Abstract
The B2 family of short interspersed elements is transcribed into non-coding RNA by RNA polymerase III. The ~180 nt B2 RNA has been shown to potently repress mRNA transcription by binding tightly to RNA polymerase II (Pol II) and assembling with it into complexes on promoter DNA, where it keeps the polymerase from properly engaging the promoter DNA. Mammalian Pol II is a ~500 kD complex that contains 12 different protein subunits, providing many possible surfaces for interaction with B2 RNA. We found that the carboxy-terminal domain of the largest Pol II subunit was not required for B2 RNA to bind Pol II and repress transcription in vitro. To identify the surface on Pol II to which the minimal functional region of B2 RNA binds, we coupled multi-step affinity purification, reversible formaldehyde crosslinking, peptide sequencing by mass spectrometry, and analysis of peptide enrichment. The Pol II peptides most highly recovered after crosslinking to B2 RNA mapped to the DNA binding cleft and active site region of Pol II. These studies determine the location of a defined nucleic acid binding site on a large, native, multi-subunit complex and provide insight into the mechanism of transcriptional repression by B2 RNA.
Keywords: protein-RNA crosslinking, ncRNA, MS/MS, transcription, Pol II
Introduction
The synthesis of mRNA by RNA polymerase II (Pol II)a is an intricate process that is tightly regulated. Many factors have been shown to alter the transcription levels of specific genes by affecting the transcription reaction at multiple steps, from regulating the accessibility of the DNA template to governing the activity of the polymerase itself.1–4 Historically, the regulation of mRNA synthesis was thought to be solely mediated by proteins; however, it has become clear that non-coding RNAs (ncRNAs) play a vital role in transcriptional regulation as well.5,6
ncRNAs have been shown to repress mRNA transcription during heat shock. Cellular stress signals such as heat shock cause an overall repression of Pol II transcription.7–9 By contrast, the ncRNAs mouse B2 and human Alu, which are transcribed from SINEs (short interspersed elements) by RNA polymerase III (Pol III), are up-regulated during heat shock.10,11 We found that these ncRNAs bind Pol II with high affinity and potently repress its transcriptional activity by preventing formation of closed pre-initiation complexes.12–14 Specifically, B2 RNA and Alu RNA prevent Pol II from properly engaging promoter DNA when the polymerase is assembled into complexes with general transcription factors at promoters.15 Experiments to delimit the functional regions of B2 RNA found that nucleotides 81-130 can bind Pol II with high affinity and potently repress transcription in vitro.13 Another SINE ncRNA, mouse B1 RNA, also binds tightly to Pol II but does not repress transcription in vitro.14,16 Interestingly, the general transcription factor TFIIF can destabilize the interaction between Pol II and B1 RNA, whereas it does not affect the stability of B2 RNA/Pol II complexes16, providing an explanation for the lack of transcriptional repression by B1 RNA.
Mammalian Pol II is a large 500 kDa complex comprised of 12 different protein subunits (RPB1-12). It contains multiple subunits that could potentially bind B2 RNA. The RPB1 C-terminal domain (CTD), which consists of 52 heptad repeats (consensus sequence YSPTSPS) that serve as binding sites for many protein factors17–19, has also been shown to bind RNA.20 In addition, the RBP4/7 submodule is known to bind RNA.21,22 Finally, Pol II has a DNA binding cleft and an RNA exit channel that interact with nucleic acids. To date, mammalian Pol II has not been amenable to reconstitution from recombinant subunits or to high resolution structural analysis. Hence, it is not feasible to identify the B2 RNA binding site on Pol II at the peptide or amino acid level using traditional methods such as co-crystallization or mutations/truncations of the protein subunits. Recent cryo-EM studies showed that when B2 RNA was added to Pol II, additional density was observed in the DNA cleft.23 Identifying peptides and amino acids directly bound to B2 RNA would provide significant insight into the mechanism of action of this ncRNA transcriptional repressor. Mass spectrometry (MS) analysis of RNA-protein reversible crosslinking affinity purifications (RCAP), has been successfully used to identify precise RNA binding sites on recombinant proteins24–26; however, has not yet been applied to multi-subunit, native complexes such as Pol II.
To define the B2 RNA binding site on Pol II, we used a MS method that identified the Pol II peptides that crosslink to the minimal functional region of B2 RNA: nucleotides 81-130. Formaldehyde was used to crosslink B2 RNA(81-130) to Pol II immunopurified from human cells, and the complex was proteolyzed with trypsin. ncRNA/peptide conjugates were isolated, crosslinks were reversed, and the peptides recovered were identified by MS/MS using a highly sensitive LTQ-Orbitrap mass spectrometer. Using a label-free peptide quantitation approach, we found that nine crosslinked peptides were significantly enriched, indicating that these peptides are in contact with B2 RNA when it is stably bound to Pol II. Mapping these peptides onto a crystal structure of yeast Pol II revealed that the binding site for B2 RNA(81-130) mainly comprises a region containing the DNA binding cleft and active site of human Pol II.
Results
Pol II binding and transcriptional repression by B2 RNA does not require the CTD
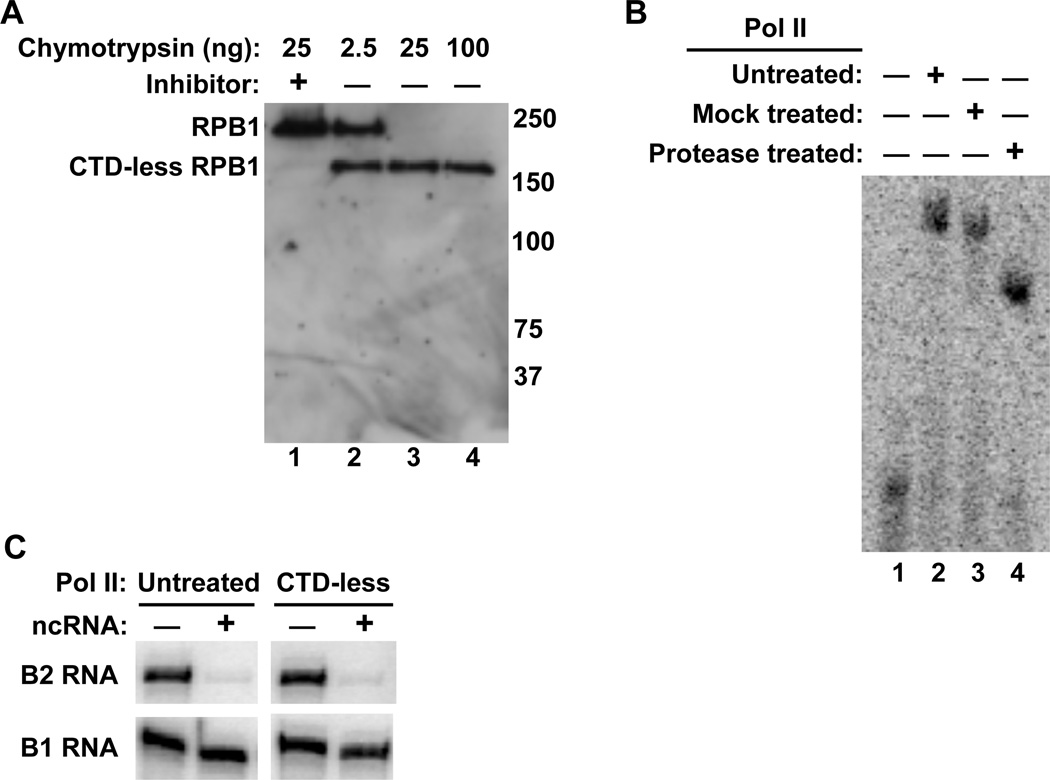
To begin to delineate the region(s) of Pol II that are involved in B2 RNA binding and transcriptional repression we removed the RPB1 CTD from purified human Pol II using limited protease digestion, as previously described.27,28 The protease used was chymotrypsin, which cleaves at aromatic side chains. The CTD heptapeptide repeats have the consensus sequence YSPTSPS, and each of the 52 repeats in human Pol II contains one chymotrypsin cleavage site (i.e. Y), making the predominantly unstructured CTD highly susceptible to fragmentation by chymotrypsin. An increasing amount of chymotrypsin was added to purified Pol II. A Western blot of the RBP1 subunit of Pol II showed that 25 ng of chymotrypsin removed the CTD from purified Pol II and the rest of RPB1 remained intact (Figure 1A). Moreover, the decrease in the size of RPB1 was blocked by the addition of chymotrypsin inhibitor (lane 1).
Figure 1.
The loss of the Pol II CTD does not alter the ability of B2 RNA to bind Pol II and repress transcription in vitro. (A) The Pol II CTD was digested by limited proteolysis with chymotrypsin. Shown is a Western blot of the RPB1 subunit of Pol II after incubation with increasing amounts of chymotrypsin. Molecular weight markers (kDa) are labeled on the left. (B) CTD-less Pol II binds B2 RNA as well as untreated Pol II. ncRNA/Pol II complexes were resolved by EMSAs. (C) Loss of the Pol II CTD does affect the ability of B2 RNA to repress transcription in vitro. Shown are the transcripts produced from in vitro reactions containing CTD-less Pol II and untreated Pol II in the absence and presence of 2.5 nM B2 RNA, or 2.5 nM B1 RNA as a negative control.
The influence of the CTD on the ability of Pol II to bind B2 RNA was investigated using electrophoretic mobility shift assays (EMSAs). Pol II, CTD-less Pol II (treated with chymotrypsin), or mock treated Pol II (treated with chymotrypsin and chymotrypsin inhibitor) were incubated with B2 RNA. As shown in Figure 1B, all three forms of Pol II bound B2 RNA to a similar extent, although the CTD-less Pol II/B2 RNA complex migrated further in the gel. Hence, the CTD is not required for B2 RNA to bind Pol II. The change in migration of the Pol II/B2 RNA complex upon removal of the CTD was large considering that ~10% of the Pol II mass was removed. A likely explanation for this is that the predominately unstructured CTD has a large conformational effect on the migration of the complex in native gels, and removal of the CTD allowed the resulting more compact complex to migrate much further in the gel.
We also asked whether the CTD contributes to transcriptional repression by B2 RNA. Repression by B2 RNA was analyzed using a TATA-containing promoter in a minimal in vitro transcription system composed of the general transcription factors TBP, TFIIB, and TFIIF, along with Pol II; the CTD is not required for transcriptional activity in this system.27,29 In vitro transcription assays were performed with CTD-less Pol II and untreated Pol II in the absence and presence of B2 RNA, or B1 RNA as a negative control. As shown in Figure 1C, B2 RNA effectively repressed transcription by the CTD-less Pol II. Together, these assays show that the CTD is not required for B2 RNA to bind Pol II and repress transcription.
Biotinylated B2 RNA(81-130) represses transcription in vitro
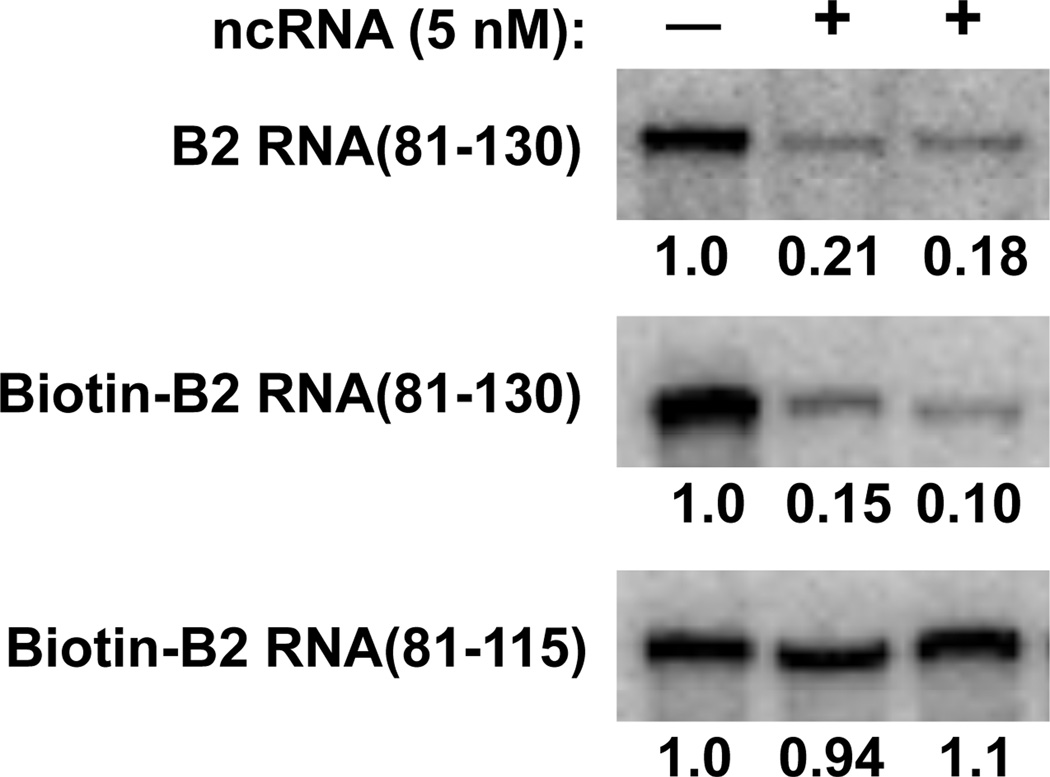
We previously identified nucleotides 81-130 as the minimal region of B2 RNA that could bind Pol II and potently repress in vitro transcription.13 When this region was further deleted, we found that B2 RNA(81-115) could neither bind Pol II nor repress transcription.13 This pair of RNAs would be useful in crosslinking studies to identify the region of Pol II specifically bound by the functional B2 RNA(81-130), with B2 RNA(81-115) serving as a negative control. The crosslinking studies we describe below required the use of 5’-biotinylated RNAs. As shown in Figure 2, 5’-biotin-B2 RNA(81-130) repressed transcription similarly to non-biotinylated B2 RNA(81-130). As anticipated, 5’-biotin-B2 RNA(81-115), did not repress transcription (bottom panel).
Figure 2.
Biotinylation of B2 RNA(81-130) does not affect its ability to potently repress transcription in vitro. B2 RNA(81-130), 5’-biotin-B2 RNA(81-130), or 5’-biotin-B2 RNA(81-115) were individually added to transcription reactions where indicated. 390 nt transcript was monitored. The values indicate the amount of transcript produced normalized to the reaction that did not contain ncRNA.
Identification of Pol II peptides that crosslink to B2 RNA
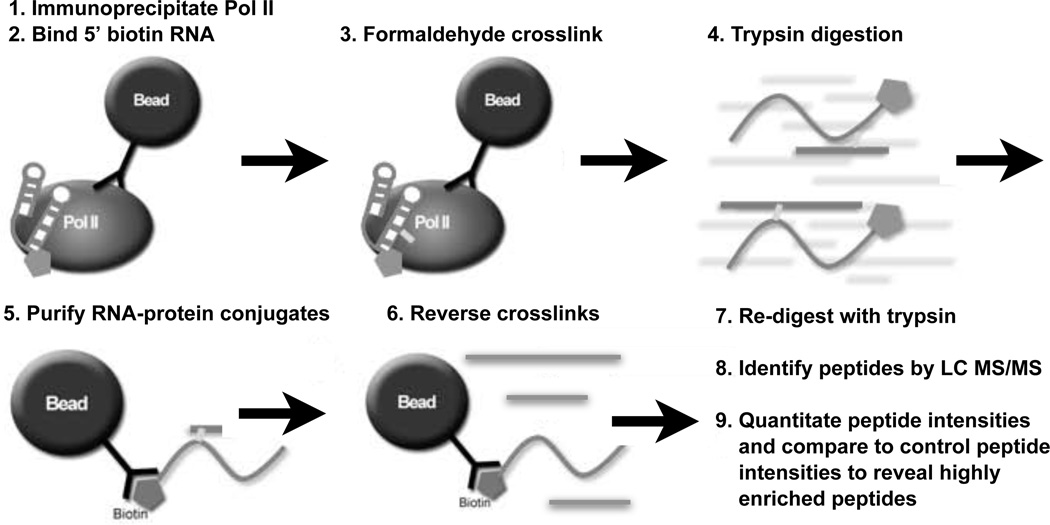
To identify the region(s) of Pol II that binds to B2 RNA(81-130) we used the approach diagrammed in Figure 3. First, human Pol II was immunoprecipitated from HeLa nuclear extracts and extensively washed. We used the antibody 8WG16, which targets the CTD and was therefore unlikely to disrupt binding of B2 RNA to Pol II (see Figure 1). Next, 5’-biotinylated RNA was bound to the immunopurified Pol II. To remove unbound RNA and disrupt any interactions between the RNA and beads, washes with competitor DNA and higher salt were performed. Following the washes, formaldehyde was added to crosslink the 5’-biotinylated RNA to Pol II. Subsequently, the sample was treated with trypsin to digest Pol II. We wanted to isolate only those peptides that crosslinked to the RNA, therefore, the biotinylated RNA was purified by streptavidin affinity chromatography. Peptides were eluted by reversing the crosslinks. Recovered peptides were treated a second time with trypsin to cleave after K or R residues that might have been protected by crosslinking. Peptides were then sequenced by LC MS/MS using a highly sensitive LTQ-Orbitrap mass spectrometer capable of detecting attomole amounts of peptides.30,31 This protocol was performed: 1) in duplicate with 5'-biotin-B2 RNA(81- 130), 2) with the negative control 5'-biotin-B2 RNA(81-115), and 3) without adding RNA. The latter two samples served as controls to ensure that recovered peptides were bound specifically to B2 RNA(81-130) and not to an RNA that does not bind with high affinity to Pol II, the streptavadin beads, or biotin.
Figure 3.
Schematic overview of the ncRNA-Pol II formaldehyde crosslinking protocol. (1) Pol II was immunoprecipitated from nuclear extracts using bead-immobilized antibody. (2) 5'-biotinylated RNA was bound to Pol II, followed by washes with a nonspecific competitor DNA and higher salt to reduce nonspecific binding. (3) Complexes were treated with formaldehyde to covalently crosslink ncRNA to Pol II. (4) The entire solution containing the RNA and bead-bound Pol II was digested with trypsin. (5) After centrifugation, the supernatant was mixed with streptavidin beads to purify the ncRNA and enrich for RNA-crosslinked peptides. (6) After centrifugation the supernatant was heated to reverse formaldehyde crosslinks. (7) Peptides were then re-digested with trypsin. (8) Recovered peptides were subjected to LC MS/MS sequencing and identified using Mascot. (9) The intensities of peptides recovered from the crosslinked samples were compared to those from the control samples as described in the text.
The MS/MS spectra were searched using Mascot against the IPI v3.65 human database to identify the recovered peptides with Mascot scores greater than 20. Peptides identified by Mascot in one biological replicate and not in the other were manually checked for their presence in the latter spectra. We identified 46 different Pol II peptides that were present in both of the crosslinked B2 RNA(81-130) samples. Ten of these peptides were also found in the B2 RNA(81-115) sample, and hence were not further considered; no peptides were identified in the control sample lacking RNA. The limited number of peptides identified in these control samples indicates that the recovered peptides were specific for crosslinking to B2 RNA(81-130) and did not result from interaction with the beads or biotin. The 36 remaining Pol II peptides that crosslinked to B2 RNA(81-130) are listed in Supplementary Table 1. Some peptides had different forms such as charge state and modifications due to sample preparation (i.e. oxidation of methionine or deamidation of asparagine or glutamine), and each identified form is listed as a separate line in Supplementary Table 1. The ten peptides identified in the control B2 RNA(81-115) sample are listed in Supplementary Table 2.
The thirty-six crosslinked peptides were distributed across diverse locations on the surface of Pol II, which perhaps is not surprising since Pol II binds nucleic acids at multiple locations, for example, the RNA exit channel, pore, and cleft. B2 RNA(81-130), which has high affinity for binding Pol II might transiently interact with such sites outside of its primary docking site. Because crosslinking can covalently capture even transient interactions, it is reasonable that a much greater number of peptides would be recovered than represented by the high affinity B2 RNA(81-130) docking site. Moreover, it is possible that forming transient interactions across the surface of Pol II is part of the mechanism by which the properly oriented and kinetically stable Pol II/B2 RNA complex forms. For these reasons, it was necessary to determine which of the crosslinked peptides were most highly enriched in the B2 RNA(81-130) samples. The crosslinking coupled to LC MS/MS identified interactions, but did not reveal the frequency with which each interaction occurred. The high sensitivity of the LTQ-Orbitrap mass spectrometer would allow even low abundance peptides to be identified. We rationalized, therefore, that determining which peptides were highly enriched in the crosslinked samples would allow us to identify the location of the kinetically stable B2 RNA docking site.
Identification of the most highly enriched peptides recovered from the crosslinked samples
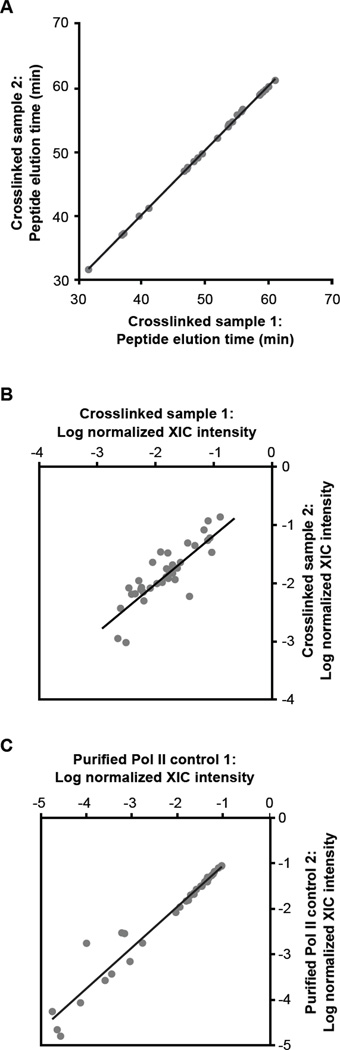
We utilized a label-free quantitation approach to determine which peptides were most highly enriched in the crosslinked B2 RNA(81-130) samples. The basis of label-free quantitation in mass spectrometry is to compare the intensity of a peptide in an experimental sample to the intensity of the same peptide in a control sample.32–34 Comparative quantitation is necessary because the unique properties of a peptide (e.g. hydrophobicity, charge, composition, etc.) affect its ability to ionize in the mass spectrometer. Therefore even if peptides are present stoichiometrically in a sample, their intensities in mass spectrometry data can be quite different.35,36 Despite large differences in ionization efficiency, the ratio of a peptide’s intensity between two samples is equal to the ratio of the corresponding protein’s abundance between the two samples.32–34,36 Here we applied label-free quantitation to determine which of the peptides recovered after crosslinking B2 RNA(81-130) to Pol II were most highly enriched compared to peptides recovered from purified Pol II control samples. Critical to the success of label-free quantitation is obtaining highly reproducible mass to charge ratios (M/Z values) and elution times of the recovered peptides since these values are critical to determining the accurate intensity for each peptide.33,37 The difference in mass (ppm error) for paired peptides in the two B2 RNA(81-130) replicates was less than 6 ppm. The times at which the peptides recovered from the B2 RNA(81-130) samples eluted from the HPLC column immediately prior to ionization were highly reproducible (Figure 4A).
Figure 4.
Properties of Pol II peptides isolated using the approach shown in Figure 3 are highly reproducible. (A) The elution times for peptides identified in the two crosslinking experiments were highly reproducible. The times at which peptides eluted from the column prior to ionization for the two B2 RNA(81-130) crosslinked samples were plotted against one another.
(B) Comparison of the total XIC values of the peptides crosslinked to B2 RNA(81-130) from two separate experiments. The total XIC value for each peptide was normalized by the sum of all XIC values from that sample, and the log of these numbers were plotted. (C) Comparison of the total XIC values of the peptides from two separate trypsin treated Pol II control samples. The total XIC value for each peptide was normalized by the sum of all XIC values from that sample, and the log of these numbers were plotted.
We first determined the intensity of each of the 36 peptides in each of the in the B2 RNA(81-130) crosslinked samples. The intensity of a peptide can be determined by quantitating the area under the peak of the extracted ion chromatogram (XIC) for the parent spectra of the peptide at a specific mass, charge, and time.36,38 We determined XIC values for each peptide, including distinct charge states and modifications due to sample preparation (Supplementary Table 1). For each peptide in each sample, the XIC values for the different states were then summed to obtain a total intensity for that peptide. We compared the total intensity of each peptide between the two samples to assess reproducibility. To do so, we normalized each peptide's total XIC value by the sum of the XIC values for all 36 peptides in a sample, then plotted the log of the normalized values from the two samples against one another (Figure 4B). The data fit to a line that has a slope of 0.86 and a Y-intercept of −0.25, both of which are similar to the expected values of 1 and 0, respectively, if the data were perfectly reproduced between the two samples.
Next, to allow comparative label-free quantitation, we trypsin digested two different samples of purified Pol II. Peptides in these control samples were subjected to LC-MS/MS using the LTQ-Orbitrap. We focused on the 36 peptides (all identified charge and modification states) that were recovered in the crosslinked samples. The difference in mass (ppm error) for paired peptides in the two control replicates was less than 2.7 ppm. 34 of the 36 peptides were found in the two control samples and were included in the analysis described below. Two of the thirty-six peptides that crosslinked to Pol II were not identified in the control Pol II samples (RPB1-8 and RPB5-2); these will be discussed in greater detail later. We determined the total intensity for each of the 34 peptides in each of the two control samples using their XIC values. To assess the reproducibility of the total XIC values from the two independent control samples we normalized each peptide's total XIC value by the total of the XIC values for all 34 peptides in a sample and plotted the log of the normalized values from the two control samples against one another (Figure 4C). The data fit to a line that has a slope of 0.91 and a Y-intercept of −0.13, both of which are similar to the expected values of 1 and 0, respectively, if the data were perfectly reproduced between the samples.
Nine peptides are present in the crosslinked samples at a significantly higher level compared to the control sample
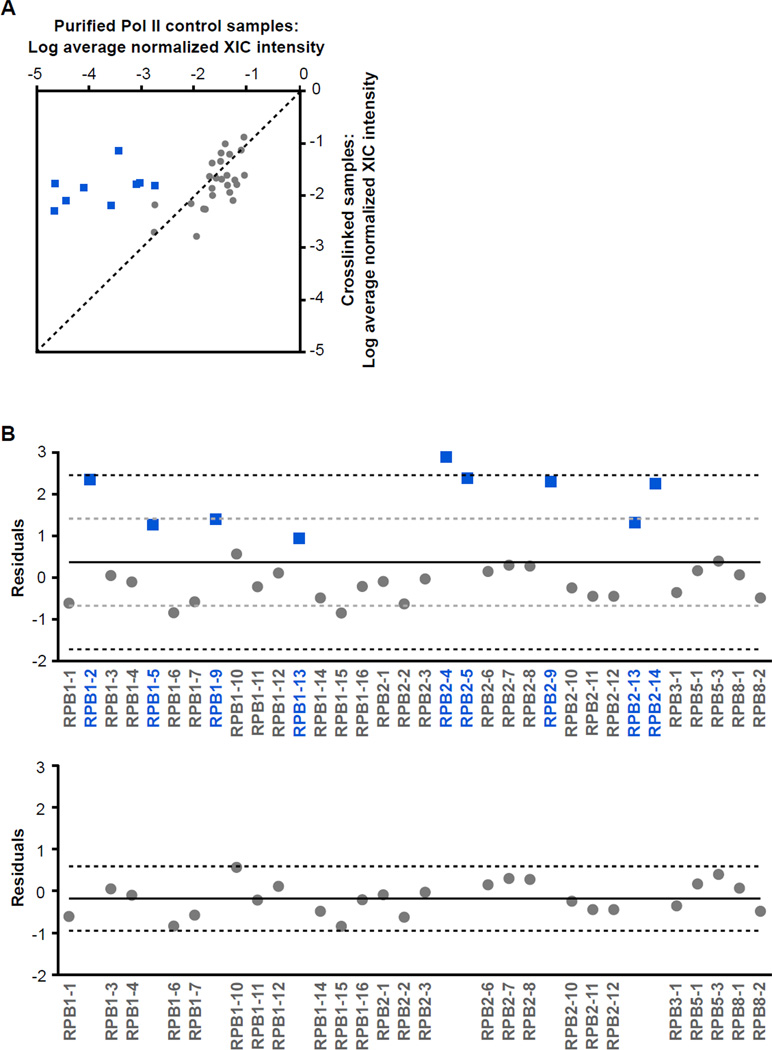
Our goal was to quantitatively compare the XIC intensities obtained from the crosslinked samples with the XIC intensities obtained from control samples for the 34 peptides. We averaged the internally normalized XIC values from the duplicate samples of each sample type (crosslinked or control), and plotted the log of these averages against one another (Figure 5A). The crosslinked samples are plotted on the Y axis and the control samples are plotted on the X axis; the color coding will be explained below. The dashed line is the theoretical line that would be obtained if the normalized intensities of each peptide in the crosslinked and control samples were identical. Peptides falling above (to the left of) the line would therefore be enriched in the crosslinked samples, and those below (to the right of) the line would be under-represented in the crosslinked samples compared to the control samples. The majority of the peptides cluster near this theoretical line, showing they were recovered in the crosslinked samples at a similar level to the control samples. Several peptides are noticeably shifted above the line, suggesting they are enriched in the crosslinked samples.
Figure 5.
Nine peptides in the crosslinked samples are significantly enriched compared to the control samples. (A) Comparison of the average XIC values of peptides crosslinked to B2 RNA(81-130) to the average XIC values from the control samples. For each peptide, the normalized total XIC values from the two crosslinked samples were averaged, and normalized total XIC values for the two control samples were averaged. The log of the average total XIC values for the crosslinked samples were plotted versus those for the control samples. The dashed line represents the theoretical line along which the peptides would fall if they were recovered at the same intensity in the crosslinked and control samples. The blue squares represent peptides found to be significantly enriched in the crosslinked samples using the statistical analysis in Figure 5B, whereas the grey circles represent peptides that were neither over- nor under-represented in the crosslinked samples versus the control samples. (B) Statistical analysis to determine which peptides were significantly enriched in the crosslinked samples. The residuals of each point from the theoretical line in panel A were plotted versus the peptide name. The solid black line represents the mean of the residuals, the grey dashed lines are one standard deviation from the mean, and the black dashed lines are two standard deviations. The bottom panel displays the points remaining after omitting points removed by the statistical analysis explained in detail in the Results section. The solid line represents the average of the remaining residuals and the dashed lines are two standard deviations.
We next statistically determined which peptides were significantly over- or under-represented in the crosslinked samples versus the control samples. First, for each peptide we calculated the difference between the log of the average normalized XIC intensities in the crosslinked and control samples (i.e. we subtracted the X-value from the Y-value for each point in Figure 5A). These residuals are plotted versus peptide name in the upper panel of Figure 5B. In this figure, the positive points are peptides over-represented in the crosslinked samples compared to the control samples, whereas the negative points are peptides that are under-represented. The solid black line represents the mean of the residuals, the grey dashed lines represent one standard deviation away from the mean, and the black dashed lines are two standard deviations. Next, we identified the point having the largest absolute value of the residual (RPB2-4 in the upper panel of Figure 5B). If this residual was greater than two standard deviations from the mean of the residuals, then we removed the point and recalculated the mean and standard deviation of the remaining residuals. We iteratively repeated this process of identifying and removing individual points that differed from the mean by more than two standard deviations, until no remaining point met this criterion.
At the completion of this process we arrived at the plot shown in the lower panel of Figure 5B. None of the points are more than two standard deviations (black dashed lines) from the mean of the remaining residuals (solid black line). All of the points shown as blue squares in the upper panel of Figure 5B were removed. During the iterative analysis, at no time did a grey point fall more than two standard deviations from the mean. We conclude that the XIC intensities for the nine peptides (blue squares) significantly differed between the crosslinked and control samples. Notably these nine peptides were all over-represented in the crosslinked samples compared to the control samples (i.e. no peptide removed by the statistical analysis was under-represented), which can be visualized in Figure 5A. We repeated the entire analysis using the XIC values from each individual crosslinked sample compared to the average of the control samples. As shown in Supplementary Figures 1 and 2, separately analyzing the two crosslinked samples resulted in the same nine peptides being identified as significantly enriched over the control samples. Sequence and position information for these nine peptides are provided in Table 1.
Table 1.
The nine significantly enriched peptides.
| Peptide | Amino acids | Human peptide sequencea |
|---|---|---|
| RPB1-2 | 33–42 | K.RMSVTEGGIK.Y |
| RPB1-5 | 213–220 | K.KILLSPER.V |
| RPB1-9 | 686–697 | H.TIGIGDSIADSK.T |
| RPB1-13 | 854–862 | K.TAETGYIQR.R |
| RPB2-4 | 200–211 | K.MATNTVYVFAKK.D |
| RPB2-5 | 437–445 | K.TRIISDGLK.Y |
| RPB2-9 | 750–762 | H.VLYYPQKPLVTTR.S |
| RPB2-13 | 1065–1078 | R.GPIQILNRQPMEGR.S |
| RPB2-14 | 1142–1150 | R.NKTQISLVR.M |
We noticed that six of the nine significantly enriched peptides contained missed trypsin cleavages (i.e. K or R at a position other than the C-terminus of the peptide). In each of these cases we detected the same missed cleavage event in the control samples. Two additional peptides in the crosslinked samples (RPB1-8 and RPB5-2) showed missed cleavages that were not identified in the control samples. Missed cleavage events might indicate that a crosslink between B2 RNA(81-130) and either a K or an R residue resulted in lack of cleavage at this position during the first trypsin treatment (i.e. prior to crosslink reversal) or incomplete cleavage during the second trypsin treatment (i.e. after crosslink reversal). To evaluate the correlation between missed cleavage events and enrichment in the crosslinked samples we changed the color-coding in the plot shown in Figure 5A, using open symbols for peptides with missed cleavages and black symbols for peptides lacking missed cleavages (Supplementary Figure 3). The squares represent peptides statistically enriched in the crosslinked samples, whereas the circles represent peptides that were neither over- nor under-represented in the crosslinked samples. Peptides with missed cleavages are spread throughout the plot. Indeed, of the 25 peptides not significantly over- or under-represented in the crosslinked samples, 11 had missed cleavages. This analysis indicates that although missed trypsin cleavage events could contribute to allowing a peptide to be identified as over-represented in the crosslinked samples, this was not the defining factor in identifying peptides enriched in the B2 RNA(81-130) crosslinked samples.
The majority of the significantly enriched peptides that crosslink to B2 RNA(81-130) cluster in the DNA binding cleft and active site region of Pol II
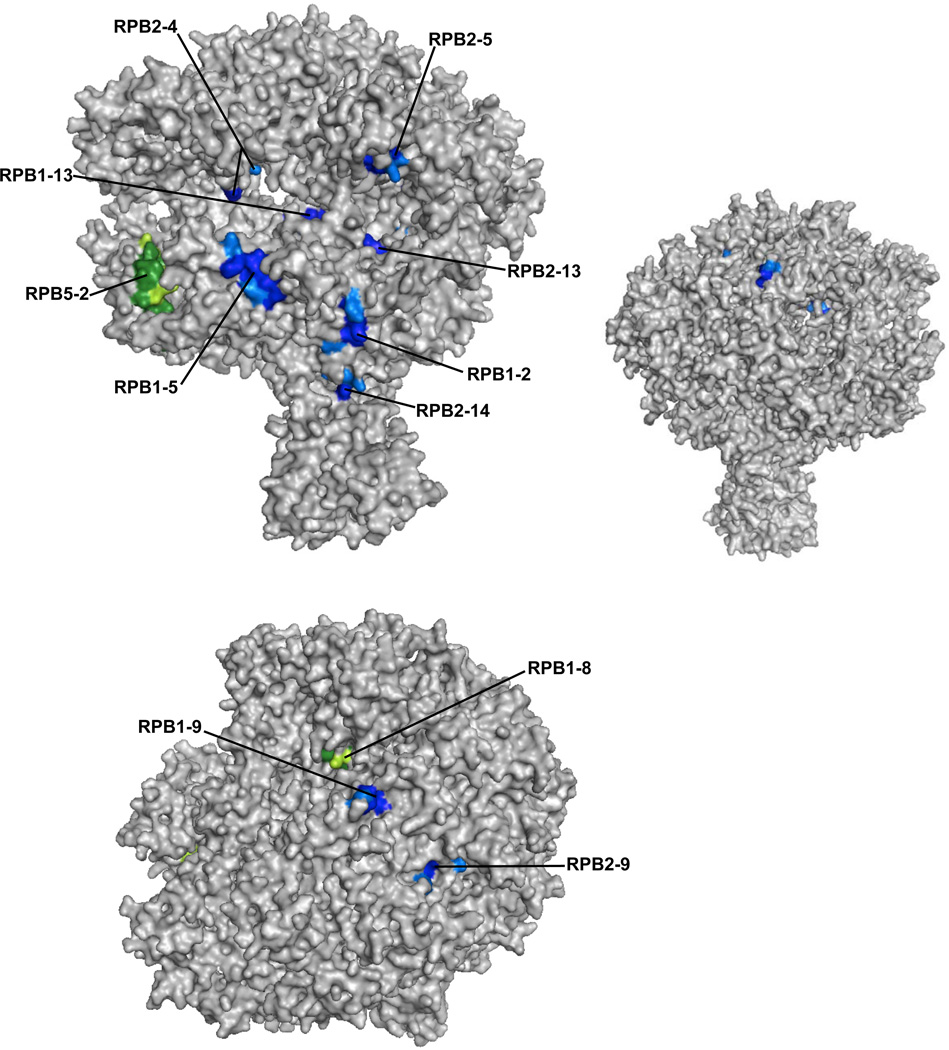
To visualize where the most highly enriched peptides localize on the surface of human Pol II we used a structure of yeast Pol II derived from X-ray crystallography. The high degree of sequence conservation makes it possible to map the peptides we identified from human Pol II onto a 12 subunit S. cerevisiae Pol II structural model.39,40 Figure 6 shows the structure of Pol II with the nine significantly enriched crosslinked peptides shown in blue, with their potential crosslinkable amino acids highlighted in lighter blue. Each peptide is exposed on the surface of Pol II, with its crosslinkable amino acids accessible to a docked ncRNA. Seven of the nine peptides map to the side of Pol II that contains the DNA binding cleft and active site region, and five of these peptides are exposed in the DNA binding cleft (upper image). These peptides extend over an area consistent with the size of a single B2 RNA(81-130) molecule.41 The remaining two peptides (from the nine) map to the top of Pol II (lower image). The two peptides identified in both crosslinked samples that were not present in the control Pol II sample are mapped in green, with their crosslinkable amino acids highlighted in lighter green. These peptides, RPB5-2 and RPB1-8, map to the cleft and near the pore of Pol II, respectively. As a point of comparison, we flipped the upper image horizontally, obtaining the smaller image to the right. This surface of Pol II is largely devoid of peptides enriched in the crosslinked samples; the three peptides partially exposed on this surface (RPB1-9, RPB2-4, RPB2-9), are more highly visible on the two other representations of the polymerase. We conclude that the peptides enriched in the crosslinked sample mainly cluster on the surface of Pol II containing the DNA binding cleft and active site region.
Figure 6.
The majority of the Pol II peptides significantly enriched in the crosslinked samples map to the surface of Pol II containing the DNA cleft and active site region. The top left panel shows a front view of Pol II, and the bottom panel shows a top view. The significantly enriched peptides are shown in blue, with crosslinkable amino acids highlighted in light blue. RPB5-2 and RPB1-8, which were not found in the control Pol II samples, are shown in green, with crosslinkable amino acids in light green. For the smaller image on the top right, the image on the top left was flipped horizontally to show the lack of peptides that map to the back side of Pol II. The images were generated using Pymol and PDB structure 3FKI.39,40
Discussion
To determine the B2 RNA(81-130) binding site on Pol II, a twelve subunit 500 kDa protein complex purified from human cells, we utilized a formaldehyde crosslinking assay followed by a label-free peptide quantitation method. First, we found that the RPB1 CTD is not required for B2 RNA binding to Pol II and repressing transcription in vitro. We then crosslinked B2 RNA(81-130) to Pol II, treated with trypsin and purified RNA-peptide conjugates under stringent conditions. Recovered peptides were identified by LC-MS/MS. 36 Pol II peptides were found reproducibly crosslinked to B2 RNA(81-130) but not the negative control B2 RNA(81-115). We determined the intensity of each identified peptide using the area under the peak of the XIC, then compared this XIC value to the XIC value for the same peptide recovered from control Pol II samples. We then statistically determined that nine peptides were significantly enriched in the crosslinked samples over the control samples. Most of these peptides cluster on the surface of Pol II containing the DNA binding cleft and active site region, which is consistent with the mechanism by which B2 RNA represses transcription, as well as recent EM studies.12,15,23
Seven of the nine enriched crosslinked peptides specifically cluster on the surface of Pol II that comprises the DNA cleft and active site (Figure 6 upper panel, blue). This image also shows that one of the peptides that crosslinked to B2 RNA but was not present in the control sample (RPB5-2, green) is also part of the DNA cleft. The enrichment of peptides on this surface is further highlighted by the backside view of Pol II (Figure 6, upper right image) and the top view (Figure 6, lower image), each of which shows portions of only three crosslinked peptides. Of these, RPB1-8 is part of the pore thought to allow entry of NTPs into the active site of the polymerase.42,43 When the structure is viewed in an orientation different from that shown, a crosslinkable amino acid within RPB1-8 can be seen deep in the cleft. It is possible that this amino acid crosslinked to B2 RNA, as opposed the amino acid that is surface exposed on the top of the polymerase. Overall, 9 of the 11 peptides mapped on the Pol II model have crosslinkable amino acids on the surface of Pol II that comprises the DNA cleft and active site.
Comparing the positions of the enriched peptides to sites on Pol II that associate with promoter DNA suggests that B2 RNA would interfere with interactions between Pol II and the promoter. For example, in the structure of a Pol II elongation complex, the DNA contacts the RPB5 jaw domain at a loop containing Pro118;42 the corresponding proline residue in human Pol II is part of the RPB5-2 peptide that crosslinked to B2 RNA. Biochemical studies from the Hahn lab have mapped promoter-protein contacts in preinitiation complexes using site-specific crosslinking approaches.44 The pathway that the DNA downstream of the TATA box follows is similar to the region to which B2 RNA crosslinks. Finding that B2 RNA docks to Pol II in regions where promoter DNA typically interacts is consistent with our previous biochemical studies of the mechanism by which B2 RNA represses transcription. We found that B2 RNA binds Pol II and assembles into complexes at promoters.12,13 Although these complexes contain the polymerase, general transcription factors, and promoter DNA, they are transcriptionally inactive because B2 RNA prevents Pol II from properly engaging the DNA. Indeed, Pol II-promoter crosslinks and DNase I protection by Pol II are disrupted by B2 RNA.15 Two potential models were consistent with these previous results: 1) B2 RNA binds the DNA cleft and blocks promoter DNA from properly binding Pol II and 2) B2 RNA interacts with Pol II away from the DNA cleft and elicits a conformational change that prevents Pol II from interacting with DNA. The studies presented here support the former model.
Crosslinking studies of preinitiation complexes have also mapped protein-protein interactions. Interestingly, the areas of Pol II over which TBP, TFIIB, and TFIIF crosslink are largely devoid of B2 RNA crosslinks, suggesting that in the presence of the ncRNA, the network of protein-protein interactions in a minimal preinitiation complex could still form.45–47 This is consistent with the results of our previous biochemical experiments.12 Moreover, by binding in the DNA cleft, B2 RNA allows the polymerase to maintain its interactions with other general transcription factors, which themselves can still bind promoter DNA. These protein-protein contacts and general transcription factor-DNA contacts are likely integral for holding Pol II within inhibited complexes containing B2 RNA, since most of the polymerase/DNA contacts are not present within these complexes.15 Our crosslinking data suggest, however, that B2 RNA binding to Pol II could interfere with TFIIE and TFIIH interactions with the forming preinitiation complex. TFIIE crosslinks to the RPB1 clamp domain of Pol II47, where crosslinked peptides RPB1-5 and RPB1-2 are also located. TFIIH lies adjacent to TFIIE, enclosing the promoter DNA further downstream in the cleft;48 the crosslinked RBP5-2 peptide is in this region. It is possible that proper contacts with TFIIE and TFIIH would not occur in the presence of B2 RNA. Interestingly, we previously found that B2 RNA inhibits phosphorylation of serine 5 residues in the Pol II CTD by the TFIIH kinase.49 These studies support a model in which the B2 RNA/Pol II interaction causes a conformational change in promoter bound complexes such that TFIIH cannot access the CTD as substrate for phosphorylation (i.e. an allosteric model for inhibition)
The location at which B2 RNA(81-130) docks to Pol II is consistent with a previous crystal structure of yeast Pol II bound to an RNA aptamer that represses transcription. The FC aptamer was selected for binding yeast Pol II and found to inhibit the initiation of transcription.50 Later studies showed that the FC aptamer could compete with B2 RNA for binding to yeast Pol II.41 A co-crystal structure of yeast Pol II bound to FC revealed that the aptamer occupies the DNA cleft of the polymerase.41 The studies presented here show that indeed B2 RNA and the FC aptamer bind Pol II at overlapping sites.
Our studies are also consistent with recent cryo-EM reconstructions of human Pol II complexed with B2 RNA(81-130), which showed the RNA bound within the DNA cleft of Pol II.23 The crosslinking studies extend the cryo-EM work by identifying peptides that make direct contact with B2 RNA(81-130) within the DNA cleft. It remains to be determined which peptides in Pol II crosslink to Alu RNA, B1 RNA, and full-length B2 RNA. The cryo-EM studies show that these ncRNAs also localize to the DNA cleft of Pol II.23 It would be informative to determine the additional peptides that crosslink to Alu RNA, B1 RNA, and full-length B2 RNA compared to B2 RNA(81-130). This would provide insight into the specific contacts that distinguish binding to Pol II from the ability to repress transcription, as well as which contacts are unique to each ncRNA. Moreover, identifying such contacts could provide insight into mechanisms by which the ncRNA/Pol II interaction is regulated; for example, TFIIF can destabilize the B1 RNA/Pol II interaction but not the B2 RNA/Pol II interaction. Finally, the crosslinking-MS approach we describe here could be useful for determining nucleic acid binding sites on other large protein complexes that might otherwise be difficult to investigate.
Materials and Methods
Preparation of RNA
B2 RNA and B1 RNA were prepared as previously described.13 5’ Biotinylated RNAs were purchased from IDT. Non-biotinylated B2 RNA(81-130) was prepared by annealing template and non-template strand oligonucleotides (Invitrogen) containing the T7 RNA polymerase promoter upstream of B2 RNA(81-130), and using transcription by T7 RNA polymerase as previously described.51 RNAs were folded prior to use by incubating them for 3 min at 95°C in Buffer A (10% glycerol, 10 mM Tris (pH 7.9), 10 mM HEPES (pH 7.9), 50 mM KCl, 4 mM MgCl2, 1 mM DTT), and immediately putting them on ice for 3 min.
Preparation of CTD-less Pol II
Limited proteolysis of Pol II was performed as previously described.27,28 Briefly, Pol II was digested in Buffer A using a 1:20 mass ratio of chymotrypsin to Pol II. The increased mass ratio (typically 1:1600) was due to protease inhibitors present in our purified Pol II sample. Reactions were terminated after 20 min with 50 µg/ml soybean chymotrypsin inhibitor (Sigma) and 0.8 mM phenylmethylsulfonyl fluoride (PMSF). The RBP1 subunit was resolved by 4% SDS PAGE followed by western blotting with antibody sc899 (Santa Cruz Biotechnology).
In vitro transcription assays and EMSAs
Recombinant human TBP, TFIIB, TFIIF, and native human Pol II were prepared as previously described.52 Negatively-supercoiled plasmid containing the adenovirus major late promoter (−40 to +10) fused to a 380-bp G-less cassette was used as the DNA template. 20 µL reactions were assembled in Buffer A plus 25 µg/mL BSA as described.13 Factors were used at the following final concentrations: 3.5 nM TBP, 10 nM TFIIB, 2 nM TFIIF, 2–4 nM Pol II and 1 nM DNA template. DNA templates were pre-incubated with TBP at 30°C for 3 min and folded ncRNA was added to the mixture. TFIIB, TFIIF and Pol II were pre-incubated in a separate tube at 30°C for 3 min. The contents of these two tubes were mixed and incubated at 30°C for 20 min. 625 µM ATP, 625 µM UTP, 25 µM (5 µCi alpha-32P[CTP]) were added, then the reactions were stopped after 20 min. Transcript RNA was ethanol precipitated and resolved by 6% denaturing PAGE.
EMSAs were performed as previously described.12 Briefly, 0.015 nM 32P-body labeled ncRNA was incubated with 4nM Pol II for 10 min at 30°C in 20 µL Buffer A. Complexes were subjected to electrophoresis on 4% native gels containing 0.5× TBE, 4% glycerol, and 5 mM magnesium acetate.
Immunoprecipitation of RNA polymerase II
Nuclear extract was prepared from 1×109 HeLa cells as previously described.53 10 µL of the antibody 8WG16 was immobilized on 250 µL Protein A magnetic beads (Dyna A beads, Invitrogen) in Buffer D (50 mM Tris (pH 7.9), 0.1 mM EDTA, 20% glycerol, 1 mM DTT, 0.1 mM PMSF, 0.1% NP40, 0.1 M ammonium sulfate). The beads were incubated with HeLa nuclear extract overnight at 4°C. After removing the supernatant, the beads were washed three times with 10 column volumes Buffer D plus 0.5 M KCl, two times with Buffer D plus 0.1 M KCl, and three times with Buffer A containing 0.1% NP40.
Formaldehyde crosslinking assays
1 pmol of folded 5’-biotinylated RNA (81-130 or 81-115) was added to immunoprecipitated Pol II in 500 µL Buffer A and incubated at 30°C for 15 min in low retention eppendorf tubes. A control was also performed with no RNA added. 1250 ng of polynucleotide dGdC was added as a non-specific competitor for an additional 5 min. After removing the supernatant the beads were washed three times with Buffer A containing 0.25 M KCl and resuspended in 250 µL Buffer A. Formaldehyde (1%) was added and reactions were incubated for 10 min at 30°C. Glycine (10 mM) was added for an additional 5 min to quench the crosslinking reaction. After removing the supernatant, the immobilized Pol II-RNA crosslinked complexes were then washed three times with 100 mM ammonium bicarbonate and digested with trypsin overnight (0.02 g/L) at 37°C. The eluate was collected and put on 20 µL streptavidin beads (MyOne T1, Invitrogen) and nutated at room temperature for 30 min. After removing the supernatant, the beads were washed three times with 100 mM ammonium bicarbonate and three times with 50 mM ammonium bicarbonate. The beads were resuspended in 50 µL of 50 mM ammonium bicarbonate and incubated at 70°C for 1.5 hr to reverse the formaldehyde crosslinks. The supernatant was removed and re-digested with 3 ng trypsin for 4 hr at 37°C. Formic acid was added to stop the digestion and the sample volume was reduced to 10 µL using a speed vac.
For the non-crosslinked sample, Pol II was purified from HeLa nuclear extract as previously described.54 Purified Pol II was digested with trypsin overnight in 50 mM ammonium bicarbonate followed by the addition of formic acid and the sample volume was reduced to 10 µL using a speed vac.
LC-MS/MS
Samples acquired from the crosslinking experiments using B2 RNA(81-130) (two replicates), B2 RNA(81-115), the no RNA control, as well as from purified Pol II alone (two replicates) were analyzed by LC/MS/MS using an LTQ-Orbitrap mass spectrometer interfaced with a Waters nanoAcquity UPLC. Chromatographic separation was performed with a BEH C18 reversed phase column (25 cm × 75 µm i.d., 1.7 µm, 100 Å, Waters), using a linear gradient from 95% Buffer B (0.1% formic acid) to 40% Buffer C (0.1% formic acid, 80% acetonitrile) over 60 minutes at a flow rate of 300 nl/min. The top five most intense precursor ions were targeted for MS/MS sequencing using monoisotopic precursor selection, rejecting singly charged ions. Dynamic exclusion was used with a repeat count of one, a repeat duration of 30 s, and an exclusion mass width of 20 ppm. The maximum injection time for Orbitrap parent scans was 500 ms with a target AGC of 1 × 106. The maximum injection time for LTQ MS/MS scans was 250 ms with a target AGC of 1 × 104. The normalized collision energy was 35% with an activation Q of 0.25 for 30ms.
Mascot searches
MS/MS spectra acquired from the LTQ-Orbitrap were searched in Mascot using the IPI v3.65 human database. The following parameters were used to identify the peptides: monoisotopic parent mass tolerance of 20 ppm (1 C13 peak), peptide MS/MS mass tolerance of 0.6 Da, at most two missed cleavages, and the variable modifications oxidation (M), deamidation (NQ), and a minimum score of 20. Peptides identified by Mascot in one biological replicate and not another were manually checked for their presence in the latter spectra. All MS/MS spectra listed were then manually validated prior to inclusion in Supplementary Table 1.
Quantitation and comparative analysis
Abundance values for each peptide were calculated using the area peak intensity of the extracted ion chromatogram (XIC). This was determined using XCalibur software on the RAW files generated for each sample. Area under the curve ion values were determined using a 20 ppm m/z mass window on the monoionizable peak at a given elution time as previously described.34
For each peptide in the crosslinked or non-crosslinked sample, its total intensity value was determined by summing the intensities of individual ion peak areas for each charge state and for each modification induced by sample preparation (e.g. oxidation and deamidation). By means of example, consider peptide RPB1-1. As shown in Supplementary Table 1, this peptide was identified in two charge states, RPB1-1a and RPB1-1b. Therefore, the total XIC value for peptide RPB1-1 in crosslink sample 1 equals 1.20 × 106 (the sum of the XIC values for RPB1-1a and RPB1-1b, 4.61 × 105 + 7.41 × 105). Subsequent steps in data analysis are explained in detail in the Results section.
Pymol
Peptide sequences were converted to their respective S. cerevisiae sequences by aligning the human Pol II amino acid sequence to the S. cerevisiae Pol II sequence for each subunit. The converted peptide sequences were then mapped onto the S. cerevisiae Pol II crystal structure, Protein Database Bank (PDB) number 3FKI.39,40
Supplementary Material
Highlights.
The non-coding B2 RNA potently represses mRNA transcription by binding tightly to mammalian RNA polymerase II (Pol II); however, the precise sites on Pol II to which B2 RNA binds was unknown.
We used reversible formaldehyde crosslinking coupled to mass spectrometry to identify the Pol II peptides that bind directly to the minimal functional region of B2 RNA.
A label-free peptide quantitation approach revealed which of the crosslinked peptides were most highly enriched.
Mapping these peptides onto a crystal structure of yeast Pol II showed that B2 RNA associates with a surface of Pol II containing the DNA binding cleft and active site region, providing fundamental insight into how B2 RNA represses transcription.
Acknowledgements
This work was supported by a Public Health Service grant (R01 GM068414) from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: CTD (C-terminal domain of RPB1), EMSA (electrophoretic mobility shift assay), MS (mass spectrometry), ncRNA (non-coding RNA), PAGE (polyacrylamide gel electrophoresis), Pol II (RNA polymerase II), Pol III (RNA polymerase III), RCAP (reversible crosslinking affinity purifications), RPB (subunit of RNA polymerase II), SINE (short interspersed element), TBP (TATA binding protein), TFII- (transcription factor of RNA polymerase II), XIC (extracted ion chromatogram
References
- 1.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 2.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 4.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 5.Barrandon C, Spiluttini B, Bensaude O. Non-coding RNAs regulating the transcriptional machinery. Biol. Cell. 2008;100:83–95. doi: 10.1042/BC20070090. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit. Rev. Biochem. Mol. Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spradling A, Penman S, Pardue ML. Analysis of Drosophila mRNA by in situ hybridization: sequences transcribed in normal and heat shocked cultured cells. Cell. 1975;4:395–404. doi: 10.1016/0092-8674(75)90160-9. [DOI] [PubMed] [Google Scholar]
- 8.Sonna LA, Gaffin SL, Pratt RE, Cullivan ML, Angel KC, Lilly CM. Effect of acute heat shock on gene expression by human peripheral blood mononuclear cells. J. Appl. Physiol. 2002;92:2208–2220. doi: 10.1152/japplphysiol.01002.2001. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez J, Pauli D, Tissieres A. Transcriptional regulation in Drosophila during heat shock: a nuclear run-on analysis. Chromosoma. 1993;102:233–248. doi: 10.1007/BF00352397. [DOI] [PubMed] [Google Scholar]
- 10.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucl. Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 12.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 13.Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA. 2007;13:583–596. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner SD, Kugel JF, Goodrich JA. TFIIF facilitates dissociation of RNA polymerase II from noncoding RNAs that lack a repression domain. Mol. Cell. Biol. 2010;30:91–97. doi: 10.1128/MCB.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko S, Manley JL. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3' end formation. Mol. Cell. 2005;20:91–103. doi: 10.1016/j.molcel.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 21.Orlicky SM, Tran PT, Sayre MH, Edwards AM. Dissociable Rpb4-Rpb7 subassembly of rna polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem. 2001;276:10097–10102. doi: 10.1074/jbc.M003165200. [DOI] [PubMed] [Google Scholar]
- 22.Meka H, Werner F, Cordell SC, Onesti S, Brick P. Crystal structure and RNA binding of the Rpb4/Rpb7 subunits of human RNA polymerase II. Nucl. Acids Res. 2005;33:6435–6444. doi: 10.1093/nar/gki945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassube SA, Fang J, Grob P, Yakovchuk P, Goodrich JA, Nogales E. Structural insights into Transcriptional Repression by ncRNAs that bind to Human Pol II. J. Mol. Biol. 2012 doi: 10.1016/j.jmb.2012.08.024. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han YT, Hsu YH, Lo CW, Meng M. Identification and functional characterization of regions that can be crosslinked to RNA in the helicase-like domain of BaMV replicase. Virology. 2009;389:34–44. doi: 10.1016/j.virol.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Gorman W, Thomas B, Kwek KY, Furger A, Akoulitchev A. Analysis of U1 small nuclear RNA interaction with cyclin H. J. Biol. Chem. 2005;280:36920–36925. doi: 10.1074/jbc.M505791200. [DOI] [PubMed] [Google Scholar]
- 26.Kim YC, Russell WK, Ranjith-Kumar CT, Thomson M, Russell DH, Kao CC. Functional analysis of RNA binding by the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:38011–38019. doi: 10.1074/jbc.M508145200. [DOI] [PubMed] [Google Scholar]
- 27.Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL. The Cterminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc. Nat. Acad. Sci. USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corden JL, Cadena DL, Ahearn JM, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. Natl. Acad. Sci. USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim WY, Dahmus ME. The major late promoter of adenovirus-2 is accurately transcribed by RNA polymerases IIO. IA, and IIB. J. Biol. Chem. 1989;264:3169–3176. [PubMed] [Google Scholar]
- 30.Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 31.Macek B, Waanders LF, Olsen JV, Mann M. Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol. Cell. Prot. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong JWH, Sullivan MJ, Cagney G. Computational methods for the comparative quantification of proteins in label-free LCn-MS experiments. Brief. Bioinform. 2008;9:156–165. doi: 10.1093/bib/bbm046. [DOI] [PubMed] [Google Scholar]
- 35.Tang H, Arnold RJ, Alves P, Xun Z, Clemmer DE, Novotny MV, Reilly JP, Radivojac P. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–e488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- 36.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Prot. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Meth. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 38.Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J. Prot. Res. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 39.Meyer PA, Ye P, Zhang M, Suh MH, Fu J. Phasing RNA Polymerase II Using Intrinsically Bound Zn Atoms: An Updated Structural Model. Structure. 2006;14:973–982. doi: 10.1016/j.str.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Meyer PA, Ye P, Suh MH, Zhang M, Fu J. Structure of the 12-subunit RNA polymerase II refined with the aid of anomalous diffraction data. J. Biol. Chem. 2009;284:12933–12939. doi: 10.1074/jbc.M809199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat. Struct. Mol. Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- 42.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 A Resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 43.Cramer P, Bushnell DA, Kornberg RD. Structural Basis of Transcription: RNA Polymerase II at 2.8 A Resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 44.Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat. Struct. Mol. Biol. 2006;13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 47.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grunberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat. Struct. Mol. Biol. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA represses TFIIH phosphorylation of RNA polymerase II. Transcr. 2011;2:45–49. doi: 10.4161/trns.2.1.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J. Biol. Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- 51.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 52.Weaver JR, Kugel JF, Goodrich JA. The sequence at specific positions in the early transcribed region sets the rate of transcript synthesis by RNA polymerase II in vitro. J. Biol. Chem. 2005;280:39860–39869. doi: 10.1074/jbc.M509376200. [DOI] [PubMed] [Google Scholar]
- 53.Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 54.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Metz B, Kersten GFA, Hoogerhout P, Brugghe HF, Timmermans HAM, De Jong AD, Meiring H, ten Hove J, Hennink WE, Crommelin DJA. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J. Biol. Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- 56.Metz B, Kersten GFA, Baart GJE, de Jong A, Meiring H, ten Hove J, van Steenbergen MJ, Hennink WE, Crommelin DJA, Jiskoot W. Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconj. Chem. 2006;17:815–822. doi: 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.