Membranes were blocked using 5% milk powder solution in tris-buffered saline solution (20 mM Tris/HCl, pH 7.7, 150 mM NaCl, 0.1% triton X100) for 30 min before being incubated overnight at 4°C in blocking solution containing the appropriate secondary antibody. Following extensive washing in tris-buffered saline solution, membranes were incubated for 1 h at room temperature in blocking solution containing HRP-labeled secondary antibodies. Proteins were detected by enhanced chemiluminescence (GE Healthcare, RPN2209).
Fluorescence microscopy
Cells were fixed in 2% formaldehyde (EM grade, TAAB, F003), cytospun for 5 min at 800 rpm and permeabilized with 0.1% Triton X100. Cells were then immunolabeled, stained with DAPI (0.1 µg/ml) and mounted in Mowiol supplemented with DABCO (25 mg/ml, D2522). Wide-field fluorescence images were obtained using an Olympus IX-71 inverted microscope (Olympus UK Ltd.) (60× Uplan Fluorite objective 0.65 to 1.25 NA, at maximum aperture) fitted with a CoolSNAP HQ CCD camera (Photometrics) driven by MetaMorph software (Molecular Devices). Confocal images were obtained using a Leica AOBS SP2 microscope (Leica Microsystems UK) (63× PLABO objective 1.4 NA) with 0.2 μm to 0.4 μm z-steps. Automated autophagosome puncta scoring was performed using the TopHat morphology filter in MetaMorph, selecting for round objects of diameter 5 pixels (= 1 µm) or less.
High-pressure freezing transmission electron microscopy (TEM)
Up to 5x106, samples were resuspended in a small volume of medium and fixed in 100 µm carriers using a Leica EM PACT2 + RTS high-pressure freezer (Leica Microsystems UK). Freeze-substitution was performed with 1% osmium-0.1% uranyl acetate in a FSP Leica AFS. Samples were dehydrated with acetone and infiltrated with increasing amounts of resin (25%, 50%, 75% Epon-acetone for 1 h each followed by 2 h in 100% Epon). They were finally embedded in fresh Epon and left to harden for 48 h at 60°C. 70 nm sections were counter stained with 3% aqueous uranyl acetate and 80 mM lead citrate. Cells were observed using a Tecnai 12-FEI 120kV BioTwin Spirit transmission electron microscope (FEI Europe, Eindhoven, Netherlands) and images of randomly selected cells (minimum 22 cells per sample) were recorded with an FEI Eagle 4 k × 4 k CCD camera. For analysis, cells and organelles were outlined and measured (surface area and/or perimeter in pixels) using MetaMorph.
Supplementary Material
Acknowledgments
We acknowledge the support of the Medical Research Council in providing an Infrastructure Award to establish the School of Medical Sciences Cell Imaging Facility at Bristol University, and the Wolfson Foundation for recent funds to modernize and expand the facility. We thank Dr. Paul Verkade for invaluable advice on the identification and interpretation of membrane compartments by electron microscopy, and for training and advice on the technique of high-pressure freezing. The authors also wish to acknowledge the assistance of Dr. Andrew Herman for cell sorting, and the University of Bristol Faculty of Medical and Veterinary Sciences Flow Cytometry Facility. This work is funded by a project grant from the NHS Blood and Transplant (Ref: PG07/1).
Glossary
Abbreviations:
- ATG
autophagy-related
- GFP
green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3 (MAP1LC3)
- GABARAP
gamma-aminobutyric acid receptor-associated protein
- GABARAPL1/2
gamma-aminobutyric acid receptor-associated protein-like 1/2
- Bnip3
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3
- TEM
transmission electron microscopy
- HPF
high-pressure freezing/freeze substitution, LAMP, lysosomal associated membrane protein
- q-RT-PCR
quantitative real time polymerase chain reaction
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/24172
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24172
References
- 1.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009;15:217–24. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariño G, Uría JA, Puente XS, Quesada V, Bordallo J, López-Otín C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 7.Mariño G, Fernández AF, Cabrera S, Lundberg YW, Cabanillas R, Rodríguez F, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 9.Kang YA, Sanalkumar R, O’Geen H, Linnemann AK, Chang CJ, Bouhassira EE, et al. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012;32:226–39. doi: 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–20. [PubMed] [Google Scholar]
- 11.Simpson CF, Kling JM. The mechanism of mitochondrial extrusion from phenylhydrazine-induced reticulocytes in the circulating blood. J Cell Biol. 1968;36:103–9. doi: 10.1083/jcb.36.1.103. [DOI] [PubMed] [Google Scholar]
- 12.Takano-Ohmuro H, Mukaida M, Kominami E, Morioka K. Autophagy in embryonic erythroid cells: its role in maturation. Eur J Cell Biol. 2000;79:759–64. doi: 10.1078/0171-9335-00096. [DOI] [PubMed] [Google Scholar]
- 13.Fader CM, Colombo MI. Multivesicular bodies and autophagy in erythrocyte maturation. Autophagy. 2006;2:122–5. doi: 10.4161/auto.2.2.2350. [DOI] [PubMed] [Google Scholar]
- 14.Heynen MJ, Verwilghen RL. A quantitative ultrastructural study of normal rat erythroblasts and reticulocytes. Cell Tissue Res. 1982;224:397–408. doi: 10.1007/BF00216882. [DOI] [PubMed] [Google Scholar]
- 15.Kent G, Minick OT, Volini FI, Orfei E. Autophagic vacuoles in human red cells. Am J Pathol. 1966;48:831–57. [PMC free article] [PubMed] [Google Scholar]
- 16.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A. 2010;107:832–7. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–64. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 19.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migliaccio AR, Masselli E, Varricchio L, Whitsett C. Ex-vivo expansion of red blood cells: how real for transfusion in humans? Blood Rev. 2012;26:81–95. doi: 10.1016/j.blre.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths RE, Kupzig S, Cogan N, Mankelow TJ, Betin VM, Trakarnsanga K, et al. Maturing reticulocytes internalize plasma membrane in glycophorin A-containing vesicles that fuse with autophagosomes before exocytosis. Blood. 2012;119:6296–306. doi: 10.1182/blood-2011-09-376475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southcott MJ, Tanner MJ, Anstee DJ. The expression of human blood group antigens during erythropoiesis in a cell culture system. Blood. 1999;93:4425–35. [PubMed] [Google Scholar]
- 26.Leberbauer C, Boulmé F, Unfried G, Huber J, Beug H, Müllner EW. Different steroids co-regulate long-term expansion versus terminal differentiation in primary human erythroid progenitors. Blood. 2005;105:85–94. doi: 10.1182/blood-2004-03-1002. [DOI] [PubMed] [Google Scholar]
- 27.McDonald KL, Morphew M, Verkade P, Müller-Reichert T. Recent advances in high-pressure freezing: equipment- and specimen-loading methods. Methods Mol Biol. 2007;369:143–73. doi: 10.1007/978-1-59745-294-6_8. [DOI] [PubMed] [Google Scholar]
- 28.Singleton BK, Burton NM, Green C, Brady RL, Anstee DJ. Mutations in EKLF/KLF1 form the molecular basis of the rare blood group In(Lu) phenotype. Blood. 2008;112:2081–8. doi: 10.1182/blood-2008-03-145672. [DOI] [PubMed] [Google Scholar]
- 29.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–66. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, et al. A comprehensive glossary of autophagy-related molecules and processes (2nd edition) Autophagy. 2011;7:1273–94. doi: 10.4161/auto.7.11.17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–9. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair U, Yen WL, Mari M, Cao Y, Xie Z, Baba M, et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8:780–93. doi: 10.4161/auto.19385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn WA., Jr. Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–45. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–68. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell. 2004;15:3132–45. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–97. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann-Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–6. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 39.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 40.Abeliovich H, Dunn WA, Jr., Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–34. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 42.Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–54. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol. 2011;192:839–53. doi: 10.1083/jcb.201008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sagiv Y, Legesse-Miller A, Porat A, Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kittler JT, Rostaing P, Schiavo G, Fritschy JM, Olsen R, Triller A, et al. The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol Cell Neurosci. 2001;18:13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- 46.Cheng JP, Betin VM, Weir H, Shelmani GM, Moss DK, Lane JD. Caspase cleavage of the Golgi stacking factor GRASP65 is required for Fas/CD95-mediated apoptosis. Cell Death Dis. 2010;1:e82. doi: 10.1038/cddis.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Betin VMS, MacVicar TDB, Parsons SF, Anstee DJ, Lane JD. A cryptic mitochondrial targeting motif in Atg4D links caspase cleavage with mitochonrial import and oxidative stress. Autophagy. 2012;8:664–76. doi: 10.4161/auto.19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.