Abstract
Lactosylated gramicidin-containing lipid nanoparticles (Lac-GLN) were developed for delivery of anti-microRNA-155 (anti-miR-155) to hepatocellular carcinoma (HCC) cells. MiR-155 is an oncomiR frequently elevated in HCC. The Lac-GLN formulation contained N-lactobionyl-dioleoyl phosphatidylethanolamine (Lac-DOPE), a ligand for the asialoglycoprotein receptor (ASGR), and an antibiotic peptide gramicidin A. The nanoparticles exhibited a mean particle diameter of 73 nm, zeta potential of +3.5 mV, anti-miR encapsulation efficiency of 88%, and excellent colloidal stability at 4°C. Lac-GLN effectively delivered anti-miR-155 to HCC cells with a 16.1- and 4.1-fold up-regulation of miR-155 targets C/EBPβ and FOXP3 genes, respectively, and exhibited significant greater efficiency over Lipofectamine 2000. In mice, intravenous injection of Lac-GLN containing Cy3-anti-miR-155 led to preferential accumulation of the anti-miR-155 in hepatocytes. Intravenous administration of 1.5 mg/kg anti-miR-155 loaded Lac-GLN resulted in up-regulation of C/EBPβ and FOXP3 by 6.9- and 2.2- fold, respectively. These results suggest potential application of Lac-GLN as a liver-specific delivery vehicle for anti-miR therapy.
Keywords: nanoparticles, hepatocellular carcinoma, asialoglycoprotein receptor, gramicidin, drug targeting
1. Introduction
Hepatocellular carcinoma (HCC) accounts for a majority of liver cancers. It is one of the fast growing causes of cancer deaths, affecting more than 500,000 people each year [1]. MicroRNAs (miRs) are non-coding RNAs that regulate gene expression by translational repression through the RNA-induced silencing complex (RISC). Aberrant expression of miRs, either a reduction or an elevation, has been shown to be involved in various diseases, such as inflammation [8], cardiovascular disease [9], and cancers [10–12]. MiRs and anti-miRs have emerged as potential therapeutic agents. MiR-155, a hepatic oncogenic miR (oncomiR), is induced by a broad range of inflammatory mediators including proinflammatory cytokines, and is trans-activated by nuclear factor kappa B (NF-κB) [1]. Increased expression of miR-155 has been shown in human inflammatory cells and tumors [2–6]. Recent studies have shown that there is a correlation between the expression level of miR-155 and histopathological changes in choline-deficient and amino acid-defined (CDAA) diet-induced HCC and in primary human HCC. This suggests miR-155 as a potential therapeutic target in early stages of tumorigenesis [7].
The function of miRs can be inhibited by anti-miRs, which are oligonucleotides with complimentary sequences. Recently, the anti-miR strategy has been validated against several oncomiRs [13–17]. In this study, an anti-miR-155 was designed and synthesized specifically to inhibit miR-155 function. Free anti-miRs are unstable in the plasma and face barriers such as nucleases and renal clearance. Non-specific tissue uptake as well as inefficient delivery limit their potential application as a therapeutic agent. Lipid nanoparticles (LNs) have been shown to be effective for nucleic acid delivery [18–23].
HCC cells are derived from hepatic parenchymal cells with expression of the asialoglycoprotein receptor (ASGR) [24, 25]. ASGR is capable of recognizing galactose-terminated glycoproteins and glycoconjugates, therefore, is useful as a cellular marker for targeted delivery to hepatocytes and to HCC cells [26–29].
In the present study, a lipophilic ASGR targeting ligand, composed of lactobionic acid (LA), bearing a galactose moiety and linked to a phospholipid, was synthesized and incorporated into LNs for liver-specific delivery of anti-miR-155. In addition, gramicidin A, a hydrophobic peptide, was incorporated to facilitate endosomal release of the anti-miR following endocytosis [30–32]. This formulation was named lactosylated gramicidin-based LN (Lac-GLN). Its potential on hepatocyte targeting was evaluated in HepG2 cells and in mice. The physicochemical properties, cellular uptake, in vitro and in vivo delivery efficacy were investigated.
2. Materials and methods
2.1. Chemicals and reagents
1,2-Dioleoyl-3-dimethylammonium-propane (DODAP), and L-α-dioleoyl phosphatidylethanolamine (DOPE) were purchased from Avanti Polar Lipids (Alabaster, AL); 1, 2-dimyristoyl-sn-glycerol and methoxypolyethylene glycol (DMG-PEG) were purchased from NOF America Corporation (Elysian, MN); 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were from Thermo Scientific (Rockford, IL). Monomethoxy polyethylene glycol 2000-distearoyl phosphatidylethanolamine (mPEG-DSPE) was obtained from Genzyme Pharmaceuticals (Cambridge, MA). Cholesterol, lactobionic acid, gramicidin A and all other reagents were purchased from Sigma-Aldrich (St. Louis, MO) without further purification. Firefly Luciferase (GL2 + GL3) siRNA (Luci-siRNA) (AM 4629), negative scrambled control (AM 17010), and Lipofectamine 2000 were purchased from Invitrogen (Grand Island, NY). Anti-miR-155 (sequence: 5′-A*C*CCCUAUCACGAUUAGCAUU*A*A-3′, containing phosphorothioate linkages (*) and 2′-O-Methylation, Cy3-labeled anti-miR-155 (Cy3-anti-miR-155), and Cy5.5-labeled anti-miR-155 (Cy5.5-anti-miR-155) were synthesized by Alpha DNA (Montreal, Canada). The Taqman kits for real-time RT-PCR assay of miR-155 (002623) and RNU6B (001093) were purchased from Applied Biosystems (Carlsbad, CA).
2.2. Preparation of anti-miR-155 containing Lac-GLN
The targeting ligand was synthesized as described previously [33]. Briefly, lactobionic acid was activated by EDC and converted to its NHS ester, which is then reacted with DOPE to yield n-lactobionyl-DOPE (Lac-DOPE). The product was characterized by Fourier transform infrared (FTIR) spectrometry on a Nexus 470 FTIR Spectrometer (Thermo Scientific, Rockford, IL). Lac-GLNs were prepared by the ethanol injection method. The lipid mixture, composed of DODAP, Lac-DOPE, DOPE, DMG-PEG and gramicidin A at a molar ratio of 50:10:28:2:10, was dissolved in ethanol, and rapidly injected into RNAse- and DNAse-free HEPES buffered solution (20mM, pH 7.4). The resulting lipid nanoparticles were sonicated for 2 min by a bath sonicator and dialyzed against RNAse- and DNAse-free water for 4 hr at room temperature to remove ethanol using a molecular weight cut-off (MWCO) 10,000 Dalton Float-A-Lyzer (Spectrum Laboratories Inc., Ranco Dominguz, CA).
The anti-miR-155 containing Lac-GLN was prepared by adding an equal volume of anti-miR-155 dissolved in RNAse- and DNAse-free HEPES buffer to Lac-GLN, followed by brief vortexing for 10 sec and incubation at room temperature for 10 min. The weight ratio of lipids: anti-miR was fixed at 10: 1, and the concentration of anti-miR-155 was 1 μg/mL. The resulting nanoparticles were sterilized using 0.22 μm filters (Fisher Scientific, Pittsburgh, PA). Control formulations were prepared by the same method.
2.3. Size, surface charge, and encapsulation efficiency measurements
The particle size of anti-miR-155 containing Lac-GLN was determined by dynamic light scattering on a Model 370 NICOMP Submicron Particle Sizer (NICOMP, Santa Barbara, CA) in the volume-weighted distribution mode. Particles were dispersed in cell culture medium. The morphology of Lac-GLN was examined by a FEI Tecnai G2 Bio TWIN transmission electron microscope (FEI Company, OR, USA). Briefly, samples were prepared as described above. A drop of the sample was negatively stained with uranyl acetate for 1 min on a perforated carbon grid for analysis. Images were recorded using a Gatan 791 MultiScan CCS camera and processed by the Digital Micrograph 3.1 software package.
The zeta potential of anti-miR-155 containing Lac-GLN was examined in 20mM HEPES buffer using ZetaPALS zeta potential analyzer (Brookhaven Instruments Corp., Holtsville, NY).
Encapsulation efficiency of Lac-GLN was determined by Quant-iT RiboGreen RNA Kit (Invitrogen, Grand Island, NY) following the manufacturer’s protocol, and the fluorescence intensity (FI) was determined using a luminescence spectrometer (KS 54B, Perkin Elmer, UK) at an excitation of 480 nm and an emission of 520nm. The encapsulation efficiency was calculated by the following equation.
2.4. Colloidal and serum stability of Lac-GLN
The colloidal stability of anti-miR-155 containing Lac-GLN was determined by monitoring changes in its particle size over a 30-day period during storage at 4°C or 25°C. A serum stability test was performed to investigate the ability of Lac-GLN to protect anti-miR from serum nuclease degradation. Briefly, anti-miR-155-lac-GLN and free anti-miR-155 were exposed to 50% fetal bovine serum (FBS) and incubated at 37°C for various time periods. Aliquots of each sample were then loaded onto a 1.5% (w/v) agarose gel containing ethidium bromide.
2.5. Cell culture and in vitro transfection studies
Human HCC SK-Hep-1 and HepG2 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2.
For Luci-siRNA transfection, 2 × 104 SK-Hep-1 cells stably expressing luciferase, were seeded per well in 800 μl culture medium in 48-well plates and allowed to grow overnight at 37°C under 5% CO2 atmosphere. Next day, the culture medium was replaced with medium containing 0%, 10% and 20% FBS, and cells were transfected with Luci-siRNA containing Lac-GLN and various control formulations at 100 nM for 4 hr. After transfection, the medium was replaced with fresh medium containing 10 % FBS. At 48 hrs post transfection cells were washed with PBS and luciferase activity in cell lysates was determined using Luciferase Assay Kit (Promega, Madison, WI) following manufacturer’s instruction. Briefly, the total amount of protein of each well was determined using BCA Assay Kit (Pierce, Rockford, IL), and luciferase activity was determined by normalization to the total amount of protein. Luciferase down-regulation was then calculated as a relative value compared to the untreated negative control.
For anti-miR-155 transfection, HepG2 cells were plated at 2 × 105 cells per well in 6-well plates with 2 ml cultured medium, and incubated overnight at 37°C under 5% CO2 atmosphere. The culture medium was then replaced with fresh medium, and cells were transfected with 100 nM anti-miR-155 using Lipofectamine 2000, Lac-GLN, and control formulations and after 4 hr incubation, the medium was replaced with fresh medium. Cells were incubated for an additional 48 hr at 37°C, then miR-155 and its target gene expression level was determined by real time RT-PCR analysis. As a positive control, cells transfected with Luc-siRNA and anti-miR-155 using Lipofectamine 2000 were performed following manufacturer’s protocol. Untreated cells and empty Lac-GLN were used as negative controls.
2.6. Cytotoxicity study by MTS assay
The cytotoxicity of Lac-GLN was evaluated by MTS assay (Promega, Madison, WI). HepG2 cells were seeded in 96-well plates at a density of 1 × 104 cells per well. After overnight incubation, cells were treated with empty Lac-GLN, negative control RNA alone, anti-miR-155 alone, negative control RNA containing Lac-GLN, and ant-miR-155 containing Lac-GLN at an RNA concentration of 100 nM for 24 hr. MTS reagent (20 μl) was then added to each well, and cells were incubated for another 2hr. The optical density (OD) at 490 nm of each well was measured using a Multiskan Ascent automatic plate reader. Untreated cells were used as control and defined as 100% viable. Cell viability was calculated as a percentage of the untreated cells.
2.7. Cellular uptake of Lac-GLN by confocal microscopy and by flow cytometry
Analysis of the cellular uptake of Lac-GLN was performed by delivery of fluorescent Cy3-anti-miR-155 into HepG2 cells, evaluated by confocal microscopy and by flow cytometry.
For confocal microscopy, 2 × 105 HepG2 cells per well were seeded in 6-well plates containing a sterile glass coverslip at the bottom of each well (Fisher Scientific, 12-545-82, Pittsburgh, PA) and allowed to grow overnight. Cells were then treated with 100 nM Cy3-anti-miR-155 containing GLN, Lac-GLN, or Lac-GLN with 20 mM lactose and 1% BSA for 1 hr at 37°C, followed by a wash step with PBS five times. Cells were fixed with 4% paraformaldehyde for 15 min, and stained with Hoechst 33342 (Invitrogen, Grand Island, NY) and Alexa-488 phalloidin (Invitrogen, Grand Island, NY) for 10 min each at room temperature. The glass coverslip with the cells was then detached from the plates and covered with a regular glass slide. Confocal analysis was performed on an Olympus FV1000 Filter Confocal Microscope (Olympus Optical Co., Tokyo, Japan).
For the flow cytometric analysis, 2 × 105 HepG2 cells were treated with 100 nM Cy3-anti-miR-155 containing GLN, Lac-GLN, GLN with 20 mM lactose and 1% BSA, or Lac-GLN with 20 mM lactose and 1% BSA for 1 hr at 37°C. Cells were suspended using 0.25% trypsin, washed with PBS five times, and fixed with 4% paraformaldehyde. The fluorescent intensity was measured on a Becton Dickinson FACScalibur Flow Cytometer (Franklin Lakes, New Jersey), and a total of 10,000 events were collected for each sample.
2.8. Assessment of gene silencing by real-time RT-PCR
Total RNA from transfected cells or tissue extracts was isolated by TriZol reagent (Invitrogen, Grand Island, NY) and purified by following the standard protocol. The miR-155 cDNA was synthesized using TaqMan MicroRNA reverse transcription Kit (Applied Biosystems, Carlsbad, CA), and the cDNA was amplified and quantified using the TaqMan MicroRNA Kit (Applied Biosystems, Carlsbad, CA). The cDNA of C/EBPβ and FOXP3 was synthesized using the first-strand cDNA synthesis kit (Invitrogen, Grand Island, NY) and resulting cDNA was amplified and quantified using SYBR Green method (Applied Biosystems, Carlsbad, CA).
Primers were designed by the Primer Express Program (Applied Biosystems), C/EBPβ. (forward: 5′-AGAAGACCGTGGACAAGCACAG-3′, reverse: 5′-TTGAACAAGTTCCGCAG GGTGC-3′), FOXP3 (forward: 5′-AATGGCACTGACCAAGGCTTC-3′, reverse: 5′-TGTG GAGGAACTCTGGGAATGTG-3′) and GAPDH (forward: 5′-CCCCTGGCCAAGGTCATC CATGACAACTTT-3′, reverse: 5′-GGCCATGAGGTCCACCACCCTGTTGCTGTA-3′). miR-155 level was normalized to that of RUN6B, while C/EBPβ and FOXP3 levels were normalized to that of GAPDH. Their expression levels were calculated using the 2−ΔCT approach [34].
2.9. In vivo biodistribution studies by confocal microscopy and IVIS imaging
Fluorescent Cy3-anti-miR-155 containing GLN and Lac-GLN were used in this study for confocal microscopy analysis. Male C57BL/6 mice were given Cy3-anti-miR-155 (50 μg) containing GLN or Lac-GLN intravenously with a total injection volume of 200 μl. After 4 hr, mice were sacrificed and tissues were collected. Harvested tissues were fixed in 4% paraformaldehyde for 6 hr and soaked in 30% sucrose overnight at 4 °C. Tissues were then transferred to block holders, embedded with O.C.T. freezing medium (Fisher Scientific, Pittsburgh, PA), and frozen in liquid nitrogen. Tissue samples were processed by sectioning, and were stained with Hoechst 33342 (Invitrogen, Grand Island, NY) and Alexa-488 phalloidin (Invitrogen, Grand Island, NY) for 10 min each at room temperature. The Fluorescent images were obtained using an Olympus FV1000 Filter Confocal Microscope (Olympus Optical Co., Tokyo, Japan).
Fluorescent Cy5-anti-miR-155 containing GLN and Lac-GLN were used for measuring in vivo uptake in different tissues by IVIS imaging. The same treatment conditions as described above were used for this experiment. Whole tissues were harvested and fixed in 4% paraformaldehyde for 6 hr and immersed in 30% sucrose for 12 hr at 4°C. Whole tissue Cy5 fluorescent signals were measured using Xenogen IVIS-200 Optical In Vivo Imaging system (Caliper Life Sciences, Hopkinton, MA).
2.10. In vivo delivery of anti-miR-155 containing Lac-GLN
Negative control RNA or anti-miR-155 containing Lac-GLN and other controls were given to male C57BL/6 mice by intravenous injection at a dose of 1.5 mg/kg. At 48 hr post administration, the mice were anesthetized, and liver tissues were harvested and immediately frozen in liquid nitrogen. RNA extraction and RT-PCR were performed as described above.
2.11. Statistical analyses
Results were reported as mean ± standard deviation, and a minimum of triplicates were used in each experiment. Comparisons between the groups were analyzed by Student’s t test for two groups and ANOVA for multiple groups. Results were considered statistically significant when p values were <0.05. All statistical analyses were performed by Microsoft Excel 2003 software.
3. Results
3.1.Synthesis of Lac-DOPE
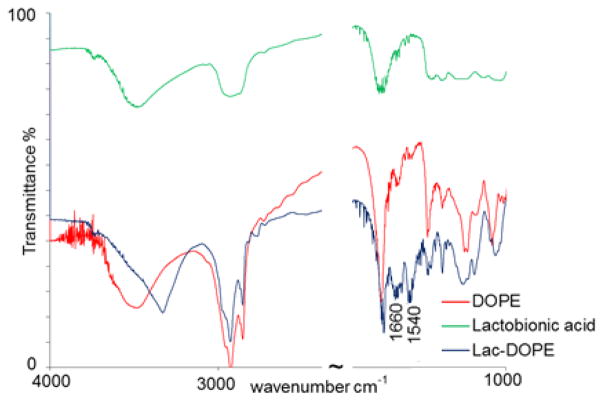
This targeting ligand has been synthesized in our lab previously [33]. In the current study, the product was further characterized by FTIR to confirm the identity of the conjugate. As shown in Fig. 1, the absorption peaks of lac-DOPE in blue at 1660 cm−1 and 1540 cm−1 indicate amide bond formation. The spectra of reactants, DOPE and lactobionic acid, were indicated in red and green, respectively.
Figure 1.
FTIR spectra of Lac-DOPE (blue), DOPE (red) and lactobionic acid (green).
3.2. Characterization of anti-miR-155 containing Lac-GLN
The particle size and zeta potential of GLN with various molar percentages of Lac-DOPE were evaluated and presented in Fig. 2A. The formulation with 10% Lac-DOPE had an average diameter of 72.66 nm and a zeta potential of +3.49 mV. This composition was selected as the delivery vehicle for the subsequent experiments and termed Lac-GLN. The size and morphology of Lac-GLN was further examined by TEM. The image in Fig. 2B showed a spherical shape and a relatively uniform size distribution of Lac-GLN with < 100 mm in diameter, which was consistent with data obtained by DLS.
Figure 2.
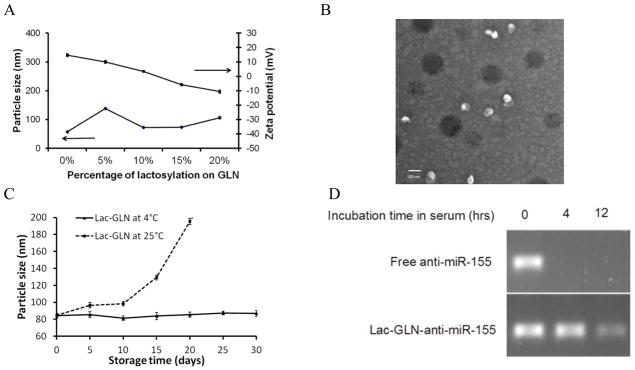
Characterization of Lac-GLN. A. Particle size and zeta potential of GLN with varying degrees of lactosylation. Each value represents the mean ± SD of five measurements. B. Morphology of anti-miR-155-Lac-GLN by TEM. Scale bar represents 100 nm. C. Colloidal stability of Lac-GLN. Lac-GLN-anti-miR-155 was stored at 4 °C or 25°C and particle size was measured over time. Results are the mean of three separate experiments. Error bars stand for standard deviations. D. Serum stability of anti-miR-155-Lac-GLN. Anti-miR-155 alone or anti-miR-155-Lac-GLN were mixed with 50% FBS at 37°C for 0 hr, 4 hr, and 12 hr. Samples were then analyzed gel electrophoresis analysis.
The encapsulation efficiency was determined as described in Materials and Methods. As shown in Table 1, the encapsulation efficiency of Lac-GLN was >85%.
Table 1.
Physicochemical properties of anti-miR-155 containing GLN and Lac-GLN. Values are mean ± SD (n=5).
| Formulation | Particle size (nm) | Zeta potential (mV) | Encapsulation efficiency (%) |
|---|---|---|---|
| GLN | 57.39 ± 2.73 | 14.72 ± 1.15 | 87.5 ± 1.9 |
| Lac-GLN | 72.66 ± 3.14 | 3.49 ± 0.77 | 88.9 ± 2.2 |
The colloidal stability was determined by monitoring the change in particle size over time. As shown in Fig. 2C, the average diameter of Lac-GLN remained unchanged over a 30 day period at 4°C, but a significant increase in the average diameter was observed under storage at 25°C.
The ability of Lac-GLN to protect anti-miR was evaluated by a serum stability test. In this study, free anti-miR and anti-miR-155-Lac-GLN were mixed with FBS and incubated at 37°C for different time periods. As shown in Fig. 2D, Lac-GLN was able to protect ant-miR-155 from nuclease degradation for up to 12 hrs. Meanwhile free anti-miR-155 was completely digested within 4 hr. This result demonstrated good serum stability for Lac-GLN.
3.3. Cellular uptake of GLN and Lac-GLN
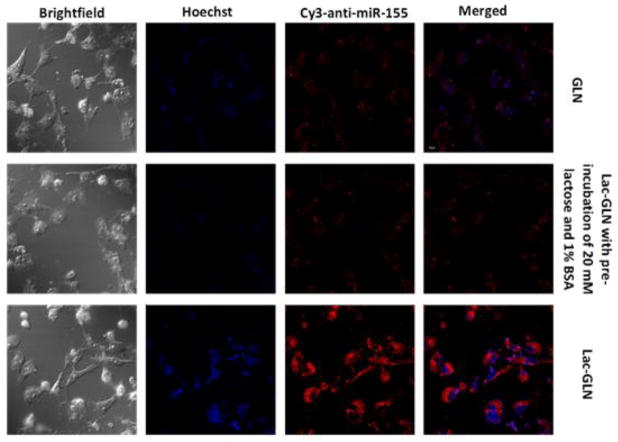
Uptake of GLN and Lac-GLN by HCC HepG2 cells, which has highASGR expression was determined using Cy3-anti-miR-155 containing formulations. The pre-incubation with 20 mM lactose and 1% BSA was used to block ASGR-mediated and non-specific uptake, respectively. Cells were evaluated by confocal microscopy. As shown in Fig. 3, cells treated with Lac-GLN showed a significantly stronger fluorescence signal than those treated with non-targeted GLN. This uptake enhancement was reduced in cells pre-treated with lactose, an ASGR blocking agent, which demonstrated that the cellular uptake of Lac-GLN was ASGR-specific.
Figure 3.
Cellular uptake of Cy3-anti-miR-155 containing Lac-GLN and other control formulations in HepG2 cells as determined by confocal microscopy.
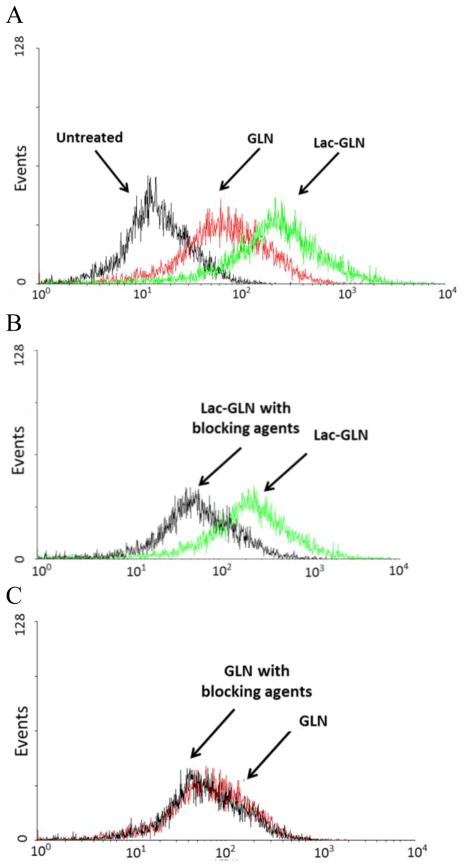
Cellular uptake of GLN and Lac-GLN was further quantified by flow cytometry. As shown in Fig. 4A, the uptake of Lac-GLN was about 3.58-fold higher than that of non-targeted GLN in HepG2 cells. The fluorescence signal did not reduce significantly in the GLN pretreated cells with 20 mM lactose and 1% BSA, suggesting ASGR-independent uptake of GLN by HCC cells (Fig. 4C). However, the uptake of Lac-GLN was reduced by 3.51-fold in cells pre-incubated with lactose. While in absence of lactose the uptake levels of GLN and Lac-GLN treated cells were comparable (Fig. 4B). This result further confirmed that ASGR-targeted delivery improved cellular uptake of Lac-GLN in HCC cells.
Figure 4.
HepG2 cells were treated with GLN, Lac-GLN or Lac-GLN pre-incubated with 20 mM lactose and 1% BSA. Fluorescence signals were measured on a FACSCalibur flow cytometer. A. HepG2 cells were treated with GLN or Lac-GLN. B and C. The effect of pre-incubation with 20 mM lactose and 1% BSA on Lac-GLN and GLN, respectively. Results are shown in the histogram with the X- and Y-axis indicating the fluorescence intensity and the cell count, respectively.
3.4 In vitro delivery of Lac-GLN
HCC SK-Hep1 cells, stably expressing firefly luciferase mRNA, were used to determine the transfection efficiency of different delivery vehicles and the effects of several factors, including targeting ligand, gramicidin A, and serum, on transfection efficiency, using siRNAs for luciferase as a reporter molecule. Result shows significantly reduced expression of luciferase in Lac-LN treated group (78.95%) compared to that in the LN treated group (96.35%) in FBS-free medium. This confirmed an advantage for the ASGR-targeted strategy. In addition, addition of 20% FBS reduced transfection efficiency by only 6%. Treatment with the commercial Lipofectamine 2000 caused 7.84% reduction in luciferase expression, close to the LN treated group, and the transfection was strongly inhibited by serum at high concentration. Moreover, the effect of increasing concentration of gramicidin A was analyzed. Surprisingly, in the Lac-GLN treated groups, neither 5% nor 10% gramicidin A was affected by the presence of FBS during transfection. In contrast, transfection activities of previously reported formulations were sensitive to the presence of serum [35–37]. Similar results were obtained in HepG2 cells (data not shown). This is significant since serum is a major barrier for in vivo delivery in a clinical setting.
Cytotoxicity of the anti-miR formultions was investigated on HCC cells. HepG2 cells were treated with equal amount of empty Lac-GLN, negative control RNA alone, anti-miR-155 alone, negative control RNA-Lac-GLN, or anti-miR-155-Lac-GLN. No significant change in cell viability was observed between treated cells and untreated cells (data not shown). This result suggested a low cytotoxicity of Lac-GLN in HepG2 cells.
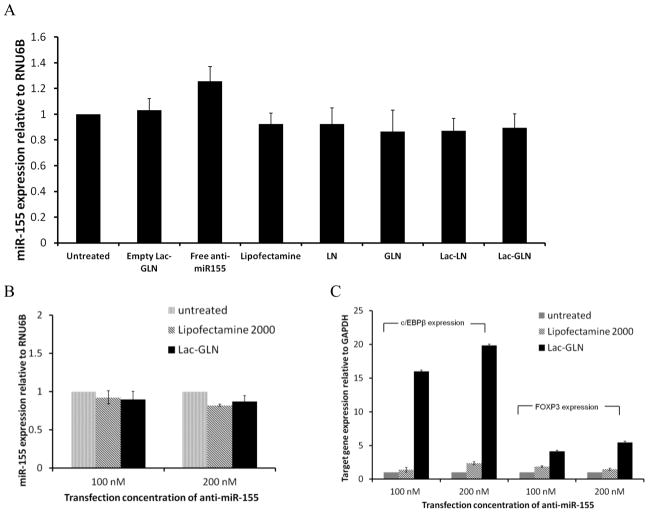
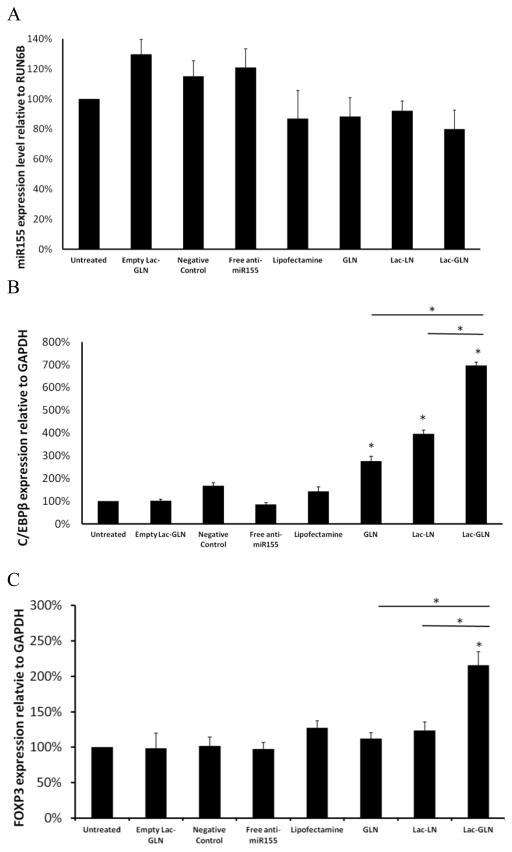
Next, the effects of Lac-GLN containing anti-miR-155 on miR-155 and its downstream targets expression were evaluated in HepG2 cells. Cells were treated with anti-miR-155 containing Lac-GLN and other control formulations for 4 hr, and the miR-155 and its target gene expression were measured 48 hr after transfection by real time RT-PCR. Fig. 5A shows the miR-155 expression levels from different treatment groups relative to the untreated group. The positive control, treated with Lipofectamine 2000, had 92.4% miR-155 expression of the untreated. Meanwhile, LN, GLN, Lac-LN and Lac-GLN treated groups exhibited a similar miR-155 expression level to that of Lipofectamine 2000 treated group. Differences among these groups were small.
Figure 5.
In vitrodelivery of anti-miR-155-Lac-GLN. The results are the mean of three repeats. Error bars stand for standard deviations. A. HepG2 cells were transfected with anti-miR-155 containing Lac-GLN at the concentration of 100 nM for 4 hr, and miR-155 expression was evaluated 48 hr after transfection. B. Evaluation of different concentrations of anti-miR-155 treatments on miR-155 expression. HepG2 cells were transfected with 100 nM or 200 nM anti-miR-155 containing Lipofectamine2000 and Lac-GLN for 4 hr, and miR-155 expression was evaluated 48 hr after transfection. C. Evaluation of miR-155 targeting gene expressions. C/EBPβ and FOXP3 gene expression were evaluated 48 hr after HepG2 cells were transfected with positive control or Lac-GLN containing 100 nM and 200 nM anti-miR-155.
When the anti-miR-155 concentration was doubled from 100 nm to 200 nM, as shown in Fig. 5B, the miR-155 expression in the Lipofectamine 2000 treated group changed from 92.4% to 89.5%. This difference was still not statistically significant. In the Lac-GLN treated group, a similar trend was observed, where the expression of miR-155 was 87.2% and 82.9% in 100 nM and 200 nM anti-miR-155 treatments, respectively. These suggested anti-miR-155 delivery did not lead to miR-155 degradation.
To further examine the delivery efficiency, the expression of miR-155 target genes, C/EBPβ and FOXP3, were evaluated. The results are summarized in Fig. 5C. In contrast to the steady expression level of miR-155 (Fig. 5B), there were a 16.1- and 4.1-fold increase in C/EBPβ and FOXP3 expression, respectively, in the Lac-GLN 100 nM anti-miR-155 treatment group. Only a 1.4-, 1.9-fold increase of C/EBPβ and FOXP3 expression was observed in cells transfected with Lipofectamine 2000, respectively. Furthermore, doubling the anti-miR-155 concentration resulted in an increase in up-regulation of C/EBPβ and FOXP3 expression, clearly demonstrating that the miR-155 targeting gene expression was dependent on anti-miR-155 concentration. Thus, the delivery of anti-miR-155 most likely resulted in functional inhibition of miR-155 rather than its degradation, as is often the case for high affinity anti-miRs. In addition, these results indicated Lac-GLN’s superiority over the commercial available agent in anti-miR delivery.
3.5. In vivo biodistribution of Lac-GLN
In order to assess the in vivo delivery efficiency and tissue specificity of Lac-GLN, tissue distribution study was performed in C57BL/6 mice that were administrated Cy5-anti-miR-155 containing GLN or Lac-GLN intravenously at a dose of 1.5 mg/kg. After 4 hr, organs were harvested and fluorescence signals were compared. As shown in Fig. 6, lung, spleen and liver were the major organs exhibiting fluorescence signals when mice were injected with non-targeted GLN. In contrast, maximal fluorescence signals accumulated in the liver when mice were treated with Lac-GLN with very weak signals in the spleen and kidney and no detectable signal in lung. These results suggested that the delivery of Cy5-anti-miR-155 by Lac-GLN was liver-specific and that Lac-GLN was able to minimize off-target uptake, thus improving the overall delivery efficiency to the liver.
Figure 6.
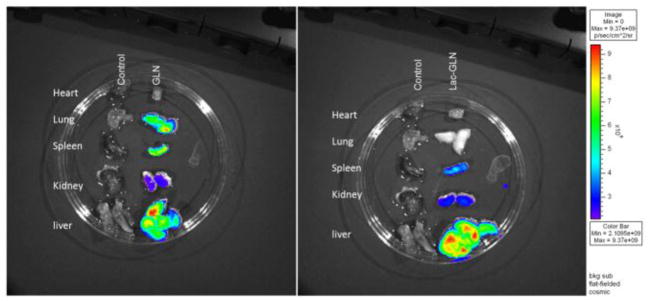
Tissue distribution of Cy5-anti-miR-155 containing GLN and Lac-GLN. Heart, lung, spleen, kidney and liver were harvested from C57BL/6 mice 4 hr after intravenous administration of Cy5-anti-miR-155 containing GLN or Lac-GLN. Cy5 fluorescence signals were measured by IVIS Imaging system.
Confocal microscopy was performed on the liver and other organs to further evaluate the delivery properties of GLN and Lac-GLN. Besides hepatocytes, the liver also contains a large population of Kupffer cells, which are residential macrophage. As shown in Fig. 7A, a major proportion of fluorescence signals was taken up by hepatocytes rather than Kupffer cells in liver when mice were treated with Lac-GLN, while the uptake was predominantly by Kupffer cells in the non-targeted GLN-Cy5-anti-miR-155 treated liver.
Figure 7.
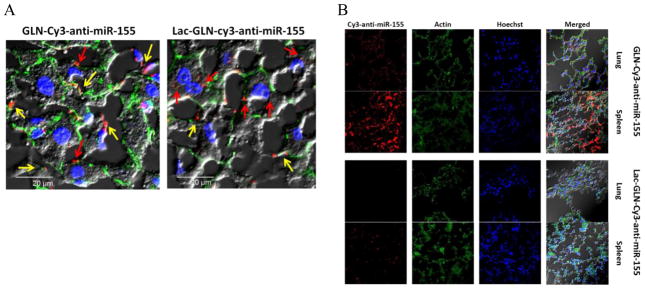
Confocal microscopy of Cy3-anti-miR-155 containing GLN and Lac-GLN in liver (A) and other organs (B). Liver, lung, and spleen were harvested from C57BL/6 mice after 4 hr intravenous administration of Cy3-anti-miR-155 containing GLN or Lac-GLN. Cy3 fluorescence signals were visualized on an Olympus FV1000 Filter Confocal Microscope. In (A), the red and yellow arrows indicate the uptake of Cy3-anti-miR-155 by hepatocytes and Kupffer cells, respectively.
The distribution of fluorescence signals in lung and spleen were also examined to evaluate Lac-GLN delivery. As shown in Fig. 7B, fluorescence signals accumulating in lung and spleen in the Lac-GLN treated mice were less than those in the GLN treated mice, suggesting high specificity of Lac-GLN for delivery to the liver.
3.6.In vivo delivery of anti-miR-155 containing Lac-GLN
Next, we sought to determine the delivery efficiency of Lac-GLN-anti-miR-155 in C57BL/6 mouse liver. For this purpose, mice were injected a single dose of 1.5 mg/kg anti-miR-155 formulated in Lipofectamine 2000, GLN, Lac-LN or Lac-GLN through the tail vein. Injections of empty Lac-GLN, negative control RNA containing Lac-GLN or free anti-miR-155 were used as negative controls. At 48 hr post administration, the mice were sacrificed and livers were harvested. The expression of miR-155 and its target, C/EBPβ, was evaluated by real time RT-PCR (Fig. 8A). As expected, miR-155 level was not altered in the negative control groups. A slight decrease in miR-155 expression by 13% and 20% was observed when anti-miR-155 was delivered using Lipofecamine 2000 and Lac-GLN, respectively, compared to the untransfected control. However, the differences among GLN, Lac-LN and Lac-GLN were not significant. On the contrary, the delivery efficiency reflected by C/EBPβ expression varied considerably among these groups as demonstrated by a 2.8-, 3.7- and 6.9-fold increases in its expression in GLN, Lac-LN and Lac-GLN treated groups, respectively, compared to the untreated group (Fig. 8B). No significant changes were observed in the negative control groups, and the Lipofectamine 2000 treated group only exhibited a 1.4-fold up-regulation of C/EBPβ. In addition, another miR-155 target gene, FOXP3 expression, was increased by 1.1-, 1.2-, and 2.1-fold in GLN, Lac-LN and Lac-GLN treated groups, respectively (Fig. 8C). These data demonstrate superior delivery efficiency by Lac-GLN and agree with the results of our in vitro experiments (Fig. 5).
Figure 8.
In vivo evaluation of anti-miR-155 treatments on miR-155 and target gene expressions. C57BL/6 mice were treated with 1.5 mg/kg anti-miR-155 containing Lac-GLN and control formulations. 4 hr after intravenous administration, liver tissues were harvested and RNA was extracted. Each value represents the mean ± SD of three measurements. A. The expression of miR-155 was analyzed by real time RT-PCR. B and C. The expression of the miR-155 downstream targets, C/EBPβ and FOXP3, were analyzed by real time RT-PCR. Each value represents the mean ± SD of three measurements.
4. Discussion
Aberrant miR expression is observed in almost all types of cancer. Since miRs repress multiple target mRNAs and regulate various cellular pathways, extensive studies have been focused on the biological function of miRs and their potential applications in cancer therapies. In HCC, dysregulated miR expression profiles are under investigation, and miR-155 has been recently identified as a potential oncogenic miR in CDAA diet-induced hepatocarcinogenesis and in human primary HCC [7, 38–39]. The overexpression of miRs can be specifically diminished by anti-miRs that are complimentary to the sequence of target miRs, thus silencing the activity of targeted miRs. Therefore, anti-miR-155 was designed and delivered to specifically inhibit the function of miR-155. Traditional methods for oligonucleotide delivery include electroporation and commercial lipid-based delivery agent such as Lipofecamine 2000. Chemical modification to the backbone of oligonucleotides is also used to enhance the stability against enzymatic degradation, and is well known for its ability for direct delivery to cells without the help of transfection agents. However, these methods often exhibit limited delivery efficacy in in vivo applications due to various barriers such as serum nucleases, lysosomal degradation, and off target uptake. In this study, a novel HCC-targeted LN with the capability of overcoming in vivo delivery barriers was designed and synthesized, and the delivery efficiency was evaluated in both HCC cells and in mice.
To increase the delivery efficiency, a hepatocyte targeting ligand was first designed and introduced to the formulation. Mammalian hepatic parenchymal cells are well known for their highly specific expression of ASGR on the surface of cell membranes. These receptors are capable of binding galactose moieties and internalize them through receptor-mediated endocytosis. Consequently, LA bearing a galactose group was employed as the recognition moiety to ASGR, and was attached to DOPE through an amide bond to form the lipophilic ligand (Fig. 1). The long hydrocarbon tails of DOPE facilitated stable insertion into the LN. Because this synthesized ligand was a negatively charged phospholipid-based compound, it was able to reduce the surface charge of cationic LN to some degrees (Fig. 2). Neutral LN has been shown in several studies to be advantageous in lipid-based delivery systems to the liver due to favorable adsorption from or in endogenous ApoE [40–41]. Therefore, a 10% Lac-DOPE was chosen as the molar ratio in the Lac-GLN formulation based on the resultant neutral surface charge and optimal particle size (Table 1 and Fig. 2). The delivery efficiency was further improved by the incorporation of a hydrophobic peptide, gramicidin A. The general function of gramicidin A is ion channel formation in a lipid bilayer. This ionophore enables transportation of monovalent ions across the membrane. During endocytosis processes, endosomal escape is known to be a limiting step for lipid-based delivery system [42–43]. Therefore, gramicidin A was incorporated in the formulation to improve endosomal escape. Presumably, this ionophore could provide additional ion transport and increase membrane permeability, in turn causing the swelling of the endosome and release of the trapped substances into cytoplasm, preventing lysosomal degradation [44–45].
The combination of synthesized ASGR targeting ligand and gramicidin A in the formulation seem to contribute to improved cellular uptake (Fig. 3), low cytotoxicity (data not shown), and enhanced transfection efficiency compared to commercial transfection agent (Fig. 5C). When Lac-GLN were delivered in vivo, they favored localization to the liver and diminished the off-target uptake from other tissues to a great extent (Fig. 6 and Fig. 7B). Kupffer cells are liver macrophages that are found next to hepatocytes in liver. The phagocytic activity of these non-parenchymal cells leads to non-specific uptake of LN. A targeted strategy employing Lac-GLN mediated delivery ensured a successful active ASGR targeting to hepatocytes and reduced the uptake by Kupffer cells significantly (Fig. 7A). Besides the barriers to endosomal escape, another dominant barrier in in vivo LN delivery is the serum inhibition, which cannot be ignored in clinical application. The inhibitory effect of serum occurs when serum proteins bind to nanoparticles and prevent their delivery to target cells. Indeed, this Lac-GLN exhibited greatly reduced serum-sensitivity in vitro compared to Lac-LN without gramicidin in the formulation (Fig. 8B).
An interesting observation in this study involved the expression level of miR-155 after the delivery of anti-miR-155. The miR-155 expression was not affected by anti-miR-155 transfection in both HepG2 cells and mouse liver, and its expression remained unaffected with increasing dose of anti-miR-155 (Fig. 5A, Fig. 5B and Fig. 8A). Conversely, the miR-155 target gene expression levels were strongly modulated by anti-miR-155 delivery in a concentration-dependent manner (Fig. 5C, Fig. 8B and Fig. 8C). One possible explanation would be that the delivery of anti-miR-155 blocked miR-155 function without triggering its degradation. Indeed, the binding affinity between antisense oligonucleotide and its targeted miR has been reported as the critical determinant of anti-miR activity. While low affinity binding promotes miR degradation, high affinity binding represses miR function by forming a stable complex without inducing degradation [46].
To our knowledge, there are no previous reports focusing on the development of a targeted lipid-based delivery system, in combination with a peptide in the formulation, for anti-miR delivery to liver. In this study, an ASGR-targeted peptide-based LN was synthesized for specific delivery of anti-miR-155 to HCC cells and liver. This Lac-GLN exhibited favorable physiochemical properties, preferential uptake by targeted cells and tissues, and the ability to overcome serum inhibition. The silencing potency of Lac-GLN containing anti-miR-155 was confirmed by the significant up-regulation of target genes that are known to be repressed by miR-155 both in vitro and in vivo. These findings suggest potential application of the formulation in anti-miR therapies in liver disease. The delivery efficacy of Lac-GLN in HCC tumor models will be carried out in in future studies.
Acknowledgments
This work was supported by the grants DK088076 (K.G) and CA152969 (K.G., S.T.J., L. J. L. and R. L.) from National Institutes of Health. The authors wish to thank Richard Montione from OSU CMIF for the help on TEM imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA- 155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertzog PJ, O’Neill LA, Hamilton JA. The interferon in TLR signaling: more than just antiviral. Trends Immunol. 2003;24:534–539. doi: 10.1016/j.it.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, Poppema S. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2006;37:20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 6.Tam W, Dahlberg JE. MiR-155/BIC as an oncogeneic miRNAGenes. Chromosomes Cancer. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- 7.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal k, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connel RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA–cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 13.Love TM, Moffett HF, Novina CD. Not miR-ly small RNAs: big potential for microRNAs in therapy. J Allergy Clin Immunol. 2008;121:309–19. doi: 10.1016/j.jaci.2007.12.1167. [DOI] [PubMed] [Google Scholar]
- 14.Stenvang J, Lindow M, Kauppinen S. Targeting of microRNAs for therapeutics. Biochem Soc Trans. 2008;36:1197–200. doi: 10.1042/BST0361197. [DOI] [PubMed] [Google Scholar]
- 15.Xiao J, Yang B, Lin H, Lu Y, Luo X, Wag Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination of the pacemarker channel genes HCN2 and HCN4. J Cell Physiol. 2007;212:285–92. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]
- 16.Su J, Baigude H, McCarroll J, Rana TM. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. 2011;39(6):e38. doi: 10.1093/nar/gkq1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JK, Kogure T, Nuovo GJ, Jiang J, He L, Kim JH, Phelps MA, papenfuss TL, Croce CM, Patel T, Schmittgen TD. miR-221 silencing blocks hepatocellular carcinoma and promotes curvival. Cancer Res. 2011;71:7608–7616. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YH, Liu F, Choi JS, Kim SW, Park JS. Characterization of a targeted gene carrier, lactose-polyethylene glycol-grafted poly-L-lysine and its complex with plasmid DNA. Human gene therapy. 1999;10:2657–2665. doi: 10.1089/10430349950016690. [DOI] [PubMed] [Google Scholar]
- 19.Toncheva V, Wolfert MA, Dash PR, Oupicky D, Ulbrich K, Seymour LW, Schacht EH. Novel vectors for gene delivery formed by self-assembly of DNA with poly (L-lysine) grafted with hydrophilic polymers. Biochim Biophys Acta. 1998;138(3):354–368. doi: 10.1016/s0304-4165(98)00004-x. [DOI] [PubMed] [Google Scholar]
- 20.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv Deliver siRNA and miRNA for cancer Therapy. Mol Ther. 2010;18:650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petcot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2010;28:172–6. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferber D. Gene therapy: safer and virus-free. Sience. 2001;294:1638–1642. doi: 10.1126/science.294.5547.1638. [DOI] [PubMed] [Google Scholar]
- 24.Wall DA, Wilson G, Hubbard AL. The galactose-specific recognition system of mammalian liver: the route of ligand internalization in rat hepatocyts. Cell. 1980;21:79–93. doi: 10.1016/0092-8674(80)90116-6. [DOI] [PubMed] [Google Scholar]
- 25.Weigel PH, Oka JA. Endocytosis and degradation mediated by the asialoglycoprotein receptor in isolated rat hepatocytes. J Biol Chem. 1982;257:1201–1207. [PubMed] [Google Scholar]
- 26.Gao SY, Chen JN, Xu XR, Ding Z, Yang YH, Hua ZC, Zhang JF. Gatactosylated low molecular weight chitosan as DNA carrier for hepatocyte-targeting. Int J Pharm. 2003;255:57–68. doi: 10.1016/s0378-5173(03)00082-6. [DOI] [PubMed] [Google Scholar]
- 27.Wu GY, Wu CH. Receptor-mediated gene delivery and expression in vivo. J Biol Chem. 1988;263:14621–14624. [PubMed] [Google Scholar]
- 28.Hara T, Aramaki Y, Takada S, Koike K, Tsuchiya S. Receptor-mediated transfer of pSV2CAT DNA to human hepatoblastoma cell line HepG2 using asialofetuin-labeled cationic liposomes. Gene. 1995;159:167–174. doi: 10.1016/0378-1119(95)00100-k. [DOI] [PubMed] [Google Scholar]
- 29.Sato A, Takagi M, Shimamoto A, Kawakami S, Hashida M. Small interferin RNA delivery to the liver by intravenous administration of galactosylated cationic liposomes in mice. Biomaterials. 2007;28:1434–1442. doi: 10.1016/j.biomaterials.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Kelkar DA, Chattopadhyay A. The gramicidin ion channel: A model membrane protein. Biochim Biophys Acta. 2007;1768:2011–2025. doi: 10.1016/j.bbamem.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Lundbaek JA, Collingwood SA, Ingolfsson HI, Kapoor R, Andersen OS. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. J R Soc Interface. 2010;7:373–395. doi: 10.1098/rsif.2009.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CH, Choi HK. Effect of gramicidin on percutaneous permeation of a model drug. AAPS PharmSciTech. 2000;1(2):E13. doi: 10.1208/pt010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Zhang M, Yung B, Li H, Zhou C, Lee LJ, Lee RJ. Lactosylated liposomes for targeted delivery of doxorubicin to hepatocellular carcinoma. Int J Pharm. 2012;7:1–10. doi: 10.2147/IJN.S33965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X, Klibanov AL, Huang L. Lipophilic polylysines mediate efficient DNA transfection in mammalian cells. Biochim Biophys Acta. 1991;1065:8–14. doi: 10.1016/0005-2736(91)90003-q. [DOI] [PubMed] [Google Scholar]
- 36.Crook K, Stevenson BJ, Dubouchet M, Porteous DJ. Inclusion of cholesterol in DOTAP transfection complexes increases the delivery of DNA to cells in vitro in the presence of serum. Gene Ther. 1998;5:137–143. doi: 10.1038/sj.gt.3300554. [DOI] [PubMed] [Google Scholar]
- 37.Yang JP, Huang L. Time-dependent maturation of cationic liposome- DNA complex for serum resistance. Gene Ther. 1998;5:380–387. doi: 10.1038/sj.gt.3300596. [DOI] [PubMed] [Google Scholar]
- 38.Varnholt H. The role of microRNAs in primary liver cancer. Ann Hepatol. 2008;7:104–113. [PubMed] [Google Scholar]
- 39.Vissone R, Petrocca F, Croce CM. Micro-RNA in gastrointestinal and liver disease. Gastroenterology. 2008;135:1866–1869. doi: 10.1053/j.gastro.2008.10.074. [DOI] [PubMed] [Google Scholar]
- 40.Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Rajeev KG, Cantley WL, Dorkin JR, Butler JS, Qin L, Racie T, Sprague A, Fava E, Zeigerer A, Hope MJ, Zerial M, Sah DW, Fitzgerald K, Tracy MA, Manoharan M, Koteliansky V, Fougerolles A, Maier MA. Targeted delivery of RNAi therapeutics with endogenous and exogenousligand-based mechanisms. Mol Ther. 2010;18:1357–64. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan X, Kuipers F, Havekes LM, Dontje B, Poelstra K, Scherphof GL, Kamps JA. The role of apolipoprotein E in the elimination of liposomes from blood by hepatocytes in the mouse. Biochem Biophys Res Commun. 2005;328:57–62. doi: 10.1016/j.bbrc.2004.12.137. [DOI] [PubMed] [Google Scholar]
- 42.Xie FY, Woodle MC, Lu PY. Harnessing in vivo siRNA delivery for drug discovery and therapeutic development. Drug Discov Today. 2006;11:67–73. doi: 10.1016/S1359-6446(05)03668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signaling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 44.Behr JP. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–36. [Google Scholar]
- 45.Bolkent S, Zierold K. Effects of the ionophores valinomycin, ionomycin and gramicidin A on the element compartmentation in cultured rat hepatocytes. Toxicology in Vitro. 2002;16:159–165. doi: 10.1016/s0887-2333(01)00108-4. [DOI] [PubMed] [Google Scholar]
- 46.Torres AG, Fabani MM, Vigorito E, Gait MJ. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA. 2011;17:933–943. doi: 10.1261/rna.2533811. [DOI] [PMC free article] [PubMed] [Google Scholar]