Abstract
Leaf senescence is an important process in the developmental life of all plant species. Senescence efficiency influences important agricultural traits such as grain protein content and plant growth, which are often limited by nitrogen use. Little is known about the molecular mechanisms regulating this highly orchestrated process. To enhance our understanding of leaf senescence and its regulation, we have undertaken the structural and functional characterization of previously unknown proteins that are involved in the control of senescence in barley (Hordeum vulgare L.). Previous microarray analysis highlighted several barley genes whose transcripts are differentially expressed during senescence, including a specific gene which is greater than 40 fold up-regulated in the flag leaves of early- as compared to late-senescing near-isogenic barley lines at 14 and 21 days past flowering (anthesis). From inspection of its amino acid sequence, this gene is predicted to encode a glycine-rich RNA-binding protein herein referred to as HvGR-RBP1. HvGR-RBP1 has been expressed as a recombinant protein in E. coli, and preliminary NMR data analysis has revealed that its glycine-rich C-terminal region [residues: 93–162] is structurally disordered whereas its N-terminal region [residues: 1–92] forms a well-folded domain. Herein, we report the complete 1H, 13C, and 15N resonance assignments of backbone and sidechain atoms, and the secondary structural topology of the N-terminal RNA Recognition Motif (RRM) domain of HvGR-RBP1, as a first step to unraveling its structural and functional role in the regulation of barley leaf senescence.
Keywords: Barley, Hordeum vulgare L. barley strain, HvGR-RBP1 barley protein, NMR, Protein structure-function, RNA-Binding Protein, RNA Recognition Motif, Plant Senescence
Biological Context
Leaf senescence is a complex, highly regulated developmental process in the life of plants and is critical for overall plant fitness. Senescence results in the coordinated degradation of macromolecular polymers (proteins, nucleic acids, fatty acids, carbohydrates) and the subsequent remobilization of nutrients from leaves to surviving plant structures, i.e. seeds in annual plant species; stems and roots in perennial species. Due to the strong correlation between senescence and crop quality, agronomical importance has stimulated the need for a better understanding of the molecular mechanisms regulating crop plant senescence (Gregersen et al. 2008).
As a principal feed and malting cereal crop, a plant species of interest is barley (Hordeum vulgare L.), whose grain protein content (GPC) is strongly influenced by the timing of senescence. Near iso-genic barley lines Karl (high-GPC) and 10–11 (low-GPC) differ in allelic state on a major chromosome locus shown to influence both timing of senescence and grain protein content (See et al. 2002). These two plant cultivars undergo senescence at different times in their growth cycle, and exhibit a measurable difference in chlorophyll content and loss of protein by degradation (Jukanti et al. 2008). Transcriptomic analysis revealed that several genes are differentially expressed in early senescing (10–11) versus late-senescing (Karl) germplasms (Jukanti et al. 2008). The transcriptomics study aimed to identify senescence-associated genes (SAGs) essential for the regulation and timing of the senescence process in barley plants. During the onset of senescence (e.g. 14 and 21 days past flowering (i.e. past anthesis)) of barley line 10–11, several genes were found to be significantly up regulated (e.g. > 10 fold increase in transcript expression), and encoded proteins belonging to diverse functional groups such as several transcription factors including WRKY, NAM, ATAF1/2 and CUC2, the latter three belonging to a family of plant-specific transcriptional factors referred to as NAC; cell signaling and protein kinases and phosphatases involved in signal transduction; a putative glycine-rich RNA-binding protein (HvGR-RBP1); lipases, proteases, nucleases involved in macromolecular degradation and nutrient recycling; membrane transport proteins; and proteins involved in plant defenses against pathogens infections (Jukanti et al. 2008). Of the largest differences in transcript abundance between 10_11 and Karl barley lines, the gene labeled as ‘contig 17116_at’ stood out because its expression is more than 40 fold up-regulated in barley line 10_11 compared to Karl (Jukanti et al. 2008). Examination of the protein coding sequence of ‘contig 17116_at’ (referred to as HvGR-RBP1) revealed sequence homology to a large class of RNA binding proteins comprised of RNA recognition motifs (RRMs). In addition to an RRM, HvGR-RBP1 is comprised of a C-terminal glycine rich region containing a series of RGG repeats that are also thought to participate in RNA binding (Corbin-Lickfett et al. 2010). Proteins with RRM motifs have been shown to bind RNA in a variety of ways (Clery et al. 2008; Maris et al. 2005). They are ubiquitous in eukaryotes (e.g. 2% gene products in humans encode RRMs), and are also highly represented in proteins from other domains of life, highlighting the evolutionary importance of the RRM fold (Maris et al. 2005). Most interestingly, although RRMs have a highly conserved structural motif, they do regulate a wide range of post-transcriptional processes including pre-mRNA processing, mRNA stability, RNA editing, mRNA export, pre-rRNA complex formation, translation regulation and RNA degradation (Clery et al. 2008; Maris et al. 2005). Overall, the presence of RRM and RNA binding elements in HvGR-RBP1 supports the notion that this protein plays an important role in the regulation of barley plant senescence.
To assess the validity of this hypothesis, we have undertaken the structural and functional characterization of HvGR-RBP1. Herein we describe the cloning, protein expression and purification of the recombinant full-length [residues: 1–162] HvGR-RBP1 protein, and of its N-terminal domain that contains its postulated RRM motif [residues: 1–92], referred to as NHvGR-RBP1. We report the 15N, 13C, and 1H resonance assignments for the backbone and sidechain atoms of N-HvGR-RBP1, report spectral differences in the 2D 1H-15N correlation spectra of full-length HvGR-RBP1 and N-HvGR-RBP1, and describe the sequence topology of secondary structural elements that make up the N-terminal RRM domain of HvGR-RBP1.
Methods and experiments
cDNA Cloning
The coding sequence of HvGR-RBP1 (contig17116_at, 22-k Barley1 DNA microarray Affymetrix Santa Clara, CA, USA) was amplified from barley RNA and cloned into a pDrive plasmid from barley mRNA extract using SuperScriptIII™ Platinum Two-Step qRT-PCR kit (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA) and QIAGEN™ PCR Cloning kit (QIAGEN, Valencia, CA, USA). One sequence specific forward and two reverse primers: [(F)- 5’-GACGACGACAAGATGGCAGAGTCGGACGGCGCCG-3’] and [(R)-FL-HvGR-RBP1: 5’-GAGGAGAAGCCCGGTTCAGTATCCGACGGAGTTGCCTCCGCG-3’/ (R)-NHvGR- RBP1: 5’-GAGGAGAAGCCCGGTTCAGCCGCCGTACCCGCGAGACTGG-3’] were used to PCR amplify both the gene encoding the full length HvGR-RBP1 protein (FL- HvGRRBP1] (e.g. residues Met1-Tyr 162] and the N-terminal segment of the HvGR-RBP1 gene encoding amino acid residues Met1-Gly92 (N- HvGR-RBP1). Each synthesized PCR product was gel purified and ligated into pET46 Ek/LIC vector (Novagen Inc. Billerica, MA, USA). The resulting engineered pET46 vectors code for either FL-HvGR-RBP1 or an N-HvGR-RBP1 protein products with a fused N-terminal 6x His-Tag adding the following 14 residues MAHHHHHHVDDDDK to the native sequence. The vector also contains an ampicillin resistance gene for strain selection. Escherichia coli strain DH5-α cells (Invitrogen Life Technologies Corporation, Carlsbad, CA, USA) optimized for DNA plasmid amplification were transformed with resulting pET46-FL-HvGR-RBP1 or pET46-N-HvGR-RBP1 plasmids, and cells were plated on LB agar containing ampicillin. A single colony of each construct was picked and grown overnight at 37°C in Luria broth (LB) and used for plasmid purification with the QIAGEN™ Mini Prep Kit (QIAGEN, Valencia, CA, USA).
Protein expression and purification
For protein expression, E. coli strain BL21-(DE3)-RIL cells (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA) were transformed with purified pET46-FL-HvGR-RBP1 and pET46-N-HvGR-RBP1 plasmids. Resulting cells were screened by colony based PCR and further verified by DNA sequencing (Iowa State University DNA Facility, Ames, IA, USA) to ensure that the cells contained the correct DNA template encoding the correct full length (FL) or N- HvGR-RBP1 proteins. A single positive clone was chosen to inoculate 50 mL of LB, grown overnight and subsequently used to inoculate a 1 L culture of M9 minimal media supplemented with 15NH4Cl (99% 15N enriched, CIL, Cambridge, MA, USA) and uniformly 13C-labeled glucose (i.e. D-glucose-13C6, 99% 13C-enriched, CIL) as sole source of nitrogen and carbon, respectively. The latter were used to produce uniformly 15N or 15N/13C labeled N- HvGR-RBP1 protein samples. Following inoculation, the cells were allowed to grow at 37°C to an OD600nm of ~ 0.6, prior to induction of protein expression with 1 mM IPTG, and then allowed to grow for an additional 10 hours. Cells were then harvested by centrifugation at 5,000 × g (Sorvall, RC2-B) and the resulting cell pellet stored at -20°C until further use.
Cells were thawed and resuspended in 5 ml/gram of lysis buffer (1 M NaCl, 250 mM Na2HPO4/NaH2PO4, 10 mM imidazole, pH 8) with freshly prepared phenylmethylsulfonyl fluoride (0.1 mM PMSF) and Complete Mini Protease Inhibitor cocktail™ (Roche Applied Science, Indianapolis, IN, USA) to minimize protein loss by protease activity. The cells were then lyzed using an M-110L microfluidizer (Microfluidics, Newton, MA, USA). The resulting cell lysate was kept at 4°C and clarified from resulting cell debris by centrifugation at 20,000 × g for 25 minutes. The resulting supernatant was applied to a nickel affinity column containing 3–5 ml bed volume of HisPur-N-NTA™ resin (Thermo Scientific, Rockford, IL, USA). The nickel affinity purification step was followed by a second anion exchange and a third size exclusion chromatography (SEC) purification step, using an anion-exchange Q column (GE Life Sciences, Piscataway, NJ, USA) and a size exclusion Superdex 75 column (GE Life Sciences, Piscataway, NJ, USA), respectively. Protein-containing fractions were pooled together and dialyzed against NMR buffer (300 mM NaCl, 50 mM Na2HPO4/NaH2PO4, 95% H2O/5% (v/v) D2O (or 100% D2O for the 13C-edited TOCSY and NOESY experiments), 0.05% sodium azide, 0.1 mM PMSF, pH 6.5) and concentrated to 1 mM protein concentration as determined by OD280nm readings. Prior to NMR data collection, the oligomerization state and molecular weight of the sample were verified by SEC, SDS-PAGE, and electrospray ionization (ESI) micro-TOF mass spectrometry, using a Bruker MS instrument housed in the Proteomics & Metabolomics Mass Spectrometry Research Facility of Montana State University.
NMR Spectroscopy
All NMR spectra were acquired at 298 K on a four-channel Bruker DRX-600 MHz spectrometer, with an inverse detection triple resonance (1H, 13C, 15N) conventional NMR probe equipped with triple axis gradients, as previously described for other proteins of interest (Schlenker et al. 2012). All data were processed and analyzed using NMRPipe Spectral Processing and Analysis System (Delaglio et al. 1995) and Sparky NMR Assignment and Integration Software (Goddard and Kneller, SPARKY 3, University of California, San Francisco, USA). Sequential 15N/1H/13C backbone and side chain resonance assignments were extracted from standard heteronuclear (1H, 15N, 13C) multidimensional NMR experiments (HNCA, HNCACB, CBCA(CO)NH, C(CO)NH, HBHA(CO)NH, HC(CO)NH, 13C-edited 1H-1H TOCSY, 13C-1H HSQC, and 13C-edited and 15N-edited 1H-1H NOESY). 1H, 15N, and 13C chemical shift dimensions were indirectly referenced to DSS.
NMR Analysis of Full Length HvGR-RBP1
Inspection of the 2D 1H-15N HSQC of the full length HvGR-RBP1 (FL-HvGR-RBP1) protein revealed only ~117 well defined resonances along with a large cluster of strong signals in the random coil region of the spectrum. Additionally, FL-HvGR-RBP1 appeared to show signs that the protein in solution aggregates at the mM protein concentration necessary for multidimensional heteronuclear NMR data collection with a conventional room temperature TCI NMR probe. Since the C-terminus of the HVGR-RBP1 contains 40 glycine residues as well as repetitive Tyr and Arg residues (reducing the likelihood of sequential assignments) and is predicted using secondary structure prediction programs to be essentially random coil, we engineered a truncated HvGR-RBP1 protein construct (N-HvGR-RBP1) missing this C-terminal disordered region. The resulting 2D- 1H-15N HSQC spectrum of this 92-residue N-HvGR-RBP1 protein exhibits 91 well-resolved 1HN/15N NMR signals. Overlay of the 2D 1H-15N HSQC spectra of N-HvGR-RBP1 and FL-HvGR-RBP1 indicated that the well-dispersed resonances seen in the 2D-1H-15N HSQC spectrum of N-HvGR-RBP1 had essentially same chemical shifts (i.e. within 0.27 ppm) in the 2D 1H-15N HSQC spectrum of FL-HvGR-RBP1 (see Supplementary Figure S2), indicating clearly that the presence of the C-terminal (residues 93–162) segment of the protein does not influence to a significant extent the structural fold of the N-HvGR-RBP1 domain at least as assessed by 2D 1H-15N correlation spectroscopy. The extensive spectral dispersion observed in the 2D 1H-15N HSQC spectrum of N-HvGR-RBP1 indicated that this region of the protein spanning residues 1–92 is well folded and amenable to structural characterization, thus motivating complete resonance assignments of N-HvGR-RBP1.
Extent of NMR Assignments and Data Deposition
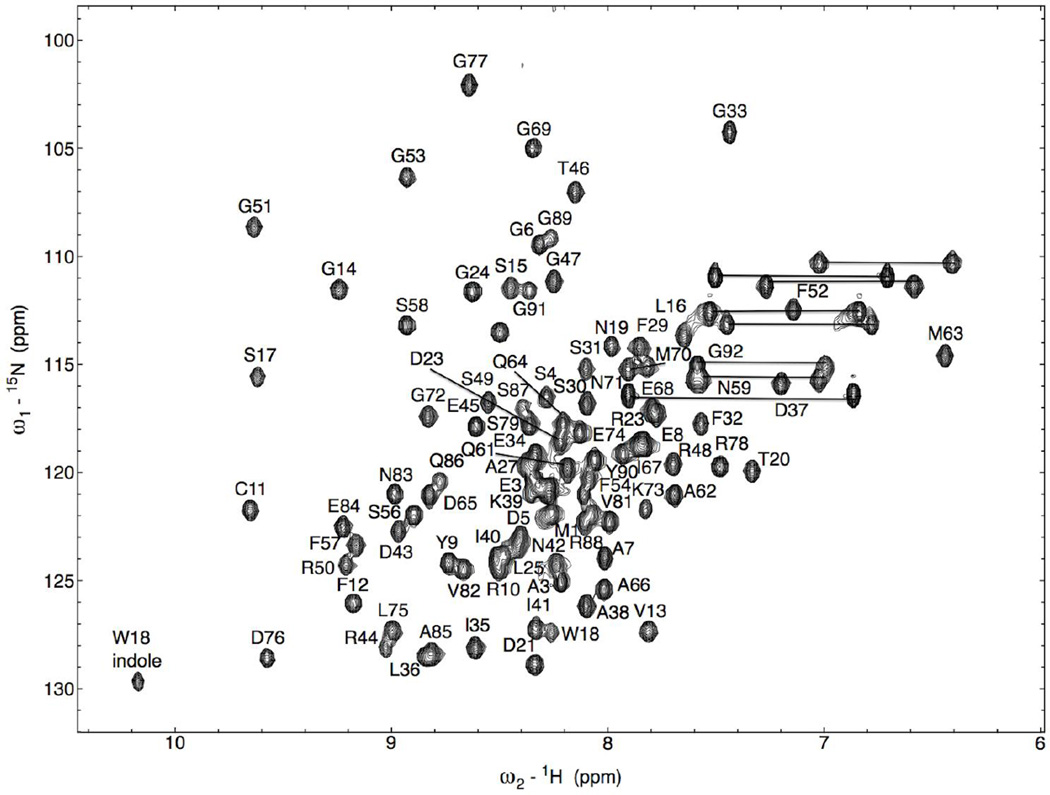
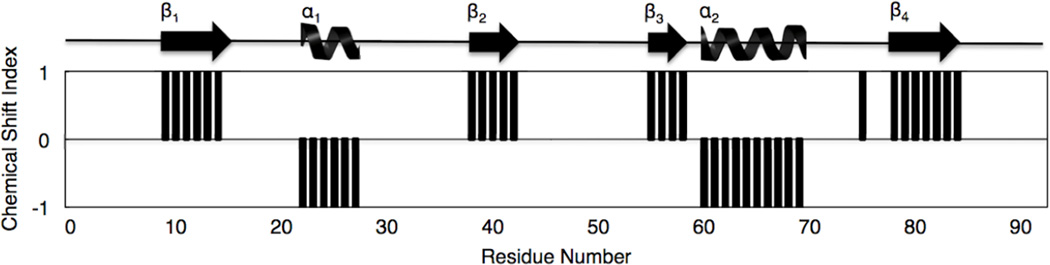
NMR data collected on N-HvGR-RBP1 enabled the assignments of all NH resonances except for the NH of V55 that was unobservable in the 2D 1H-15N HSQC spectrum of N-HvGRRBP1 (see Fig. 1). In total, 91 out of a total of 92 NH resonances were assigned for N-HvGRRBP1 (which contains no proline). Complete assignments for Cα/Cβ carbon chemical shifts were achieved for all residues of N-HvGR-RBP1. Assignment of Hα and Hβ resonances was > 98% and assignment of aliphatic sidechain carbons and protons was achieved to > 96% of all assignable resonances. In addition, several sidechain resonances corresponding to the aromatic carbons and hydrogen atoms of Tyr, Phe, and Trp were assigned, although not to completion. Backbone and side chain (1H, 15N, 13C) NMR resonance assignments for N-HvGR-RBP1 have been deposited in the BioMagResBank (BMRB) of the University of Wisconsin-Madison under accession number: 18776. The 2D 1H-15N-HSQC spectrum of N-HvGR-RBP1 is shown in Figure 1. The spectral dispersion of chemical shifts and the 1H-15N resonance line widths are consistent with N-HvGR-RBP1 being monomeric in solution. Chemical shift index (CSI) analysis (Wishart et al. 1998) of N-HvGR-RBP1’s NMR data (see Fig. 2) reveals the presence of two α-helices and four β-strands in the protein, with helices 1 and 2 spanning residues: 22–30; 60–70, respectively, and β-strands 1, 2, 3, 4 spanning residues: 10–14; 48–56; 72–74; 78–84, respectively. The β1α1β2β3α2β4 secondary structural topology of N-HvGR-RBP1 is consistent with the topological sequence arrangement of secondary structural elements in canonical RRM domains. Further chemical shift analysis of all assigned NMR resonances of N-HvGR-RBP1 revealed no unusual chemical shifts compared to expected chemical shifts in the NMR database, or chemical shifts of comparable residues observed in other RRM containing protein structures solved by NMR. However, it remains to be determined whether the 3D structural architecture of the RMM domain of HvGR-RBP1 maps onto the 3D folds other RRM-containing proteins in three-dimensional space. Ultimately this will be accomplished by determining the 3D structure of N- HvGR-RBP1 in solution, which is underway.
Fig. 1.
2D 1H-15N- correlation-HSQC spectrum of a 1mM uniformly [1H-15N]-labeled N-HvGRRBP1 sample dissolved in buffer containing 300 mM NaCl, 50 mM Na2HPO4/NaH2PO4, 5 % (v/v) D2O, 0.05% sodium azide, 0.1 mM PMSF, at pH 6.5. The spectrum was acquired at 298 K on a 14.1 Tesla (600 MHz 1H Larmor frequency) Bruker solution NMR spectrometer. NMR signals connected by a thin line correspond to resonances from side chain NH2 groups of glutamine and asparagine residues. NH resonance assignments are indicated by the one-letter code of amino acids and their number position in the protein amino acid sequence on each NMR peak.
Fig. 2.
Chemical shift index (CSI) plot derived from consensus chemical shift indexing (i.e. based on the analysis of Hα, Cα, Cβ chemical shift deviations from random coil values) delineating elements of secondary structure as a function of residue number. Alpha helices and beta strands are depicted as coils and arrows, respectively above the CSI versus residue number plot.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by funds from the National Science Foundation (Grant number IOS-0918037 to A. Fischer (PI) and V. Copié (Co-PI)). The NMR experiments were recorded at Montana State University on a DRX600 Bruker solution NMR spectrometer, purchased in part with funds from the NIH Shared Instrumentation Grant (SIG) (Grant Number 1S10-RR13878-01), and recently upgraded to an AVANCE III console and cryogenically cooled TCI probe (Grant Number 1S10-RR026659-01). Support for the Mass Spectrometry Facility has been provided by The Murdock Charitable trust, NIH INBRE grant P20-RR-16455-08, and NIH Grants P20-RR-020185 and 1P20-RR-024237 from the NIH Center of Biomedical Research Excellence (CoBRE) Programs.
Abbreviations
- CSI
Chemical shift index
- ESI
micro-TOF-MS, electrospray ionization micro-time-of-flight mass spectrometry
- FL- HvGR-RBP1
full-length glycine-rich RNA binding protein 1
- HSQC
heteronuclear single quantum coherence
- GPC
grain protein content
- micro-TOF-MS
micro-time-of-flight mass spectrometry
- MS
mass spectrometry
- NAC family of transcription factors
NAC refers to the first letter of the following three NAM, ATAF1/2, and CUC2 transcription factor family members
- N- HvGR-RBP1
N-terminal domain of Hordeum vulgare L.-glycine rich- RNA binding protein 1
- NMR
nuclear magnetic resonance
- PMSF
Phenylmethylsulfonyl fluoride
- DSS
4, 4-dimethyl-4-silapentane-1-sulfonic acid
- RRM
RNA recognition motif
- SAGs
Senescence-associated genes
- SEC
size exclusion chromatography
REFERENCES
- Clery A, Blatter M, Allain F. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Corbin-Lickfett KA, Souki SK, Cocco MJ, Sandri-Goldin RM. Three Arginine Residues within the RGG Box Are Crucial for ICP27 Binding to Herpes Simplex Virus 1 GC-Rich Sequences and for Efficient Viral RNA Export. J Virology. 2010;84:6367–6376. doi: 10.1128/JVI.00509-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Gregersen PL, Holm PB, Krupinska K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol (Stuttg) Suppl. 2008;1:37–49. doi: 10.1111/j.1438-8677.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- Jukanti A, Heidlebaugh N, Parrott D, Fischer I, McInnerney K, Fischer A. Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol. 2008;177:333–349. doi: 10.1111/j.1469-8137.2007.02270.x. [DOI] [PubMed] [Google Scholar]
- Kuwasako K, Takahashi M, Tochio N, Abe C, Tsuda K, Inoue M, Terada T, Shirouzu M, Kobayashi N, Kigawa T, Taguchi S, Tanaka A, Hayashizaki Y, Guntert P, Muto Y, S, Y Solution Structure of the Second RNA Recognition Motfi (RRM) Domain of Murine T Cell Intracellular Antigen-1 (TIA-1) and Its RNA Recognition Mode. Biochemistry. 2008;47:6437–6450. doi: 10.1021/bi7024723. [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain F. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Schlenker C, Goel A, Tripet B, Menon S, Willi T, Dlakic M, Young M, Lawrence M, Copie V. Structural Studies of E73 from a Hyperthermophilic Archaeal Virus Identify the, RH3, alpha Domain, an Elaborated Ribbon, Helix, Helix Motif Involved in DNA Recognition. Biochemistry. 2012;51:2899–2910. doi: 10.1021/bi201791s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See D, Kanazin V, Kephart K, Blake T. Mapping genes controlling variation in barley grain protein concentration. Crop Sci. 2002;42:680. [Google Scholar]
- Tripet B, Goel A, Copié V. Internal dynamics of the tryptophan repressor (TrpR) and two functionally distinct TrpR variants, L75F-TrpR and A77V-TrpR, in their l-Trp-bound forms. Biochemistry. 2011;23:5140–5153. doi: 10.1021/bi200389k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D, Nip A. Protein chemical shift analysis: a practical guide. Biochem Cell Biol. 1998;76:153–163. doi: 10.1139/bcb-76-2-3-153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.