Abstract
Background
Perturbations in the function of core circadian clock components such as the Period (Per) family of genes are associated with alcohol use disorder, and disruptions in circadian cycles may contribute to alcohol abuse and relapse. This study tested ethanol consumption, reinforcement, and metabolism in mice containing functional mutations in Per1 and/or Per2 genes on an ethanol-preferring background, C57BL/6J mice.
Methods
Mice were tested in: (A) free-access intake with ascending concentrations of ethanol (2–16% v/v); (B) conditioned place preference using ethanol (2 g/kg for males; 2.5 g/kg for females) vs. saline injections; (C) recovery of the righting reflex following a 4 g/kg bolus of ethanol; and (D) blood ethanol levels 1 hour after a 2 g/kg bolus of ethanol.
Results
All Per mutant (mPer) mice showed increased ethanol intake and condition place preference compared to controls. There were also genotypic differences in blood ethanol concentration: in males, only mPer1 mice showed a significantly higher blood ethanol concentration than WT mice, but in females, all mPer mice showed higher blood ethanol levels than WT mice.
Conclusions
Mutation of either Per1 or Per2, as well as mutations of both genes, increases ethanol intake and reinforcement in an ethanol-preferring mouse model. In addition, this increase in ethanol seeking behavior seems to result both from a change in ethanol metabolism and a change in reward responding to ethanol, but not from any change in sensitivity to ethanol’s sedating effects.
Keywords: Alcoholism, Clock, Addiction, Genetics
1.1 Introduction
Circadian rhythms evolved to allow organisms to adapt to the daily rotation of our planet. The circadian system keeps time independently of any environmental cue, but a variety of stimuli known as zeitgebers or “timegivers” (e.g. light, food) function to keep internal time, synchronized with external time. In mammals, the molecular clock is composed of a network of genes that form a transcriptional/translational feedback loop in which the transcription factors Brain and Muscle ARNT-like (BMAL)1 and CLOCK comprise the positive arm of the feedback loop and drive the expression of two families of negative arm proteins, PERIOD (PER) and CRYPTOCHROME. This circuit, in conjunction with post-transcriptional and post-translational regulatory mechanisms, controls the expression of virtually all clock-controlled genes. On a physiological level, the circadian clock governs the timing of many homeostatic functions such as metabolism, body temperature, and the sleep/wake cycle and disruptions in these functions are associated with a wide variety of physical, mental, and emotional disorders, including substance abuse and dependence (Falcon and McClung, 2009). Specifically, there are direct correlations of both sleep disruptions and changes in circadian gene expression with increases in alcohol drinking behaviors and increased sensitivity to alcohol (Agapito et al.; Benca et al., 1992; Falcon and McClung, 2009; Kovanen et al., 2010; Perreau-Lenz et al., 2009; Spanagel et al., 2005; Wirz-Justice et al., 2001). Thus, recent work in this field has begun to assess the role of specific clock genes in alcohol addiction phenotypes. Animals with targeted knockouts or mutations of clock genes show altered behavioral and neural responses to drugs of abuse, such as a sensitized response to cocaine in Per2 mutants as a result of changes in glutamate uptake and catecholamine breakdown (Abarca et al., 2002; Agapito et al.; Hampp and Albrecht, 2008; Perreau-Lenz et al., 2009; Spanagel et al., 2005). In addition, inhibition of one component of the molecular clock - casein-kinase-1-epsilon/delta – is sufficient to prevent ethanol relapse in mice (Perreau-Lenz et al., 2012). Work has also demonstrated that mutation of any of the three Period gene homologs leads to increased ethanol intake (Crum et al., 2004; Dong et al., 2011; Spanagel et al., 2005; Wang et al., 2012; Zghoul et al., 2007). In addition, ethanol consumption has been tied to aberrant rhythms in the hypothalamic-pituitary-adrenal axis hormone secretion and body temperature, as well as to changes in free-running periods (the activity pattern in the absence of light cues), and ethanol preference varies with circadian phenotypes (McCulley et al., 2013; Trujillo et al., 2011).
Previously characterized mice harboring a mutation in the circadian genes Period1 (mPer1Brdm1; herein mPer1) or Period2 (mPer2Brdm1; herein mPer2) express a mutant protein that lacks a key domain for normal circadian function (Cermakian et al., 2001; Zheng et al., 1999). These mice have a dysfunctional circadian oscillator as they exhibit abnormal sleep-wake cycles in the absence of light cues; mPer1 mice show a shortened sleep-wake cycle compared to wild-type (WT) mice, whereas mPer2 mice show a shortened sleep-wake cycle that slowly devolves into arhythmicity, and double mPer1Per2 mice show a complete absence of circadian rhythms as soon as the light cycle is lost (Zheng et al., 2001; Zheng et al., 1999). In addition, mPer mice also demonstrate addictive phenotypes as they drink more ethanol than WT mice (Dong et al., 2011; Spanagel et al., 2005) and show a sensitized response to cocaine (Abarca et al., 2002). Alcoholics display many characteristics associated with a dysfunctional circadian clock, such as disruptions in sleep cycles, including difficulty with sleep onset and frequent sleep disturbances (Gillin, 1994), changes in eating habits, and changes in homeostatic rhythms, such as body temperature fluctuations (Danel et al., 2001). Interestingly, polymorphisms in the human Per2 gene are also associated with variations in the ability to regulate ethanol intake (Spanagel et al., 2005). Thus, it is possible that there may be an association between the disrupted circadian rhythm phenotype and alcohol addiction phenotypes, or that ethanol may act directly on the circadian clock to affect ethanol-seeking behavior.
Previous findings indicate that mPer2 mice drink more ethanol than WT mice in any free access setting and mPer1 mice drink more than WT, but only as a response to stress (Spanagel et al., 2005; Zghoul et al., 2007). This suggests that Per2, and to a lesser extent Per1, genes are protective against high ethanol intake. However, these studies were performed in male 129SvEvBrd/C57BL/6-Tyrc-Brd mice, which are derived from a non-ethanol-preferring genetic background (Yoneyama et al., 2008) and drink relatively small amounts of ethanol over a 24-hour period (Spanagel et al., 2005). We hypothesized that Per1 mice would display an increase in ethanol intake in a free access system when backcrossed onto the ethanol preferring C57BL6 background, thus bolstering the idea that the Per genes provide a protective function from ethanolism. Furthermore, we hypothesized that this increased ethanol intake is due to reinforcement from ethanol in mutant mice compared to WT mice, rather than to any difference in ethanol metabolism. Thus, in the current study we assessed free-access ethanol intake, ethanol reinforcement, ethanol sedation, and ethanol metabolism in mPer1, mPer2, and mPer1Per2 mice backcrossed onto the ethanol-preferring C57BL/6J background.
Many studies have reported that sex can influence ethanol intake in rodents; specifically, males drink significantly less than females (Eriksson and Pikkarainen, 1968; Meliska et al., 1995; Middaugh et al., 1999). Although Per2 polymorphisms are associated with higher alcohol intake in both men and women (Spanagel et al., 2005), adolescent males show greater effects of Per2 polymorphism expression on alcohol intake than adolescent females (Comasco et al., 2010). Thus, to explore whether sex modulates the effects of Per mutations on ethanol drinking behavior, we tested both male and female mice.
2.1 Methods
2.1.1 Animals
All experiments were carried out in accordance with the National Institutes of Health guide for the care and use of laboratory animals (NIH Publications No. 8023, revised 2011) and were approved by the Dartmouth Institutional Animal Care and Use Committee.
Adult, male and female C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME) and male and female Per1Brdm1, Per2Brdm1, Per1Per2 Brdm1 mice (Zheng et al., 2001; Zheng et al., 1999) backcrossed to a C57BL/6J background were generously provided by Dr. Amita Seghal from the University of Pennsylvania.. Mice, 10–26 weeks of age (15–35g), were housed individually or in groups of 4 and given ad libitum access to food and water throughout each study. All animals were maintained on a 12:12 light:dark (LD) cycle.
2.1.2 Experiment 1: Free Access Ethanol
We first tested the effect of Per mutations on ethanol intake in C57BL/6J background mice. 69 animals (8–9 per group) were individually housed and tested for free access, chronic ethanol intake and preference in their home cage. Mice were given free access to a water bottle, a bottle containing an ascending concentration of ethanol (2–16%, v/v), and food. Within each experiment, the positions of the two drinking bottles were rotated on a daily basis to prevent positional preference. A technician, blinded to the experimental conditions, measured fluid intake every 24 hours, and measured food intake and body weight every 48 hours. After a one week acclimation period, during which ad libitum water and food intake, as well as body weight, were measured, each mouse was given access to ascending concentrations of ethanol (2–16% v/v for 5–9 days at each concentration. The concentration was increased incrementally once ethanol intake reached a steady baseline at each concentration (which required, on average, 6 days).
2.1.3 Experiment 2: Conditioned Place Preference
We next examined whether the differences in free access ethanol intake cold be explained by differences in ethanol reinforcement. All conditioning and testing occurred in a dedicated conditioned place preference (CPP) test room. Four shuttleboxes were used for CPP. Each shuttlebox consisted of two Plexiglas chambers (4″ × 4″ × 6″) separated by a narrow holding room (4″ × 1.5″ × 6″). One chamber had metal grid flooring and the other had a plastic floor with evenly spaced holes. The holding room had a plain Plexiglas floor. Guillotine doors barred movement between areas. Shuttleboxes were isolated in wooden shelves, in a room with a red house light and a sound machine generating white noise to conceal any external noises. A video recording system was used to monitor all sessions for later scoring. A pilot test demonstrated no significant preference for either chamber, and so animals were randomly assigned to mesh flooring in the left vs. right chamber and associated with saline vs. ethanol injections. The first day of training was a pre-exposure day, during which drug-naïve animals had free access to the entire apparatus with the flooring removed for 10min. Any mouse that spent greater than 75% of the time in one chamber was removed from the experiment (<5% of animals). The next 8 training days consisted of alternating days of intraperitoneal (i.p.) saline or ethanol (20% v/v in saline; 2 g/kg for males and 2.5 g/kg for females, based on pilot data in a set of WT mice) injections, with the treatment schedule randomized within groups. Ethanol injections are used to avoid confounding effects of ethanol taste. Each animal (64; 8 per group) was injected, placed into a conditioning chamber with the guillotine doors shut, and allowed to explore for 6min. In between animals, chambers were cleaned with 70% ethanol and then dried thoroughly. On test day, animals were injected with saline and then placed in the holding room. After 10sec, the guillotine doors were removed and the animal was allowed to explore freely for 30min. After testing was complete, each 30min session was scored based on the number of entries and total time spent in each chamber.
2.1.4 Experiment 3: Loss of the Righting Reflex
Although the greater CPP for ethanol in the mutant mice may underlie their greater intake of ethanol as compared to WT mice, there were no differences between the sexes in CPP. However, these differences did exist in free access ethanol intake. To determine whether sex differences in ethanol intake within the Per mutant mice could be explained by a difference in ethanol sedation, we next explored the behavioral responses to a cataplexy-inducing dose of ethanol. All testing occurred in a dedicated behavioral testing suite. Each animal (57; 7–8 per group) was injected i.p. with 4 g/kg ethanol (20%v/v in saline) and placed in an empty Plexiglas housing box. Latency to loss (LORR) and recovery of the righting reflex was recorded. LORR was determined when the animal could not right itself after being lain on its side twice in 30sec. Recovery was determined when this ability was restored.
2.1.5 Experiment 4: Blood Ethanol Concentration
As there were no genotype-specific differences in sedation in response to a high dose of ethanol, our final experiment tested whether there are differences in the blood ethanol concentrations (BECs) produced by a lower dose of ethanol that was associated with ethanol CPP in Experiment 2. Each mouse (32; 4 per group) was injected i.p. with 2 g/kg ethanol (20% v/v in saline) and then returned to their home cage. Sixty minutes later, based on a pilot study showing 60min as the peak of BEC in WT mice (data not shown), mice were euthanized by cervical dislocation and trunk blood was collected in a capillary tube (Analox Instruments, Lunenberg, MA) and spun down in a centrifuge for 2 minutes at 14,000 rev/min. The plasma was then analyzed by an AM1 Analyzer (Analox Instruments) to obtain BEC (mg/dl). To assure the accuracy of the reading, each sample was analyzed twice and a mean from the two readings was used for data analysis.
2.1.6 Data Analysis
For Experiment 1, measurements for the last four days at each ethanol concentration, of ethanol intake (g/kg), ethanol preference (ethanol intake [mL]/total fluid intake [mL]), water intake (mL/kg), food intake (g/kg), and body weight (g) data, were analyzed using 3-way repeated measures analysis of variance (RMANOVA), using sex and genotype, as independent variables. For Experiment 2, the percentage of time spent in each compartment and the total number of entries into each compartment was analyzed by 2-way ANOVA, with sex and genotype as the independent variables. For Experiments 3 and 4, LORR and recovery or BEC were each analyzed by 2-way ANOVA, with sex and genotype as the independent variables. When the analysis indicated that significant differences existed between treatments, post-hoc pairwise comparisons between groups were made using the Tukey adjustment. Post-hoc comparisons were also made to compare baseline and treatment periods across groups using paired-samples t-tests for Experiment 1. Significance was determined at p<0.05. Data are expressed as mean (M) ± standard error of the mean (SEM).
3.1 Results
3.1.1 Experiment 1: Free-Access Ethanol
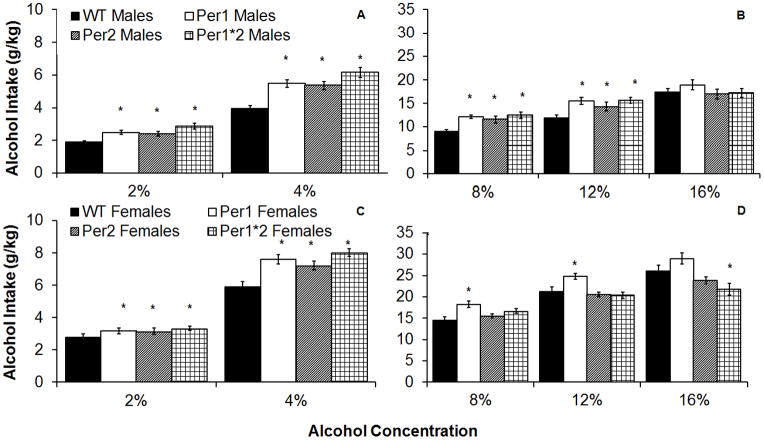
There were significant effects of ethanol concentration, F(4,1640)=2291.4, p<0.001, sex, F(1,410)=482.5, p<0.001, and genotype, F(3,408)=15.29, p<0.001, as well as interactive effects of sex by genotype, F(3,408)=4.1, p<0.01, concentration by genotype, F(12,1632)=10.8, p<0.05, and concentration by sex, F(4,1640)=96.6, p<0.001 on ethanol intake. Across genotypes, females drank more than males at all concentrations (data not shown), which is consistent with previous findings (Middaugh et al., 1999). Additionally, all mutant mice drank more than WTs from 2–12% ethanol (p<0.05), which suggests a potentially important role for the Per genes in limiting ethanol consumption and is consistent with previous findings (Spanagel et al., 2005; Zghoul et al., 2007). All male mPer1, mPer2, and mPer1Per2 mice drank more than male WTs at 12% ethanol (p<0.05; Figures 1A–B), whereas only female mPer1 mice drank more than the other female mice at 12% ethanol (Figures 1C–D).
Figure 1.
Average alcohol intake at each alcohol concentration (2–16% v/v). A–B) All mutant male mice drank more than male WT mice at 2–12% alcohol (g/kg). C–D) All female mutant mice drank more than female WT mice at 2–8% alcohol, but only female mPer1 mice drank more than other groups at 12–16% alcohol (g/kg). (Mean ± SEM; * indicates a significant difference from WTs at p<0.05).
There was no difference in ethanol intake for male mice at 16% ethanol, but the female mPer1Per2 mice drank less than female WTs at 16% ethanol (p<0.05; Figures 1B and D). There were also significant effects of ethanol concentration, F(4,1640)=95.6, p<0.001, sex, F(1,410)=13.5, p<0.001, and genotype, F(3,408)=35.9, p<0.001, as well as interactive effects of concentration by genotype, F(12,1632)=14.7, p<0.001, and concentration by sex, F(4,1640)=8.7, p<0.001 on ethanol preference. Similar to what was observed with ethanol intake, females showed higher ethanol preference than males; however, this was limited to the higher concentrations of ethanol (8–16% ethanol; Figure 2A). Furthermore, all mutant mice showed higher ethanol preference than WTs at 2–8% ethanol, whereas only male and female mPer1 and male mPer1Per2 mice showed higher ethanol preference at 12% ethanol, and the only difference at 16% ethanol was decreased preference in female mPer1Per2 mice compared to WT females (p<0.05; Figures 2B and 2C).
Figure 2.
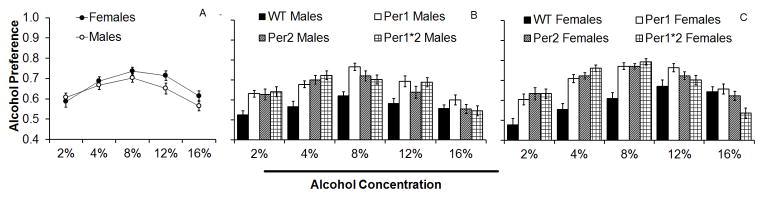
Average alcohol preference at each alcohol concentration (2–16% v/v). A) Females showed higher alcohol preference than males across genotype at 8–16% alcohol. B) All male mutant mice showed higher alcohol preference than male WT mice at 2–8% alcohol, but only malePer1 and Per1Per2 mice showed higher preference at 12% alcohol. C) All female mutant mice showed higher alcohol preference than female WT mice at 2–8% alcohol, but only female Per1 mice showed higher preference than other groups at 12% alcohol (Mean ± SEM; * indicates significant differences between groups or from WTs at p<0.05).
Overall, mPer mice preferred ethanol more than WT mice even though the volume of ethanol solution consumed decreased at higher concentrations (Data not shown). The overall reduction in consumption across genotype observed is possibly as the result of taste aversion to more concentrated ethanol solutions. Thus, mPer1 and mPer2 mice that have been backcrossed on an ethanol-preferring mouse line demonstrated increased ethanol intake and preference, especially at moderate concentrations of ethanol that are comparable to the concentrations in most alcoholic beverages. Our findings also agree with previous work demonstrating that female mice have a higher preference for ethanol as opposed to water than male mice.
Increased ethanol intake was generally associated with decreased water intake throughout the experiment, suggesting that the Per mutations do not increase ethanol intake simply as the result of increased fluid intake. The one exception was that female mice drank more water than male mice, demonstrating higher overall fluid intake (p<0.01; data not shown). Females also ate more food than males at all concentrations of ethanol, and Per2 mice ate less than WTs at all concentrations (p<0.05). Since the Per mutations did not increase food intake concurrent to increased ethanol intake, the increased ethanol intake is unlikely to represent a shift in caloric intake (data not shown). Females weighed less than males throughout the study (p<0.05; Table 1). The mPer2 mice weighed more than WTs throughout the study (p<0.05), whereas the mPer1 and mPer1Per2 mice weighed significantly less than WTs (p<0.05; Table 1). Despite the higher caloric intake per body weight in females, they consistently weighed less than males. Interestingly, the Per2 mutation was associated with higher body weights, whereas the Per1 mutation and Per1Per2 double mutation were both associated with lower weights compared to WTs.
Table 1.
Body weight (g) during Experiment 1. Across genotypes, male mice weighed more than female mice. mPer2 mice weighed significantly more than all other genotypes, while mPer1 and mPer1Per2 mice weighed significantly less than WT mice from 4%–16% alcohol (Mean ± SEM).
| 2% | 4% | 8% | 12% | 16% | |
|---|---|---|---|---|---|
| Females | 20.4±0.3 | 21.3±0.3 | 21.8±0.2 | 22.3±0.2 | 22.7±0.3 |
| Males | 25.3±0.3 | 25.9±0.4 | 26.2±0.4 | 26.8±0.4 | 27.2±0.4 |
| WT | 22.4±0.2 | 23.6±0.2 | 24.1±0.2 | 24.9±0.2 | 25.3±0.2 |
| Per1 | 22.1±0.3 | 22.5±0.4 | 22.8±0.3 | 23.4±0.3 | 23.7±0.3 |
| Per2 | 24.7±0.3 | 25.2±0.4 | 25.7±0.3 | 26.2±0.4 | 26.7±0.4 |
| Per1*2 | 22.2±0.4 | 23.0±0.4 | 23.3±0.4 | 23.7±0.4 | 24.0±0.4 |
3.1.2 Experiment 2: Conditioned Place Preference
All mice showed a significant preference for (F(1,127)=342.9, p<0.001) and more entries into (F(1,127)=5.5, p<0.05) the ethanol-paired chamber as compared to the saline-paired chamber. There was a main effect of sex on the number of chamber entries, F(1,127)=6.8, p<0.05, such that females were significantly less active than males, regardless of genotype or chamber stimulus (p<0.05; data not shown). There was also a significant effect of genotype on the number of chamber entries, F(3,125)=9.3, p<0.001, such that the mPer1 mice were significantly more active than all other lines, regardless of sex or chamber stimulus (p<0.05; Figure 3A). Finally, there was a significant effect of genotype on chamber preference, F(3, 125)=3.3, p<0.05, with all mutant mice showing a stronger preference for the ethanol-paired chamber than the WT mice (p<0.05; Figure 3B).
Figure 3.
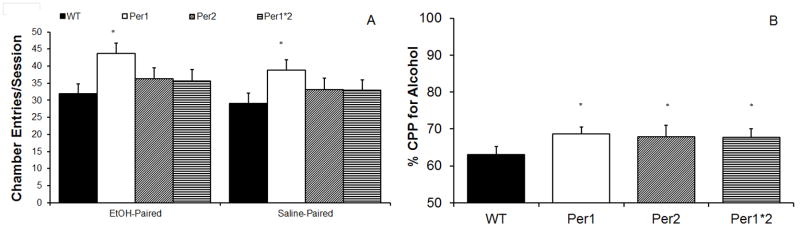
Behavior in alcohol conditioned place preference. A) Only the mPer1 mice made more chamber entries than other lines. B) Across sex, all lines of mPer mice showed a greater preference for the alcohol-paired chamber than WT mice (Mean ± SEM; * indicates significant differences from WTs at p<0.05).
3.1.3 Experiment 3: Loss of the Righting Reflex
There were no significant effects of genotype on either latency to loss of recovery of the righting reflex. There was, however, an effect of sex on the recovery of the righting reflex, F(1,56)=35.4, p<0.001, with females showing a significantly longer latency to recovery of the righting reflex compared to males (data not shown).
3.1.4 Experiment 4: Blood Ethanol Concentration
There were significant main effects of sex, F(1,31)=74.3, p<0.001, and genotype, F(3,29)=43.4, p<0.001, as well as a significant sex by genotype interaction, F(3,29)=30.1, p<0.001, on blood ethanol concentration. Overall, males had a lower BEC than females (p<0.05; Figure 4A–B); however, when each genotype was examined individually, female WT mice showed lower BACs than male WT mice, but in each mutant line, the female mice showed higher BACs then males (p<0.05, Figure 4A–B). In addition, mPer1 mice had the highest BEC, significantly higher than mPer2 mice, which in turn had significantly higher BEC than WT or double mutant mice. Within males, only the mPer1 mice had significantly higher BEC than the WT mice (p<0.05; Figure 4A), but in females, all three lines of mutant mice had significantly higher BEC than WT mice (p<0.05, Figure 4B).
Figure 4.
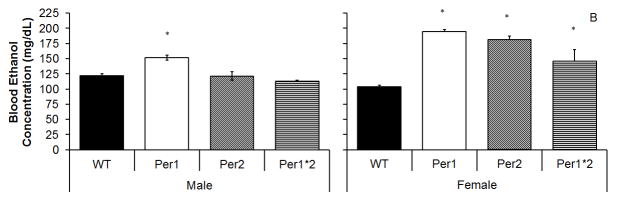
Genotype differences in blood alcohol concentration. A) In males, on mPer1 mice showed a significantly higher blood alcohol concentration than WT mice. B) In females, all mPer mice showed higher blood alcohol levels than WT mice (Mean ± SEM; * indicates significant differences between from WTs at p<0.05).
4.1 Discussion
The major finding of this study is that functional mutation of either Per1 or Per2 genes, as well as mutations in both, results in a generalized increase in ethanol intake and conditioned place preference, which suggests a protective function for these Per genes in alcohol use disorder. Our result in section 3.1.1, showing that mPer2 mice drink more ethanol than wild-type mice, is consistent with previous work (Spanagel et al., 2005) and the levels of ethanol intake in mice backcrossed onto a C57BL/6J background is also consistent with pervious findings (Brager et al., 2011). However, additional results in section 3.1.1 demonstrating an increase in voluntary ethanol intake and preference in mPer1 mice differs from previous research, which showed no difference between wild type and mPer1 mice in free access ethanol drinking (Spanagel et al., 2005; Zghoul et al., 2007). The disparity in mPer1 alcohol consumption and preference may be explained by the different genetic backgrounds used to carry the Per1 mutations; as previously mentioned, our experiments used Per1Brdm1 mice backcrossed onto the ethanol-preferring C57BL6/J line, while the previous study used the mixed 129SvEvBrd/C57BL/6-Tyrc-Brdmice. Importantly, our Per mutant colony was SNP mapped to confirm genetic purity with a homology of 97% for Per1Brdm1, 95% for Per2Brdm1, and 94% for the double mutants to C57BL6/J (data not shown).
Ethanol intake and preference can be impacted by a number of factors including reinforcement, metabolism, sedation, and taste aversion to ethanol. As a result, different mouse lines show dramatically different levels of ethanol intake (Yoneyama et al., 2008). Therefore, depending on the genetic background of the line being studied, the Per1 mutation may not adequately increase pharmacological reward to overcome factors such as taste aversion. However, Per1mice show greater ethanol consumption than WT mice in response to social defeat stress (Dong et al., 2011), suggesting that there is some predisposition to ethanol reinforcement and that the aversion to ethanol can be overcome by environmental factors. By crossing our mPer mutant onto an ethanol-preferring background, we can examine the effects of these mutations on ethanol intake and reinforcement independent of confounding factors such as taste aversion.
Recent work has shown that mPer2 mice have increased bouts of ethanol consumption over a 24-hour period in normal LD 12:12 conditions, which may be due to a 2-hour extension in nocturnal activity patterns beginning in the last 2 hours of the preceding light phase (Brager et al., 2011). Brager and colleagues reported that an earlier onset of activity in the mPer2 mice appears to correlate with an increase in ethanol intake. However, our CPP data in section 3.1.2 demonstrate a moderate but significant increase in ethanol reinforcement after systemic administration and we see increased ethanol intake in mPer1 and mPer1Per2 mice, which do not show changes in activity patterns from WT a normal L:D cycle (Shiromani et al., 2004). This suggests that there must be some factor other than an increased period of activity underlying the increased alcohol intake of these mice. It should also be noted, howver, that CPP is not the most sensitive measure of ethanol reinforcement – indeed, operant self-administration paradigms, in which animals must perform a behavior such as nose-poking or lever-pressing in order to receive an ethanol reward, show stronger, more consistent correlations with free access ethanol intake than does CPP (Green and Grahame, 2008). While we hope to address the question of motivation for ethanol through operant self-administration studies in the future, our positive CPP data represent a starting point for studying how the Per genes are related to ethanol reinforcement.
As the mPer1 mice drink more in the free-access paradigm it is surprising that the double mutant mice do not drink at similar levels to the mPer1 mice overall, or at higher levels due to an additive effect with the loss of mPer2 function. In fact the double mutant mice resembled the mPer2 mice in all other experiments. However, it is important to note that the mPer1Per2 mice did show overall higher levels of intake at 2–4% as compared to the other lines, although this difference was lost as ethanol concentration was increased, possibly due to differences in taste aversion. Furthermore, it has been shown previously that chronic ethanol intake can alter Per gene expression (Perreau-Lenz and Spanagel, 2008); therefore, it is possible that chronic intake of ethanol alters clock gene expression differently in the double mutant lines, than in the single mutants. Assessment of clock gene levels in the reward circuit of the mPer mice is a critical next step in understanding the genotypic differences uncovered here.
All of the female mutants showed higher blood ethanol levels than female WT mice in section 3.1.4, with mPer1 mice showing the greatest blood ethanol concentration. In males, only the mPer1 mice showed greater blood ethanol concentrations than WT mice. Therefore, decreased metabolism – and thus, greater potency - of ethanol in the mPer1 male mice and in all mutant female mice may be one factor underlying their increased ethanol intake and preference, especially at low concentrations of ethanol. However, there must be additional factors underlying increased ethanol intake and preference in mutant mice that do not show higher blood ethanol levels than WT mice (i.e. the mPer2 and mPer1Per2 male mice). In addition, it has been shown that there is no diurnal variation in blood ethanol metabolism in male WT mice (Perreau-Lenz et al., 2009), suggesting that the effects of Per mutation on blood ethanol levels may be the result of pleiotropy rather than a change in circadian function due to the mutation of the Per genes. Furtherwork is needed to asses the mechanism by which Per gene mutation disrupts alcohol metabolism.
As documented in the literature, female mice showed greater ethanol intake and preference than male mice. This effect may be attributed to differences in body water content or metabolism, as suggested by Hwa and colleagues (2011), because ethanol intake and preference remain steady and variability between females remains low despite the fluctuations in the estrous cycle. Although we found that female mice in general – and mutant mice specifically – metabolized ethanol more poorly than males, we also found that female WT mice showed lower blood ethanol levels than males after an equivalent, moderate dose of ethanol, yet females took longer to recover their righting reflex than males following an equivalent high dose of ethanol. Interestingly, this suggests that the female mice may drink more ethanol in free access conditions because they either metabolize it or absorb it more rapidly than males, yet they are more sensitive to the sedative effects of ethanol. Thus, it may be a decreased duration of ethanol-induced reward that leads female mice to drink more, or a greater anxiolytic response to ethanol, as females are more prone to stress and anxiety in both human (Gater et al., 1998) and animal studies (Adamec et al., 2006). This is also supported by our CPP data where more exploratory behavior (chamber entries) is evident in males than in females. Although the mPer1 mice also showed increased exploratory behavior despite a link between Per1 and stress-induced ethanol intake (Dong et al., 2011), no study has demonstrated a direct link between Per1 and anxiety, and these mice may actually have lower basal anxiety levels than the other lines tested.
In general, increased ethanol intake was associated with lower food intake in the mutant mice in section 3.1.1, possibly due to the higher caloric contribution from ethanol. This suggests that the differences in ethanol intake between mouse lines were not due simply to increased locomotor or consummatory behaviors in the mutants. Furthermore, ethanol preference was higher in the mutant mice than in WTs, suggesting that the increased ethanol intake was not matched by increased water intake. Finally, despite significant differences in body weight – mPer2 mice weighed more than WTs, while mPer1 and mPer1Per2 mice weighed less – these differences did not correlate with ethanol drinking behavior, as all three mutant lines showed enhanced ethanol intake and preference above WT levels.
Together, these data suggest a direct role for the Per genes in regulating ethanol consumption, possibly through effects on the neurotransmitter systems involved in the reward circuit, as seen in the CPP paradigm, as well as through decreases in ethanol metabolism, as seen in the BAC paradigm. The reward system effect of Per genes is supported by work recently published by Perreau-Lenz and colleagues, showing that inhibition of casein-kinase-1-epsilon/delta, the kinase responsible for PER2 degradation, blocks relapse to ethanol following multiple ethanol deprivation periods (Perreau-Lenz et al., 2012). Further work is needed to examine how the PER1 and PER2 proteins may act on the reward circuit to determine, for example, whether the homologs mediate drug reward through the same mechanism– such as changes in glutamatergic function (Spanagel et al., 2005) or monoamine oxidase activity (Hampp et al., 2008) –or perhaps through distinct mechanisms. In addition, based on recent work implicating the Per homolog Per3 in ethanol responding (Wang et al., 2012), this homolog should also be included in future studies aimed to elucidate the mechanisms through which the PER genes affect alcohol drinking behaviors. It is also important to note that although these mice harbor functional mutations of the Per genes, they still express these genes and studies in Per knockouts are important in demonstrating the significance of the loss of Per function on alcohol drinking behavior. Finally, it will be important to establish what role, if any, the circadian oscillator is playing in regulating alcoholic behavior. Although our data and others suggests a genetic link between the clock and alcoholism, probing the nature of the circadian oscillator under the conditions applied here is essential in establishing whether the effect of the Per genes is clock-related, or pleiotropic.
Highlights.
Mutation of circadian PERIOD1 and PERIOD2 genes increases alcohol intake in mice
Period mutation also increases conditioned place preference for alcohol
Period mutation also decreases alcohol metabolism
More work is warranted to elucidate the role of the circadian clock in alcohol addiction
Acknowledgments
This work was supported by grants from the National Institutes of Health: NIAAA F32AA20143-01 (DG) and R01GM083336 (JJL and JCD).
Footnotes
Disclosures: The authors have no conflict of interest to report
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abarca C, Albrecht U, Spanagel R. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc Natl Acad Sci U S A. 2002;99:9026–9030. doi: 10.1073/pnas.142039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamec R, Head D, Blundell J, Burton P, Berton O. Lasting anxiogenic effects of feline predator stress in mice: sex differences in vulnerability to stress and predicting severity of anxiogenic response from the stress experience. Physiology & behavior. 2006;88:12–29. doi: 10.1016/j.physbeh.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Agapito M, Mian N, Boyadjieva NI, Sarkar DK. Period 2 gene deletion abolishes beta-endorphin neuronal response to ethanol. Alcohol Clin Exp Res. 34:1613–1618. doi: 10.1111/j.1530-0277.2010.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benca RM, Obermeyer WH, Thisted RA, Gillin JC. Sleep and psychiatric disorders. A meta-analysis. Archives of General Psychiatry. 1992;49:651–668. doi: 10.1001/archpsyc.1992.01820080059010. discussion 669–670. [DOI] [PubMed] [Google Scholar]
- Brager AJ, Prosser RA, Glass JD. Circadian and acamprosate modulation of elevated ethanol drinking in mPer2 clock gene mutant mice. Chronobiol Int. 2011;28:664–672. doi: 10.3109/07420528.2011.601968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. Embo J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comasco E, Nordquist N, Gokturk C, Aslund C, Hallman J, Oreland L, Nilsson KW. The clock gene PER2 and sleep problems: association with alcohol consumption among Swedish adolescents. Ups J Med Sci. 2010;115:41–48. doi: 10.3109/03009731003597127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum RM, Ford DE, Storr CL, Chan YF. Association of sleep disturbance with chronicity and remission of alcohol dependence: data from a population-based prospective study. Alcohol Clin Exp Res. 2004;28:1533–1540. doi: 10.1097/01.alc.0000141915.56236.40. [DOI] [PubMed] [Google Scholar]
- Danel T, Libersa C, Touitou Y. The effect of alcohol consumption on the circadian control of human core body temperature is time dependent. Am J Physiol Regul Integr Comp Physiol. 2001;281:R52–55. doi: 10.1152/ajpregu.2001.281.1.R52. [DOI] [PubMed] [Google Scholar]
- Dong L, Bilbao A, Laucht M, Henriksson R, Yakovleva T, Ridinger M, Desrivieres S, Clarke TK, Lourdusamy A, Smolka MN, Cichon S, Blomeyer D, Treutlein J, Perreau-Lenz S, Witt S, Leonardi-Essmann F, Wodarz N, Zill P, Soyka M, Albrecht U, Rietschel M, Lathrop M, Bakalkin G, Spanagel R, Schumann G. Effects of the circadian rhythm gene period 1 (per1) on psychosocial stress-induced alcohol drinking. The American journal of psychiatry. 2011;168:1090–1098. doi: 10.1176/appi.ajp.2011.10111579. [DOI] [PubMed] [Google Scholar]
- Eriksson K, Pikkarainen PH. Differences between the sexes in voluntary alcohol consumption and liver ADH-activity in inbred strains of mice. Metabolism. 1968;17:1037–1042. doi: 10.1016/0026-0495(68)90011-5. [DOI] [PubMed] [Google Scholar]
- Falcon E, McClung CA. A role for the circadian genes in drug addiction. Neuropharmacology. 2009;56(Suppl 1):91–96. doi: 10.1016/j.neuropharm.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gater R, Tansella M, Korten A, Tiemens BG, Mavreas VG, Olatawura MO. Sex differences in the prevalence and detection of depressive and anxiety disorders in general health care settings: report from the World Health Organization Collaborative Study on Psychological Problems in General Health Care. Arch Gen Psychiatry. 1998;55:405–413. doi: 10.1001/archpsyc.55.5.405. [DOI] [PubMed] [Google Scholar]
- Gillin JC. Sleep and psychoactive drugs of abuse and dependence. In: Kryger MH, editor. Principles and Practice of Sleep Medicine. W.B. Saunders; Philadelphia: 1994. pp. 934–942. [Google Scholar]
- Green AS, Grahame NJ. Ethanol drinking in rodents: is free-choice drinking related to the reinforcing effects of ethanol? Alcohol. 2008;42:1–11. doi: 10.1016/j.alcohol.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp G, Albrecht U. The circadian clock and mood-related behavior. Commun Integr Biol. 2008;1:1–3. doi: 10.4161/cib.1.1.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, Perreau-Lenz S, Brunk I, Spanagel R, Ahnert-Hilger G, Meijer JH, Albrecht U. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol. 2008;18:678–683. doi: 10.1016/j.cub.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Hwa LS, Chu A, Levinson SA, Kayyali TM, DeBold JF, Miczek KA. Persistent escalation of alcohol drinking in C57BL/6J mice with intermittent access to 20% ethanol. Alcoholism, clinical and experimental research. 2011;35:1938–1947. doi: 10.1111/j.1530-0277.2011.01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovanen L, Saarikoski ST, Haukka J, Pirkola S, Aromaa A, Lonnqvist J, Partonen T. Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol Alcohol. 2010;45:303–311. doi: 10.1093/alcalc/agq035. [DOI] [PubMed] [Google Scholar]
- McCulley WD, 3rd, Ascheid S, Crabbe JC, Rosenwasser AM. Selective breeding for ethanol-related traits alters circadian phenotype. Alcohol. 2013 doi: 10.1016/j.alcohol.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meliska CJ, Bartke A, McGlacken G, Jensen RA. Ethanol, nicotine, amphetamine, and aspartame consumption and preferences in C57BL/6 and DBA/2 mice. Pharmacology, biochemistry, and behavior. 1995;50:619–626. doi: 10.1016/0091-3057(94)00354-8. [DOI] [PubMed] [Google Scholar]
- Middaugh LD, Kelley BM, Bandy AL, McGroarty KK. Ethanol consumption by C57BL/6 mice: influence of gender and procedural variables. Alcohol. 1999;17:175–183. doi: 10.1016/s0741-8329(98)00055-x. [DOI] [PubMed] [Google Scholar]
- Perreau-Lenz S, Spanagel R. The effects of drugs of abuse on clock genes. Drug News Perspect. 2008;21:211–217. doi: 10.1358/dnp.2008.21.4.1213350. [DOI] [PubMed] [Google Scholar]
- Perreau-Lenz S, Vengeliene V, Noori HR, Merlo-Pich EV, Corsi MA, Corti C, Spanagel R. Inhibition of the Casein-Kinase-1-Epsilon/Delta Prevents Relapse-Like Alcohol Drinking. Neuropsychopharmacology. 2012 doi: 10.1038/npp.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau-Lenz S, Zghoul T, de Fonseca FR, Spanagel R, Bilbao A. Circadian regulation of central ethanol sensitivity by the mPer2 gene. Addict Biol. 2009;14:253–259. doi: 10.1111/j.1369-1600.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- Shiromani PJ, Xu M, Winston EM, Shiromani SN, Gerashchenko D, Weaver DR. Sleep rhythmicity and homeostasis in mice with targeted disruption of mPeriod genes. Am J Physiol Regul Integr Comp Physiol. 2004;287:R47–57. doi: 10.1152/ajpregu.00138.2004. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- Trujillo JL, Do DT, Grahame NJ, Roberts AJ, Gorman MR. Ethanol consumption in mice: relationships with circadian period and entrainment. Alcohol. 2011;45:147–159. doi: 10.1016/j.alcohol.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mozhui K, Li Z, Mulligan MK, Ingels JF, Zhou X, Hori RT, Chen H, Cook MN, Williams RW, Lu L. A promoter polymorphism in the Per3 gene is associated with alcohol and stress response. Translational psychiatry. 2012;2:e73. doi: 10.1038/tp.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirz-Justice A, Haug HJ, Cajochen C. Disturbed circadian rest-activity cycles in schizophrenia patients: an effect of drugs? Schizophr Bull. 2001;27:497–502. doi: 10.1093/oxfordjournals.schbul.a006890. [DOI] [PubMed] [Google Scholar]
- Yoneyama N, Crabbe JC, Ford MM, Murillo A, Finn DA. Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol. 2008;42:149–160. doi: 10.1016/j.alcohol.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghoul T, Abarca C, Sanchis-Segura C, Albrecht U, Schumann G, Spanagel R. Ethanol self-administration and reinstatement of ethanol-seeking behavior in Per1(Brdm1) mutant mice. Psychopharmacology (Berl) 2007;190:13–19. doi: 10.1007/s00213-006-0592-z. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]