Abstract
Quantitative analysis of the intracellular trafficking of non-viral vectors provides critical information that can guide the rational design of improved cationic systems for gene delivery. Subcellular fractionation methods, combined with radiolabeling, produce quantitative measurements of the intracellular trafficking of non-viral vectors and the therapeutic payload. In this work, differential and density-gradient centrifugation techniques were used to determine the intracellular distribution of radiolabeled 25 kD branched polyethylenimine (bPEI)/plasmid DNA complexes (“polyplexes”) in HeLa cells over time. By differential centrifugation, [14C]bPEI was found mostly in the lighter fractions whereas [3H]DNA was found mostly in the heavier fractions. A majority of the intracellular polymer (~60%) and DNA (~90%) were found in the nuclear fraction. Polymer and DNA also differed in their distribution to heavier and denser organelles (lysosomes, mitochondria) in density-gradient centrifugation studies. An unexpected finding from this study was that between 18–50% of the DNA applied to the cells became cell-associated (either with the cell membrane and/or internalized), while only 1–6% of the polymer did so, resulting in an effective N/P ratio of less than 1. These results suggest that a significant amount of cationic polymer is dissociated from the DNA cargo early on in the transfection process.
Keywords: Plasmid delivery, subcellular fractionation, intracellular trafficking, poly(ethylenimine)
1. INTRODUCTION
Non-viral vectors, such as cationic polymers and lipids, have been extensively investigated for gene delivery1. Although non-viral vectors are generally considered safer than viral vectors, they are much less efficient at delivering nucleic acids to cells than viruses2–4. Non-viral vectors encounter several barriers to gene delivery that viruses can readily overcome, such as cellular uptake, endosomal escape, cytoplasmic translocation, and gene expression. Various moieties, such as endosomal escape peptides5,6 and nuclear localization7 sequences, have been incorporated into the design of non-viral vectors to overcome these barriers; however, there are only a few studies that extensively investigate the impacts of chemical modifications on the intracellular trafficking of synthetic vectors8–11.
The intracellular fate of synthetic gene delivery vehicles has been studied by fluorescence imaging, the use of chemical inhibitors, and subcellular fractionation. Fluorescence imaging methods, such as confocal microscopy, have been used to determine the intracellular distribution of non-viral vectors through colocalization studies with organelle labels12. While informative, microscopy methods are only semi-quantitative, although several efforts to develop quantitative microscopy techniques have been explored13–15. Chemical inhibitors have also been used to selectively inhibit various endocytic pathways16,17, although the specificity and the effect of these inhibitors on normal cellular activity have been debated18. Subcellular fractionation methods can provide quantitative data on intracellular distribution of materials and has been used to determine amount of delivered DNA to the nucleus with liposomal formulations19 and poly(ethylenimine) (PEI)20–22, and to provide rate constants for quantitative models4,23,24. Some studies have also tracked radiolabeled polyplexes in additional organelles using subcellular fractionation methods. While fluorescence microscopy can be affected by environmental factors, such as pH and temperature25, radioactivity can provide substantially increased sensitivity and robustness. Furthermore, the use of radiolabeled compounds allows for mass balance calculations, so that 100% of the material applied to the sample can be tracked. Laurent et al. demonstrated the use of differential and isopycnic centrifugation to track [35S]DNA complexed with poly(lysine) in vivo9. Colin et al. also tracked radiolabeled plasmid DNA (pDNA)/RGD-K16/Lipofectamine complexes in Percoll gradients, but horseradish peroxidase (HRP) was used to shift endosomal density10. In addition, only the DNA was tracked in these studies, limiting our understanding of how the interplay between both carrier and DNA affects intracellular polyplex trafficking.
The goal of the proposed work is to quantify the intracellular distribution of cationic polymer and pDNA complexes, or polyplexes, in native cell environments. We used differential and density-gradient subcellular fractionation methods combined with radiolabeling to track both branched poly(ethylenimine) (bPEI) and pDNA in a HeLa cells, a commonly used cultured cell line. We described here a detailed approach to intracellular polyplex quantification, in which, for the first time to our knowledge, both polymer carrier and cargo DNA are followed in major organelles involved in polyplex trafficking. Polymer and pDNA were found to differ slightly in their intracellular trafficking patterns, and thus, draws attention to the necessity of more quantitative methods to investigate polyplex trafficking. We were also able to quantify the cellular uptake, membrane association, and internalization of polymer and DNA. These studies elucidated that a surprisingly low amount of polymer was internalized into the cell relative to DNA, and suggest that further studies into the mechanism and role of polycation-facilitated gene delivery are necessary.
2. MATERIALS AND METHODS
2.1 Materials
60%(w/v) OptiPrep (iodixanol) was purchased from Axis-Shield (Norton, MA). HALT protease inhibitor cocktail was purchased from Thermo Fisher Scientific (Pittsburgh, PA). 10X Tris/glycine/SDS running buffer, polyacrylamide gels, and filter paper were purchased from Bio-Rad (Hercules, CA). PVDF membrane was purchased either from Bio-Rad (Hercules, CA) or Millipore (Billerica, MA). Horseradish Peroxidase (HRP)-conjugated goat anti-mouse (no. 554002), mouse anti-Rab5 (250 μg/mL, no. 610725), and mouse anti-CD49b (250 μg/mL, no. 611017) antibodies were purchased from BD Biosciences (San Diego, CA). Mouse anti-LAMP2 antibody was purchased from the Developmental Studies Hybridoma Bank (supernatant, no. H4B4, Iowa City, IA). All cell culture medium and supplements were purchased from Cellgro/Mediatech (Fisher Scientific, Pittsburgh, PA). Acetic anhydride[14C] was purchased from American Radiolabeled Chemicals (St. Louis, MO). 2′-Deoxycytidine-5′-triphosphate (dCTP), [5-3H] (no. MT 847A), was purchased from Moravek Radiochemicals (Brea, CA). Ultima Gold XR scintillation fluid was purchased from Perkin Elmer (Waltham, MA). All other chemical reagents, including poly(ethylenimine) (PEI, 25,000 g/mol, branched), were reagent-grade or better and were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. Endotoxin-free plasmid pCMV-Luc2 was prepared by using the pGL4.10 vector (Promega, Madison, WI) and inserting the CMV promoter/intron region from the gWiz Luciferase (Aldevron, Madison, WI). The plasmid was isolated and produced with the Qiagen Plasmid Giga kit (Qiagen, Germany) according to the manufacturer’s instructions.
2.2. Cell culture
HeLa (human cervical carcinoma) cells were grown in minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and 100 IU penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B at 37 °C, 5% CO2. Cells were passaged when they reached ~80% confluency.
2.3. Preparation of cell lysate for assessment of marker enzyme assays
For assessment of marker enzyme assays, a crude cell lysate was prepared. 20 × 106 HeLa cells were seeded in 150-mm plates (at 5 × 106 per plate) overnight at 37 °C, 5% CO2. Cells were washed twice with cold phosphate-buffered saline (PBS), gently lifted off the plates in 5 mL cold PBS, washed off the plates once with PBS, and transferred into pre-chilled conical tubes. Cells were then pelleted at 500 × g for 5 min and resuspended in 10 mL cold PBS. Cells were pelleted again and then resuspended in 5 mL cold homogenization buffer (0.25 M sucrose, 10 mM HEPES-NaOH, 1 mM EDTA, pH 7.4). After centrifugation at 1000 × g for 6 min, the resulting pellet was resuspended in 2.5x the wet pellet mass of homogenization buffer (with 1X protease inhibitors added). Cells were then lysed with six freeze-thaw cycles. The protein concentration of the cell lysate was determined using a microBCA kit (Pierce) according to the manufacturer’s instructions.
2.4. Marker enzyme assays
Samples containing organelles were analyzed for hexosaminidase A activity (lysosomes)26, lactate dehydrogenase activity (cytosol), alkaline phosphatase (plasma membrane), succinate dehydrogenase activity (mitochondria)27, and total protein content. Detailed methods for these assays are provided in supplementary material. Protein content was measured using a Bradford-based protein assay kit (Bio-Rad) according to the manufacturer’s instructions, using immunoglobulin as a standard.
2.5. Optimization of cell breakage
Since detergents can potentially displace polymer/DNA interactions, cell lysis was carried out using mechanical techniques. The optimization of mechanical shear-induced cell breakage through a 25-gauge needle was carried out as previously described27, but with minor modifications. HeLa cells (20 × 106) were seeded into four 150-mm dishes overnight at 37 °C, 5% CO2. All subsequent steps were completed at 4 °C, on ice, and with pre-chilled reagents. Cells were washed twice with PBS, gently lifted off the plates in 5 mL PBS, washed off the plates once with PBS, and transferred into pre-chilled conical tubes. Cells were then centrifuged at 500 × g for 5 min and resuspended in 10 mL PBS. Cells were pelleted again and then resuspended in 5 mL homogenization buffer (0.25 M sucrose, 10 mM HEPES-NaOH, 1 mM EDTA, pH 7.4). After centrifugation at 1000 × g for 6 min, the resulting pellet was resuspended in 2.5x the wet pellet mass of homogenization buffer (with 1X protease inhibitors added). The cell suspension was then passed through a 25-gauge needle up to 30 times, with 20 μL aliquots taken after a various number of passes. The samples were diluted with 100 μL 250 mM sucrose, vortexed for 30 s, and centrifuged at 2000 × g for 10 min. The supernatant was then used to determine lactate dehydrogenase activity, using cells treated with 0.1% (v/v) Triton X-100 as a reference for 100% cell breakage.
2.6. Acetylation of PEI with [14C]acetic anhydride
Branched polyethylenimine (bPEI, MW 25,000 g/mol) was reacted via acetylation of amines to obtain 14C-labeled polymer. 20 mg of bPEI was dissolved in 100 μL dioxane and incubated with 5 molar eq. of [14C]acetic anhydride (50 mCi/mL) and 20 molar eq. of N,N-diisopropylethylamine (DIPEA) per 25 kDa polymer for 2 h at room temperature. The reaction was quenched by adding 500 μL 0.1% glacial acetic acid (in dH2O). Unreacted acetic anhydride and DIPEA was removed by applying the reaction mixture through a desalting spin column (Zeba, 7k MWCO, Thermo Fisher Scientific, Rockford, IL). The resulting eluent was characterized for the final bPEI concentration using a Cu(II) acetate assay28 and by scintillation counting. The final polymer concentration was measured to be 3.7 g/L at ~7 × 104 cpm/μL. The polymer was diluted to a stock of 1 mg/mL in 0.1X PBS, acidified to pH 6 with 0.1 N HCl, and stored at 4 °C. Further dilution of the polymer was carried out in dH2O.
2.7. Labeling of plasmid DNA with [3H]dCTP
Plasmid DNA (pLuc2-CMV) was radiolabeled using nick translation and 2′-deoxycytidine 5′-triphosphate, [5-3H] (2.5 mCi/mL), according to manufacturer’s instructions (GE Healthcare, Pittsburgh, PA). Purification of unreacted nucleotides was completed using G50 microspin columns (Probequant, GE Healthcare). The mass recovery was assumed to be 100% with a final concentration of 9.1 μg/mL at ~9 × 104 cpm/μL.
2.8. Uptake of radiolabeled polyplexes
Cells were seeded in 24-well plates at 2 × 104 cells/mL/well 24 h prior to polyplex addition. Polyplexes were formulated at N/P 5 by mixing 10 μL of [3H]DNA/unlabeled DNA mixture (final concentration of 0.1 g/L in water at ~5 × 106 cpm/mL) with 10 μL bPEI (65.3 μg/mL) for 10 min at room temperature prior to 10-fold dilution in reduced serum media (Opti-MEM, Life Technologies, Carlsbad, CA). Cells were washed once with PBS, and then incubated with 200 μL polyplexes (in Opti-MEM) for 4 h at 37 °C, 5% CO2. After the incubation period, cells were washed once with PBS, allowed to incubate with 200 μL CellScrub (Genlantis, San Diego, CA) for 15 min at room temperature, washed twice with DPBS (without divalent cations), trypsinized, and then collected for scintillation counting. For other time points (6, 8, 12, 24 h), cells were washed once with PBS after the 4 h incubation with polyplexes and replaced with complete media. At various time points after media replacement (2, 4, 8, 20 h), cells were washed with PBS, CellScrub, DPBS, and trypsinized as above. All washes and solutions were collected and analyzed for radioactivity.
2.9. Treatment of cells with radiolabeled polyplexes for fractionation studies
For each time point, 5–20 × 106 cells were grown in 150-mm plates (5 × 106 cells per plate). Polyplexes were formulated at N/P 5 by mixing 1 mL of [3H]DNA/unlabeled DNA mixture (final concentration of 0.1 g/L in water at ~5 × 106 cpm/mL) with 1 mL [14C]bPEI (65.3 μg/mL) for 10 min at room temperature. Polyplexes were then be diluted with 18 mL Opti-MEM. Each plate was washed once with PBS and incubated with 20 mL of polyplexes in Opti-MEM for specified times at 37 °C, 5% CO2. In pulse-chase experiments, cells were treated with radiolabeled polyplexes (in Opti-MEM) for 4 h at 37 °C, 5% CO2, washed once with PBS, replenished with complete media, and incubated at 37 °C, 5% CO2 for the chase period.
2.10. Preparation of post-nuclear supernatant (PNS) for subsequent fractionation
5–20 × 106 HeLa cells were seeded in 150-mm plates (at 5 × 106 per plate) overnight at 37 °C, 5% CO2. In some experiments, cells were treated with polyplexes for various times at 37 °C, 5% CO2 prior to cell harvesting. All subsequent steps were completed at 4 °C, on ice, and with pre-chilled reagents. To remove dead/compromised cells, cells were washed twice with PBS, gently lifted off the plates in 5 mL PBS, washed off the plates once with PBS, and transferred into pre-chilled conical tubes. Cells were then pelleted at 500 × g for 5 min and resuspended in 10 mL PBS. Cells were pelleted again and then resuspended in 5 mL homogenization buffer. After centrifugation at 1000 × g for 6 min, the resulting pellet was resuspended in 2.5x the wet pellet mass of homogenization buffer (with 1X protease inhibitors added). Cells were then homogenized by passing through a 25-gauge needle until greater than 90% cell lysis was achieved, as confirmed by light microscopy. Nuclei and unbroken cells were pelleted at 1000 × g for 10 min. The pellet was resuspended in homogenization buffer and centrifuged again. The resulting post-nuclear supernatant (PNS) was combined from both washes and was subjected to various fractionation procedures.
2.11. Cellular fractionation of cytosolic and vesicular components
Separation of cytosol and vesicular organelles was carried out as previously described29, but with minor modifications. The PNS was layered on top of 10 μL cushion of 2.5 M sucrose and centrifuged 100,000 × g for 30 min at 4 °C (Beckman TLS-100.3). The supernatant, containing the cytosol, was transferred and the resulting pellet was resuspended in 500 μL homogenization buffer, layered on top of another 10 μL 2.5 M sucrose cushion, and centrifuged again at 100,000 × g for 10 min at 4 °C. The supernatants were combined and the resulting pellet was resuspended in 500 μL homogenization buffer. In order to break up the vesicular pellet, the resuspended pellet was passed gently through a 25-gauge needle. For studies without additional fractionation, the sucrose cushion was omitted. Aliquots of samples were snap-frozen in liquid nitrogen and stored at −80 °C for further analysis.
2.12. Cellular fractionation via differential centrifugation
Cells were treated with radiolabeled polyplexes for 1 h at 4 °C to allow for binding prior to incubation for 30 min or 4 h at 37 °C, 5% CO2. After polyplex incubation, a post-nuclear supernatant was prepared, and then centrifuged at 3000 × g for 10 min at 4 °C. The pellet (heavy mitochondrial fraction, or HM) was resuspended in 500 μL homogenization buffer and centrifuged again. The supernatants were combined and centrifuged at 15,000 × g for 10 min at 4 °C. The pellet (light mitochondrial fraction, or LM) was resuspended in 500 μL homogenization buffer and centrifuged again. The supernatants were combined again and centrifuged at 100,000 × g for 45 min at 4 °C (Beckman TLS-100.3). The pellet (microsomal fraction, or MF) was resuspended in 500 μL TES buffer and centrifuged again. The supernatants (cytosolic fraction, or C) were combined. All pellets were resuspended in 500 μL homogenization buffer and gently passed through a 25-gauge needle to break up remaining aggregates. Aliquots of each fraction were stored at −80 °C for subsequent analyses.
2.13. Preparation of continuous iodixanol gradients
Continuous iodixanol gradients were prepared as previously described30, with some minor modifications. A stock solution of 60% iodixanol (OptiPrep) was diluted to a working stock solution of 50% (v/v) iodixanol with a sucrose buffer (0.25 M sucrose, 6 mM EDTA, 60 mM HEPES-NaOH, pH 7.4). The working solution was further diluted to either 5% or 20% iodixanol with TES buffer (0.25 M sucrose, 10 mM triethanolamine, 1 mM EDTA, pH 7.4). 5–20% continuous iodixanol gradients were prepared by layering 5.5 mL 5% iodixanol on top of 5.5 mL 20% iodixanol in a 13 mL ultracentrifuge tube and preparing continuous gradients using a Gradient Master (Biocomp, New Brunswick, Canada).
2.14. Cellular fractionation via density-gradient centrifugation
For separation of plasma membrane, endosomes, and lysosomes, the resuspended vesicular pellet from an initial cytosolic/vesicular fractionation was layered on top of a 5–20% continuous iodixanol gradient. For each experiment, a control gradient was prepared with equal volume of homogenization buffer instead of cell lysate. The tubes were then centrifuged at 90,000 × g (Beckman SW41) for 16–18 h at 4 °C. Twenty-four 500 μL fractions were collected from the top of the gradient using an automated fraction collector (Brandel, Gaithersburg, MD). Collected fractions were placed immediately on ice, aliquoted, and stored at −80°C until further analysis.
2.15. Determination of gradient density
The density of the control gradient fractions was determined by measuring the refractive index (Reichert AR200, Depew, NY). The density of the gradient was calculated using
where ρ is the density of the gradient fraction (g/mL), η is the refractive index, A and B are coefficients for ionic and non-ionic media, and are equal to 3.459 and 3.622, respectively31.
2.16. Determination of radioactivity
For optimization studies, 10 μL of polymer (N/P 5) was added to 10 μL [3H]DNA/unlabeled pDNA mixture (final concentration of 0.1 g/L in water at ~5 × 106 cpm/mL), mixed, and allowed to incubate for 10 min at room temperature. Polyplexes were then treated with an equivolume of either calf thymus DNA (0.5 g/L in dH2O), 1 M NaOH, or 10X trypsin, and then allowed to incubate for 30 min prior to addition of scinitillation fluid. Samples from fractionation studies were diluted with an equivolume of 1 M NaOH to disrupt electrostatic interactions between the polymer and DNA. For samples collected from density-gradient fractionation, a 5-fold excess of dH2O was added to the sample to dilute out quenching effects from the iodixanol gradient media. 4–5 mL of scintillation fluid was added to each vial and vigorously mixed prior to determining radioactivity levels on a liquid scintillation counter (Beckman LS6500). The total radioactivity (counts per minute, or cpm) in each sample was determined by combining the radioactivity found all collected washes and fractions. 3H and 14C measurements were analyzed using methods described elsewhere32.
2.17. Protein precipitation and immunoblotting
To increase protein loading for SDS-PAGE, aliquots of fractions (250 μL) were subjected to trichloroacetic acid (TCA)-deoxycholate(DOC)/acetone precipitation. The pellets were then resuspended in Laemmli buffer (Bio-Rad) or reducing sample buffer (Pierce) prior to SDS-PAGE. Samples were then applied to a pre-cast 4–20% polyacrylamide gel (Bio-Rad) and electrophoresed through a standard Tris/glycine/SDS buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3). Proteins were then transferred onto a PVDF membrane using standard conditions (25 mM Tris-base, 192 mM glycine, 0.1% SDS, 10% methanol) for 1.5 h at 100 V. Non-specific binding sites were blocked by incubation in blocking buffer (Superblock, Pierce) for 1 h at room temperature. Membranes were then either probed with mouse anti-Rab5 (1:1000), anti-LAMP2 (1:500), or anti-CD49b (1:500) in blocking buffer overnight at 4 °C. After 3×10 min washes with TBS-T (20 mM Tris-HCl, 137 mM NaCl, 0.1% Tween-20), membranes were probed with HRP-conjugated goat anti-mouse antibody (1:100,000) in blocking buffer for 1 h at room temperature. Membranes were washed 3×10 min with TBS-T and developed with chemiluminescent substrate (West Femto, Pierce). Chemiluminescence was detected (10–15 min exposure) using a Kodak imager (Image Station 4000MM, Rochester, NY). Blots were stripped with stripping buffer according to manufacturer’s instructions (Restore stripping buffer, Pierce), and re-probed for the different antibodies. The density of each band was calculated using ImageJ.
3. RESULTS
3.1. Uptake of [3H]DNA/bPEI polyplexes
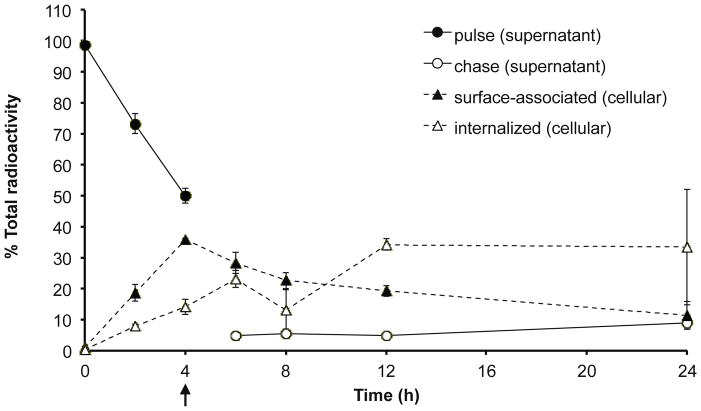
As a first approach to determine the time dependency of cellular uptake and internalization of polyplexes, a pulse-chase experiment was performed using radiolabeled polyplexes. Cells were treated with bPEI polyplexes formulated with [3H]DNA for 4 h, rinsed, and then replaced with fresh media for up to an additional 20 h. Cell-surface associated polyplexes were collected by a 15 min incubation with CellScrub buffer. During the 4 h pulse period, the rate of surface-association and cellular internalization of polyplexes, as determined by scintillation counting for the radiolabeled plasmid, appeared linear with time, with ~50% of polyplexes associating with cells after 4 h (Figure 1). During the chase period, the percentage of cell surface-associated polyplexes decreased over time, from 35.9% to 11.3%, as the percentage of internalized polyplexes increased over time, from 14.0% to 33.4%. The percentage of polyplexes also increased slightly in the chase media over time, from 4.8% after a 2 h chase to 9.1% after a 20 h chase, indicating possible gradual dissociation of polyplexes from the cell surface or polyplex exocytosis. These results suggest that the intracellular distribution of polyplexes is shifting from association with the plasma membrane to trafficking through intracellular organelles over the duration of 24 h.
Figure 1.
Distribution of [3H]DNA/bPEI polyplexes after pulse-chase in cells and cell supernatant. HeLa cells (2 × 104) were pulsed with [3H]DNA/bPEI polyplexes for 4 h and then chased (black arrow) in complete media for an additional 2, 4, 8, or 20 h. Media and washes collected during the pulse period was termed “pulse supernatant” (and similarly with the “chase supernatant”, but with media and washes collected during the chase period). CellScrub and corresponding washes collected was termed as “surface-associated” polyplexes, while trypsinized cells was termed as “internalized” polyplexes. The radioactivity counts of the pulse supernatant, chase supernatant, surface-associated, and internalized samples were summed to calculate the total radioactivity. Data are presented as mean ± S.D., n = 3.
3.2. Validation of marker enzyme assays, optimization of cell breakage, and optimization of preparation of samples for scintillation counting
Cellular fractionation studies have been used to quantitatively assess the intracellular distribution of a number of polymer/liposome conjugates10,27,30. As an initial step, marker enzyme assays were validated with cell lysate and methods for efficient cell lysis were optimized in order to evaluate the organelle distribution after fractionation and maintain intact organelles during fractionation, respectively. Various marker enzyme assays were assessed for suitable limits-of-detection (LOD) using crude cell lysate. A linear relationship was found between indicator release and amount of cell lysate in assays for alkaline phosphatase (plasma membrane marker) (Supplemental Figure 1a), hexosaminidase A (lysosomal marker) (Supplemental Figure 1b), and succinate dehydrogenase (mitochondrial marker) (Supplemental Figure 1c). Purified lactate dehydrogenase was used as a standard for lactate dehydrogenase activity (cytosolic marker) (Supplemental Figure 1d). The LOD, defined as three standard deviations from the blank33, was 0.34, 0.33, 3.9, and 0.35 μg protein in assays for alkaline phosphatase, hexosaminidase A, succinate dehydrogenase, and lactate dehydrogenase activity, respectively. An assay for 5′-nucleotidase, another commonly marker for plasma membrane, was also assessed, but the LOD was too high for use in fractionation experiments (data not shown).
Next, the minimum number of 25-gauge needle passes necessary to efficiently release organelles from whole cell suspensions was determined. A concentrated cell suspension was passed through a 25-guage needle for 0–30 passes followed by analysis of the supernatant cytosolic leakage, as determined by lactate dehydrogenase release (Supplemental Figure 2). A cell suspension treated with 0.1% (v/v) Triton X-100 was used as a reference for 100% cell breakage. Cell breakage through a 25-gauge needle was optimized at ~20 passes, which resulted in 92.1% release of lactate dehydrogenase. Attempts at gentle cell breakage using other traditional methods, such as a Dounce or Potter-Elvehjem homogenizer, yielded irreproducible and insufficient breakage (data not shown), as well as significant loss of cell lysate.
Furthermore, initial studies to determine the radioactivity in samples revealed that polyplex packaging influenced measurement readings. Therefore, poly-L-lysine (PLL) or bPEI polyplexes with [3H]DNA were treated with a final concentration of either 0.5 g/L calf thymus DNA, 0.5 M NaOH, or 5X trypsin to restore radioactivity activity (Supplemental Figure 3). Left untreated, polyplexes showed 6–12% decrease in radioactivity compared to DNA alone. Treatment with excess calf thymus DNA or trypsin did not restore measurements for both polymers; only treatment with excess base provided near 100% of the radioactivity of uncomplexed DNA. Therefore, samples were diluted with equivolume of excess base prior to radioactivity measurements.
3.3. Cytosolic and vesicular distribution of PEI polyplexes
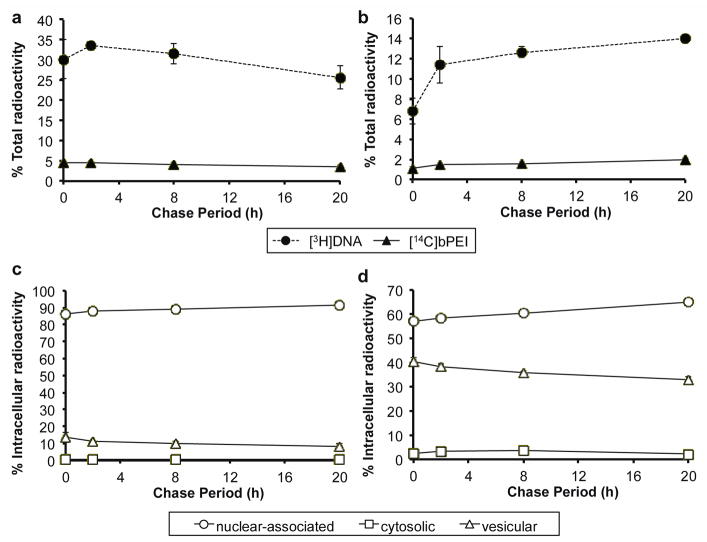
Next, a simple fractionation method was used to separate nuclear, cytosolic, and vesicular components to determine intracellular polyplex distribution in these major compartments over time. For sensitive tracking of both polymer and DNA, each component was radiolabeled, used to form polyplexes, applied to cells, and then detected in isolated organelle fractions. The partial acetylation of bPEI slightly affected polyplex condensation; complete DNA retardation was observed at N/P 3 instead of N/P 2 in gel electrophoresis assays (Supplemental Figure 4). Cells were pulsed with [14C]bPEI/[3H]DNA polyplexes for 4 h, rinsed, replaced with fresh media, and then fractionated into nuclear, cytosolic, and vesicular components after a 0, 2, 8, or 20 h chase period. The vesicular fraction contained membrane-associated organelles, such as the plasma membrane, endosomes, lysosomes, and mitochondria (data not shown). In general, the percentage of cell-associated [3H]DNA decreased slightly over the chase period, from 30.2% to 25.6% (Figure 2a). This trend was also observed in Figure 1 by summing the percent [3H]DNA in surface-associated and internalized samples. The percentage of cell-associated [14C]bPEI also decreased slightly, from 4.5% to 3.4%. Meanwhile, a gradual increase in [3H]DNA and [14C]bPEI was observed in the chase media (Figure 2b). Of the total radioactivity found in the cell-associated fractions, 86.2–91.5% of [3H]DNA (Figure 2c) and 57.9–65.0% of [14C]bPEI (Figure 2d) were present in the nuclear fraction; however, since the nuclear fraction also contained up to 10% unbroken cells, these values may be greater than the actual amount of nuclear-associated material. [3H]DNA and [14C]bPEI were present in the cytosolic fraction at low concentrations (less than 10% of the post-nuclear fractions). The percentage of [3H]DNA and [14C]bPEI in the cytosolic fraction peaked after 8 h chase to 0.79% and 3.71%, respectively, with effective N/P ratios of 3.1–4.2 throughout the chase period. The percentage of [3H]DNA and [14C]bPEI in the vesicular fraction was highest at the beginning of the chase period (0 h) at 13.3% and 40.3%, respectively, and then gradually decreased over the 20 h chase period. The effective N/P ratios in the vesicular fraction was 2.4–2.8 throughout the chase period. In contrast, the percentage of [3H]DNA and [14C]bPEI in the nuclear fraction increased over time to reach 91.5% and 65.0%, respectively, after the 20 h chase period, with effective N/P ratios ranging from 0.45 to 0.53. These results indicate that most of the polymer and DNA remain in membrane-associated organelle fractions (nuclear, vesicular) while very little material is in the soluble cytosolic fraction. These results are consistent with polyplexes that remain condensed in cytosolic and vesicular fractions, but disassemble prior to reaching the nuclear fraction.
Figure 2.
Distribution of [3H]DNA/[14C]bPEI polyplexes after pulse-chase after cytosolic and vesicular fractionation. HeLa cells (5 × 106) were pulsed with [3H]DNA/[14C]bPEI polyplexes for 4 h and then chased in complete media for an additional 2, 4, 8, or 20 h. All media, washes, and cells were collected to calculate the total radioactivity count in the sample. The percent of total radioactivity of [3H]DNA and [14C]bPEI measured in (a) cell-associated fractions and (b) chase supernatant. CellScrub and corresponding washes collected was termed as “surface-associated” polyplexes, while media and washes collected during the chase period was termed “chase supernatant”. The radioactivity counts of all samples (media, washes, cells) were summed to calculate the total radioactivity. The nuclear-associated, cytosolic, and vesicular distribution of (c) [3H]DNA and (d) [14C]bPEI as a percent of the radioactivity found in intracellular fractions (nuclear, cytosolic, vesicular). Data are presented as mean ± S.D., n = 3.
3.4 Differential centrifugation after treatment with PEI polyplexes
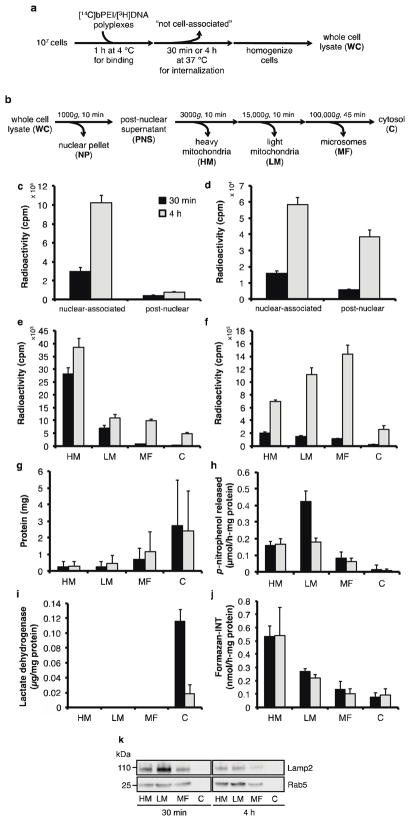
Crude separation into nuclear, cytosolic, and vesicular components only allowed a superficial understanding of polyplex distribution. Therefore, more thorough fractionation procedures were explored to further determine polyplex distribution within different organelle compartments in more detail. Differential centrifugation, in which organelles are separated on the basis of mass, is commonly used to assess the relative distribution of materials in intracellular compartments25,27,34,35. Cells were treated with dual-labeled polyplexes for 1 h at 4 °C to allow for polyplex binding to the cell surface but with minimal internalization (Figure 3a). Cells were then transferred to 37 °C for 30 min or 4 h to allow for polyplex internalization. We selected these times to investigate trafficking at an earlier time point (30 min) and at the same time point evaluated immediately after the pulse (4 h) in the aforementioned pulse-chase experiments. Afterwards, the cells were homogenized and fractionated according to the differential centrifugation schematic described in Figure 3b. After 30 min, the total percentage of [3H]DNA and [14C]bPEI associated with the cells was 15.5% and 1.2%, respectively. After 4 h, the percentage increased to 48.0% and 5.6%, respectively. After both 30 min and 4 h, a majority of [3H]DNA and [14C]bPEI in cell-associated fractions was found in the nuclear fraction (for [3H]DNA, 87.4% after 30 min, 93.0% after 4 h; for [14C]bPEI, 73.2% after 30 min, 60.3% after 4 h) (Figure 3c–d). Of the post-nuclear fractions, [3H]DNA was found mostly in the heavy mitochondrial fraction (HM) at 30 min (77.4% of total [3H]DNA found in post-nuclear fractions) (Figure 3e), and then distributed slightly throughout the lighter fractions by 4 h. In contrast, [14C]bPEI was found distributed throughout all fractions (Figure 3f), with more polymer found in the HM (40.6% of total [14C]bPEI found in post-nuclear fractions) and light mitochondrial fraction (LM) (30.9%). By 4 h, [14C]bPEI distributed to lighter fractions, with 31.9% found in the LM and 40.9% found in the microsomal fraction (MF). After 30 min, the effective N/P ratios were 0.34, 0.46, 1.4, 9.8, and 4.5 for NP, HM, LM, MF, and C fractions, respectively; after 4 h, the effective N/P ratios were 0.41, 1.3, 7.3, 10.3, and 3.9. The fold-increase of [3H]DNA from 30 min to 4 h was 3.4, 1.4, 1.5, 12.9, and 12.7 for NP, HM, LM, MF, and C, respectively, while the fold-increase of [14C]bPEI was 3.7, 3.5, 7.4, 12.5, and 10.0. In general, uptake of [3H]DNA and [14C]bPEI was significantly greater after 4 h than after 30 min. Interestingly, the fold-increase of [14C]bPEI cellular association was slightly greater than that of [3H]DNA (4.7 fold-increase for [14C]bPEI vs. 3.1 fold-increase for [3H]DNA) (Supplemental Figure 5). Polymer accumulation in the HM and LM fractions was also greater than that of DNA. Most of the [14C]bPEI did not become cell-associated (whole cell fraction, or WC) even though 48.0% of the [3H]DNA was found in the WC fraction by 4 h.
Figure 3.
Intracellular distribution of [3H]DNA/[14C]bPEI polyplexes in treated cells fractionated using differential centrifugation. (a) HeLa cells (107) were incubated with [3H]DNA/[14C]bPEI polyplexes for 1 h at 4 °C to allow for binding and then at 37 °C for 30 min (black bars) or 4 h (grey bars) to allow for internalization prior to fractionation. (b) Schematic of differential centrifugation method used to separate organelle populations. Radioactivity counts of (c) [3H]DNA and (d) [14C]bPEI measured in cell-associated fractions. Radioactivity counts of (e) [3H]DNA and (f) [14C]bPEI in post-nuclear fractions. Post-nuclear fractions were also measured for (g) protein content, (h) alkaline phosphatase (plasma membrane), (i) lactate dehydrogenase (cytosol), and (j) succinate dehydrogenase (mitochondria) activity. Data are presented as mean ± S.D., n = 3. (k) Immunoblotting for LAMP2 (lysosomes) and Rab5 (endosomes) in post-nuclear fractions. 13 μg protein was loaded into each lane, and staining was visualized using HRP-conjugated secondary antibody and a chemiluminescent substrate.
To verify organelle distribution, post-nuclear fractions were analyzed for total protein content and marker enzyme activity. Most of the protein was found in the cytosolic fraction (C) (Figure 3g). Significant overlap of marker enzyme activity between the post-nuclear fractions was observed. Alkaline phosphatase activity (plasma membrane) was found distributed mostly in the HM and LM fractions (Figure 3h). Lactate dehydrogenase activity (cytosol) was found only in the C fraction (Figure 3i), while succinate dehydrogenase activity (mitochondria) was found mostly in the HM and LM fractions (Figure 3j). Endosomal and lysosomal distribution was assessed in fractions by immunoblotting for Rab5, a small GTPase required for early endosome fusion36, and LAMP2, a lysosomal-associated membrane protein37, respectively (Figure 3k). Both proteins were found in all post-nuclear fractions except for the C fraction, with slightly increased distribution in the HM and LM fractions. Due to the incomplete separation of organelles, differential centrifugation is not an effective method for quantification of intracellular polyplex distribution. Therefore, other subcellular fractionation methods were explored to confirm these findings.
3.5. Density-gradient centrifugation after treatment with PEI polyplexes
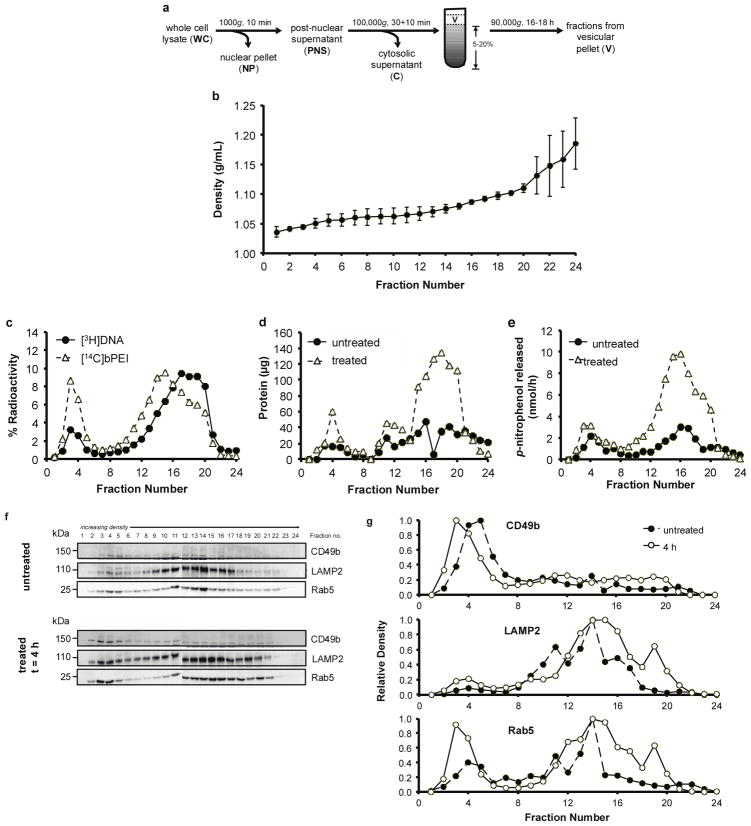
An alternative fractionation method, density-gradient centrifugation, in which organelles are separated by buoyant density, was evaluated for improved organelle separation. Density-gradient centrifugation using iodixanol as a density medium previously showed improved separation between various organelles31. Prior to density-gradient centrifugation, the cytosol was initially separated from the vesicular compartments (Figure 4a). To further investigate the polyplex distribution in the vesicular fraction, a 5–20% continuous iodixanol gradient was used to separate plasma membrane, endosomes, and lysosomes after cells were pulsed with polyplexes for 4 h, and then chased for either 0 or 20 h. The density of the control gradient (no cells) was linear over 1.04–1.19 g/mL (Figure 4b). As seen previously, more DNA than polymer was cell-associated; 27.5–33.6% of the [3H]DNA and 3.2–4.3% of the [14C]bPEI of the total radioactivity was detected in the whole cell lysate at 4 h (Supplemental Figure 6a–b). After the 20 h chase period, up to 22.2% of the [3H]DNA and 2.2% of the [14C]bPEI of the total radioactivity was detected in the chase media, indicating that about half of cell-associated materials (both DNA and polymer) is released back in the media over the 20 h chase period. Again, of the whole cell lysate, 92.9–93.4% of the [3H]DNA and 73.6–74.7% of the [14C]bPEI was found in the nuclear fraction (Supplemental Figure 6c–d). Overall, a bimodal distribution of both DNA and polymer was detected in the vesicular fraction as a function of density. The first peak, found in the less dense portion of the gradient (fractions 2–5) (Figure 4c), corresponds to fractions enriched in plasma membrane and endosomes (Figure 4f–g). The second peak (fractions 11–21) was offset between DNA and polymer; the intensities of each component peaked in different fractions (fraction 17 for [3H]DNA vs. fraction 15 for [14C]bPEI) (Figure 4c). A tailed-distribution was seen in both components; for [3H]DNA, the tail was in the less dense fractions (fraction 11–15), while for [14C]bPEI, the tail was in the denser fractions (fraction 17–21). These results were reproducibly observed in multiple experiments; similar trends were also observed after a 20 h chase period (Supplemental Figure 7).
Figure 4.
Distribution of [3H]DNA/[14C]bPEI polyplexes after 4 h pulse in cells fractionated using a 5–20% continuous iodixanol density gradient. (a) Schematic of density-gradient centrifugation method used to separate organelle populations. HeLa cells (2 × 107) were pulsed with [3H]DNA/[14C]bPEI polyplexes for 4 h, and then prepared for fractionation. A 5–20% continuous iodixanol gradient was used to separate vesicular organelles. (b) Density of control gradients. Data are presented as mean ± S.D., n = 3. (c) The percent radioactivity measured in fractions from the 5–20% gradient. 100% radioactivity is equal to the sum of the radioactivity found in all 24 fractions collected from the 5–20% gradient. (d) Total protein was measured in untreated and treated gradient fractions. (e) Hexosaminidase A (lysosome) activity was also measured in untreated and treated gradient fractions. (f) An equivolume (250 μL) of gradient fractions from untreated and treated samples was precipitated, concentrated, and probed for CD49b (plasma membrane), LAMP2 (lysosome), and Rab5 (endosome). (g) The optical density of each band was measured using ImageJ. Each time point presented is representative of duplicate experiments.
Marker enzyme assays and immunoblotting for various organelles were completed to determine the organelle distribution in the iodixanol gradient. At both time points, protein was distributed throughout the gradient in a bimodal manner (Figure 4d, Supplemental Figure 7b). Significantly higher protein amounts were found in the untreated samples after the 20 h chase period due to extensive cytotoxicity from polyplex treatment (Supplemental Figure 7b). Lysosomes (hexosaminidase A activity) were found mostly in the denser portion of the gradient (fractions 12–20) (Figure 4e, Supplemental Figure 7c). Immunoblotting for CD49b (plasma membrane), LAMP2 (lysosomes), and Rab5 (early endosomes) were also completed to confirm assay results (Figure 4f–g, Supplemental Figure 7d–e). Alkaline phosphatase activity was undetectable in fractions (data not shown). A slight difference in Rab5 distribution was observed with treated vs. untreated cells, with a slight shift of the protein to the lesser dense portion of the gradient after polyplex treatment. Most of the DNA and some polymer was also found in denser lysosomal compartments or compartments that were denser than lysosomes. Mitochondria have been found to distribute to denser portions of iodixanol gradients38; however, succinate dehydrogenase activity was undetectable in the fractions, possibly due to lack of assay sensitivity. These results also revealed that polyplex treatment affected organelle buoyancy and shifted organelle distribution in density gradients. Addition of polyplexes produced less dense endosomal fractions, potentially due to osmotic swelling of polyplex-containing endosomes39. Treatment with cationic lipoplexes and polymers has also been shown to shift organelle populations30,34,40.
4. DISCUSSION
Currently, most synthetic gene carriers in the development stage are tested by delivery of a reporter gene such as luciferase or green fluorescent protein. However, the delivery pathway is complex and reporter gene readouts only provide information about overall delivery efficiencies, leaving the details of specific steps to transfection hidden in the black box of the cell. The goal of this presented work is to improve our quantitative understanding of the intracellular trafficking of both the cargo DNA as well as the gene delivery vehicle itself. To accomplish this, we used radiolabeling and cellular fractionation strategies to track bPEI and the cargo plasmid DNA in extracellular and intracellular environments in cultured cells. Our main findings were that (1) overall cellular uptake of polymer was very low compared to the uptake of DNA; (2) nuclear association of intracellular polymer and DNA was high compared to other organelles; and (3) polymer and DNA traffic differently in intracellular vesicles.
The polycation PEI was radiolabeled by partial acetylation to gain quantitative insight into polymer cellular association and uptake, as well as intracellular distribution. Radiolabeling only slightly affected packaging ability, shifting full complexation of plasmid DNA to N/P 3 from N/P 2 (Supplemental Figure 4). Alternative radiolabeling strategies, such as polymerizing [14C]aziridine to synthesize [14C]bPEI, leaves amine density unaffected compared to unlabeled PEI but would likely generate polymers with different polydispersity and molecular weights compared to commercially available PEI, which is one of the most frequently used polycations for gene transfer. In our studies, HeLa cells were treated at an N/P ratio of 5, which was optimal for transfection efficiency (data not shown), although the actual amount of polymer associated with DNA at N/P 5 was not determined. However, several groups have studied the complexation of 25 kD bPEI with plasmid DNA, and showed that at N/P 6, <50% of the bPEI is associated with DNA41–43. Specifically, Clamme et al. estimated by fluorescence correlation spectroscopy that PEI is associated with DNA at N/P ~1 when formulated at N/P 6, and Boeckle et al. estimates this value to be ~2.8 after polyplex purification by size exclusion chromatography41,42. Surprisingly, only 1–6% of the applied polymer became associated with cells although 30–50% of the applied DNA was cell-associated. This translates to an effective N/P ratio of 0.3–0.6, which is much lower than the expected N/P 1–3 of fully condensed polyplexes. This phenomenon was seen reproducibly throughout multiple experiments (Figure 2, Supplemental Figures 5 and 6). Schaffer and Lauffenburger also noted low uptake of polyplexes by monitoring radiolabeled polycation, measuring that >90% of [125I]polylysine/DNA complexes were washed off the cell surface44; however, an effective N/P ratio was not calculated since the DNA was unlabeled. Despite an overall low uptake of polymer in our studies, the effective N/P ratios in the cytosolic and vesicular fractions was greater than 2 (Figures 2 and 3), indicating that intracellular polymer and DNA remain associated except at the nucleus. There are two possible explanations for such low overall cellular association of polymer: (1) rapid exocytosis of displaced polymer occurs after cellular uptake, or (2) polyplexes are partially unpackaged before cell uptake.
In order to gain insight into the first possibility, we analyzed radioactivity found in the collection of media from the chase periods in pulse-experiments. We found that the amount of polymer and DNA that was cell-associated after longer chase periods (i.e. 20 hours) was half that after shorter chase periods (i.e. 2 hours) (Figure 1, Figure 2, Supplemental Figure 6a–b). This indicates that polymer is released back into the media from the cell with time, but the release does not appear to be preferential compared with DNA. Although polyplex exocytosis has received little attention, previous studies have indicated that up to 65% of internalized nanoparticles may be exocytosed fairly rapidly (within 30 min) depending on the nanoparticle concentration gradient across the cell membrane45,46. In contrast, Seib et al. showed that bPEI alone did not exocytose significantly within 60 min in B16f10 melanoma cells47. Therefore, further studies to discern the role of polyplex exocytosis in cellular uptake should be pursued.
The second possibility is that polyplexes partially unpackage before cell uptake. Extracellular components, such as negatively-charged glycosaminoglycans (GAGs), have been shown to influence polyplex uptake and unpackaging by causing premature extracellular polyplex dissociation via electrostatic competition with GAGs41,43,48–51. However, previous studies using Förster resonance energy transfer (FRET) in which both the polymer carrier and cargo nucleic acid were labeled52,53 or the nucleic acid was dual-labeled54–56 showed that polyplexes remain intact, at least to some extent, in the cell. Furthermore, in our studies, a high N/P was also associated with the DNA in lighter and less dense organelles (Figures 3 and 4), indicating that the polymer and DNA separate during intracellular trafficking. Schubert et al. studied the complexation of plasmid DNA with linear PEI and found that “primary complexes” of PEI and DNA at N/P ~0.7 are first formed, which then merge into aggregates as more polymer is added57. These primary complexes are small in size (~30 nm) and can be internalized by cells. Clamme et al. also reported that in bPEI polyplexes formed at N/P 6, ~86% of the polymer existed freely in solution, and that these polyplexes were poorly compacted but can still transfect cells42. Therefore, it is possible that despite aggregate formation at N/P 5, the actual particles internalized by the cell are the primary complexes.
In addition to quantifying cellular uptake and association, intracellular polyplex nuclear distribution was measured by subcellular fractionation methods. Interestingly, most of the [3H]DNA and [14C]bPEI were found in the crude nuclear fraction, both at short times (30 min) (Figure 2c–d) and longer times (20 h) (Supplemental Figure 6c–d). The effective N/P ratio in the nuclear fraction remained less than 1 throughout multiple studies (Figures 2 and 3, Supplemental Figure 6c–d), indicating that the polymer dissociated from the DNA prior to reaching the nucleus. The percentage of cell-associated [3H]DNA in the nuclear fractions increased with time, whereas the percentage of cell-associated [14C]bPEI in these fractions decreased with time. The increase of [3H]DNA in this fraction likely also includes accumulation of degraded plasmid in the nucleus, since DNA oligonucleotides (ODNs) undergo active nuclear import58. A fractionation study by Eboue et al. also found that up to 69% of PEI/[3H]ODN complexes were found after 2.5 h in a purified nuclear fraction, indicating that a large percentage of polyplexes are nuclear-associated11. These results are in agreement with the large amount of [3H]DNA we detected in the nuclear fraction (Figure 2c–d, Figure 3c–d, Supplemental Figure 6c–d).
Incomplete separation of cytoskeletal filaments from nuclei during fractionation has been previously reported59. Cytoskeletal elements have been shown to mediate polyplex transfer to the perinuclear region60,61. Bieber et al. also showed that, by fluorescence imaging, many PEI polyplexes locate in the perinuclear space rather than the cytosol, and thus proposed that nuclear translocation remained a dominant barrier to efficient gene delivery39. Therefore, if cytoskeletal filaments separate with the nuclear fraction, this may explain why, in our studies, the cytosolic fraction contained such a small percentage of material despite the evidence for cytosolic distribution of PEI complexes15. It is difficult to discriminate polyplexes that are associated with the nuclear membrane from those that are intranuclear; Cohen et al. determined that a large amount of complexes still remained on the outer surface of the nuclear membrane using confocal microscopy despite treatment with excess cationic polymers and restriction enzymes19. Thus, the high percentage of intracellular DNA and polymer detected in our nuclear fractions does not necessarily indicate efficient delivery of intact plasmid DNA into the nucleus.
Despite a large fraction of [3H]DNA and [14C]bPEI in the nuclear fraction, differential distribution of each component was seen in post-nuclear organelles (Figures 3 and 4). In general, DNA was detected mostly in the heavier and denser fractions, which contained mostly lysosomes and mitochondria, while polymer was detected in mostly lighter and less dense fractions, which contained mostly plasma membrane, endosomes, and lysosomes. These findings may help explain the discrepancies between polymer and pDNA subcellular localization seen in other studies; in particular, ratiometric fluorescence studies demonstrated that the pH of compartments containing labeled DNA was higher than that of PEI62 and histidylated polylysine63. Reports have also shown that lysosomal pH did not increase with labeled PEI64 and that PEI in LAMP1-positive compartments also did not have increased pH65. These results may indicate an overall difference in the bulk polymer and DNA intracellular distribution, in which DNA is localized to mostly non-acidic organelles while polymer is mostly localized to acidified organelles. DNA was also seen split between two populations of denser organelles in a sucrose gradient by Laurent et al.9; they hypothesized that the non-lysosomal distribution of DNA in the denser regions of the gradient contained complexed plasmid DNA and that these structures may be phagosomes resulting from phagocytosis, a process that has been observed in HeLa cells66. The observation that polymer and DNA distribution remained largely unchanged over time indicates that polymer and/or polyplexes may be associating with vesicle-membrane lipids and traveling with these lipids during endocytosis. Since the exchange and transfer of materials through the endosomal/lysosomal pathway has been proposed to go through vesicle “kissing” and/or fusion67, free cationic polymer may be traveling with the membrane lipids while the more neutrally-charged and water-soluble polyplexes (DNA-associated) are trafficked with the soluble contents of the vesicles. This may also help explain why, in our studies, the overall effective N/P ratio in cell-associated fractions was less than 1 while fractions containing mostly endocytic organelles (endosomes, lysosomes) had effective N/P ratios of ~7–10 by 4 h (Figure 3). These results may also indicate that some polyplex unpackaging already occurs in the vesicular fractions, which was previously observed using FRET by Leong and coworkers53. In addition, poly-D-lysine, which is not degradable, exhibited similar unchanged subcellular distribution in denser organelles up to 14 h after injection into rats9. Similar trends were seen with [35S]DNA complexed with bPEI68. When [125I]bPEI alone was injected into rats, the polymer remained mostly in the lysosomal fraction after 4 and 18 h69. However, the extent of studies investigating polymer interaction with lipid membranes is focused on pore formation and membrane destabilization with non-biological lipid membranes70,71.
In summary, we demonstrate the use of subcellular fractionation methods to quantitatively assess both polymer and DNA in intracellular compartments. By radiolabeling both the synthetic carrier and the cargo DNA, we were able to quantify the amount of each component in the media, cell-associated fractions, as well as various intracellular organelles, such as the plasma membrane, nuclei, cytosol, endosomes, lysosomes, and mitochondria. These studies described general method development for the quantitative analysis of polyplex intracellular distribution, and will be applied for studying the effect of various chemical moieties on polymeric gene carriers.
Supplementary Material
Acknowledgments
This work is supported by the Center for the Intracellular Delivery of Biologics through the Washington State Life Sciences Discovery Fund Grant No. 2496490 and NIH 1R01NS064404. JS is supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-0718124 and the Howard Hughes Medical Institute/UW Molecular Medicine Graduate Student Scholarship. BC was supported by a Mary Gates Undergraduate Research Fellowship. We gratefully thank Dr. Drew Sellers for technical expertise and Dr. J. Paul Luzio (Professor of Molecular Membrane Biology, University of Cambridge), Dr. Oliver Press (Professor of Medicine, University of Washington and Fred Hutchinson Cancer Research Center), Dr. Patrick Stayton (Professor of Bioengineering, University of Washington), David Chu, and Dr. Joan Schellinger for helpful discussion.
Footnotes
Supporting Information. Supplemental figures and methods are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 2.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci US A. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akita H, Enoto K, Masuda T, Mizuguchi H, Tani T, Harashima H. Particle tracking of intracellular trafficking of octaarginine-modified liposomes: a comparative study with adenovirus. Mol Ther. 2010;18:955–964. doi: 10.1038/mt.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 5.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Funhoff AM, van Nostrum CF, Koning GA, Schuurmans-Nieuwenbroek NME, Crommelin DJA, Hennink WE. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules. 2004;5:32–39. doi: 10.1021/bm034041+. [DOI] [PubMed] [Google Scholar]
- 7.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15:1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 8.Holmes A, Dohrman A, Ellison A, Goncz K, Gruenert D. Intracellular compartmentalization of DNA fragments in cultured airway epithelial cells mediated by cationic lipids. Pharm Res. 1999;16:1020–1025. doi: 10.1023/a:1018927531003. [DOI] [PubMed] [Google Scholar]
- 9.Laurent N, Wattiaux-De Coninck S, Mihaylova E, Leontieva E, Warnier-Pirotte MT, Wattiaux R, Jadot M. Uptake by rat liver and intracellular fate of plasmid DNA complexed with poly-L-lysine or poly-D-lysine. FEBS Lett. 1999;443:61–65. doi: 10.1016/s0014-5793(98)01677-9. [DOI] [PubMed] [Google Scholar]
- 10.Colin M, Maurice M, Trugnan G, Kornprobst M, Harbottle RP, Knight A, Cooper RG, Miller AD, Capeau J, Coutelle C, Brahimi-Horn MC. Cell delivery, intracellular trafficking and expression of an integrin-mediated gene transfer vector in tracheal epithelial cells. Gene Ther. 2000;7:139–152. doi: 10.1038/sj.gt.3301056. [DOI] [PubMed] [Google Scholar]
- 11.Eboue D, Auger R, Angiari C, Le Doan T, Tenu JP. Use of a simple fractionation method to evaluate binding, internalization and intracellular distribution of oligonucleotides in vascular smooth muscle cells. Arch Physiol Biochem. 2003;111:265–272. doi: 10.1076/apab.111.3.265.23465. [DOI] [PubMed] [Google Scholar]
- 12.Watson P, Jones AT, Stephens DJ. Intracellular trafficking pathways and drug delivery: fluorescence imaging of living and fixed cells. Adv Drug Deliv Rev. 2005;57:43–61. doi: 10.1016/j.addr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Hama S, Akita H, Ito R, Mizuguchi H, Hayakawa T, Harashima H. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol Ther. 2006;13:786–794. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 17.von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E, Ogris M. The internalization route resulting in successful gene expression depends on both cell line polyethylenimine polyplex type. Mol Ther. 2006;14:745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, De Smedt SC, Sanders NN, Braeckmans K. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol Ther. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen RN, van der Aa MAEM, Macaraeg N, Lee AP, Szoka FC. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J Control Release. 2009;135:166–174. doi: 10.1016/j.jconrel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glover DJ, Leyton DL, Moseley GW, Jans DA. The efficiency of nuclear plasmid DNA delivery is a critical determinant of transgene expression at the single cell level. J Gene Med. 2010;12:77–85. doi: 10.1002/jgm.1406. [DOI] [PubMed] [Google Scholar]
- 21.Hsu CYM, Hendzel M, Uluda3 H. Improved transfection efficiency of an aliphatic lipid substituted 2 kDa polyethylenimine is attributed to enhanced nuclear association and uptake in rat bone marrow stromal cell. J Gene Med. 2011;13:46–59. doi: 10.1002/jgm.1526. [DOI] [PubMed] [Google Scholar]
- 22.Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4:438–446. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- 23.Banks GA, Roselli RJ, Chen R, Giorgio TD. A model for the analysis of nonviral gene therapy. Gene Ther. 2003;10:1766–1775. doi: 10.1038/sj.gt.3302076. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Yockman JW, Kim SW, Kern SE. Intracellular kinetics of non-viral gene delivery using polyethylenimine carriers. Pharm Res. 2007;24:1079–1087. doi: 10.1007/s11095-006-9229-5. [DOI] [PubMed] [Google Scholar]
- 25.Zheng N, Tsai HN, Zhang X, Rosania GR. The subcellular distribution of small molecules: from pharmacokinetics to synthetic biology. Mol Pharm. 2011;8:1619–1628. doi: 10.1021/mp200092v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wendeler M, Sandhoff K. Hexosaminidase assays. Glycoconj J. 2009;26:945–952. doi: 10.1007/s10719-008-9137-5. [DOI] [PubMed] [Google Scholar]
- 27.Seib FP, Jones AT, Duncan R. Establishment of subcellular fractionation techniques to monitor the intracellular fate of polymer therapeutics I. Differential centrifugation fractionation B16F10 cells and use to study the intracellular fate of HPMA copolymer - doxorubicin. J Drug Target. 2006;14:375–390. doi: 10.1080/10611860600833955. [DOI] [PubMed] [Google Scholar]
- 28.von Harpe A, Petersen H, Li Y, Kissel T. Characterization of commercially available and synthesized polyethylenimines for gene delivery. Journal of Controlled Release. 2000;69:309–322. doi: 10.1016/s0168-3659(00)00317-5. [DOI] [PubMed] [Google Scholar]
- 29.Bartz R, Fan H, Zhang J, Innocent N, Cherrin C, Beck SC, Pei Y, Momose A, Jadhav V, Tellers DM, Meng F, Crocker LS, Sepp Lorenzino L, Barnett SF. Effective siRNA delivery and target mRNA degradation using an amphipathic peptide to facilitate pH-dependent endosomal escape. Biochem J. 2011;435:475–487. doi: 10.1042/BJ20101021. [DOI] [PubMed] [Google Scholar]
- 30.Manunta M, Izzo L, Duncan R, Jones AT. Establishment of subcellular fractionation techniques to monitor the intracellular fate of polymer therapeutics II. Identification of endosomal and lysosomal compartments in HepG2 cells combining single-step subcellular fractionation with fluorescent imaging. J Drug Target. 2007;15:37–50. doi: 10.1080/10611860601010330. [DOI] [PubMed] [Google Scholar]
- 31.Plonne D, Cartwright I, Linss W, Dargel R, Graham JM, Higgins JA. Separation of the intracellular secretory compartment of rat liver and isolated rat hepatocytes in a single step using self-generating gradients of iodixanol. Anal Biochem. 1999;276:88–96. doi: 10.1006/abio.1999.4311. [DOI] [PubMed] [Google Scholar]
- 32.L’Annunziata MF. Handbook of Radioactivity Analysis. 3. Academic Press; 2012. [Google Scholar]
- 33.Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29(Suppl 1):S49–52. [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson SCW, Pattrick NG, Lavignac N, Ferruti P, Duncan R. Intracellular fate of bioresponsive poly(amidoamine)s in vitro and in vivo. J Control Release. 2010;142:78–88. doi: 10.1016/j.jconrel.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 35.Duvvuri M, Feng W, Mathis A, Krise JP. A cell fractionation approach for the quantitative analysis of subcellular drug disposition. Pharm Res. 2004;21:26–32. doi: 10.1023/b:pham.0000012148.12516.3f. [DOI] [PubMed] [Google Scholar]
- 36.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 37.Eskelinen EL. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Graham JM. Purification of a crude mitochondrial fraction by density-gradient centrifugation. Curr Protoc Cell Biol. 2001;Chapter 3(Unit 3.4) doi: 10.1002/0471143030.cb0304s04. [DOI] [PubMed] [Google Scholar]
- 39.Bieber T, Meissner W, Kostin S, Niemann A, Elsasser HP. Intracellular route and transcriptional competence of polyethylenimine-DNA complexes. J Control Release. 2002;82:441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 40.Wattiaux R, Jadot M, Laurent N, Dubois F, Wattiaux-De Coninck S. Cationic lipids delay the transfer of plasmid DNA to lysosomes. Biochem Biophys Res Commun. 1996;227:448–454. doi: 10.1006/bbrc.1996.1527. [DOI] [PubMed] [Google Scholar]
- 41.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 42.Clamme JP, Azoulay J, Mély Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys J. 2003;84:1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue Y, Jin F, Deng R, Cai J, Chen Y, Lin MCM, Kung HF, Wu C. Revisit complexation between DNA and polyethylenimine - Effect of uncomplexed chains free in the solution mixture on gene transfection. J Control Release. 2011;155:67–76. doi: 10.1016/j.jconrel.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 44.Schaffer DV, Lauffenburger DA. Optimization of cell surface binding enhances efficiency and specificity of molecular conjugate gene delivery. J Biol Chem. 1998;273:28004–28009. doi: 10.1074/jbc.273.43.28004. [DOI] [PubMed] [Google Scholar]
- 45.Panyam J, Labhasetwar V. Dynamics of endocytosis and exocytosis of poly(D,L-lactide-co-glycolide) nanoparticles in vascular smooth muscle cells. Pharm Res. 2003;20:212–220. doi: 10.1023/a:1022219003551. [DOI] [PubMed] [Google Scholar]
- 46.Park JS, Han TH, Lee KY, Han SS, Hwang JJ, Moon DH, Kim SY, Cho YW. N-acetyl histidine-conjugated glycol chitosan self-assembled nanoparticles for intracytoplasmic delivery of drugs: endocytosis, exocytosis and drug release. J Control Release. 2006;115:37–45. doi: 10.1016/j.jconrel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Seib FP, Jones AT, Duncan R. Comparison of the endocytic properties of linear and branched PEIs, and cationic PAMAM dendrimers in B16f10 melanoma cells. J Control Release. 2007;117:291–300. doi: 10.1016/j.jconrel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 48.Dai Z, Gjetting T, Mattebjerg MA, Wu C, Andresen TL. Elucidating the interplay between DNA-condensing and free polycations in gene transfection through a mechanistic study of linear and branched PEI. Biomaterials. 2011;32:8626–8634. doi: 10.1016/j.biomaterials.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 49.Bonner DK, Zhao X, Buss H, Langer R, Hammond PT. Crosslinked linear polyethylenimine enhances delivery of DNA to the cytoplasm. J Control Release. 2012 doi: 10.1016/j.jconrel.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke RS, Pun SH. Extracellular barriers to in vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjug Chem. 2008;19:693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 51.Hanzlíková M, Ruponen M, Galli E, Raasmaja A, Aseyev V, Tenhu H, Urtti A, Yliperttula M. Mechanisms of polyethylenimine-mediated DNA delivery: free carrier helps to overcome the barrier of cell-surface glycosaminoglycans. J Gene Med. 2011;13:402–409. doi: 10.1002/jgm.1587. [DOI] [PubMed] [Google Scholar]
- 52.Ho YP, Chen HH, Leong KW, Wang TH. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J Control Release. 2006;116:83–89. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Itaka K, Harada A, Yamasaki Y, Nakamura K, Kawaguchi H, Kataoka K. In situ single cell observation by fluorescence resonance energy transfer reveals fast intra-cytoplasmic delivery and easy release of plasmid DNA complexed with linear polyethylenimine. J Gene Med. 2004;6:76–84. doi: 10.1002/jgm.470. [DOI] [PubMed] [Google Scholar]
- 55.Matsumoto Y, Itaka K, Yamasoba T, Kataoka K. Intranuclear fluorescence resonance energy transfer analysis of plasmid DNA decondensation from nonviral gene carriers. J Gene Med. 2009;11:615–623. doi: 10.1002/jgm.1338. [DOI] [PubMed] [Google Scholar]
- 56.Alabi CA, Love KT, Sahay G, Stutzman T, Young WT, Langer R, Anderson DG. FRET-labeled siRNA probes for tracking assembly and disassembly of siRNA nanocomplexes. ACS Nano. 2012;6:6133–6141. doi: 10.1021/nn3013838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schubert US, Perevyazko I, Pavlov G, Hoeppener S, Schubert S, Bauer M, Fischer D. Polyelectrolyte complexes of DNA and linear PEI: formation, composition and properties. Langmuir. 2012;28:16167–16176. doi: 10.1021/la303094b. [DOI] [PubMed] [Google Scholar]
- 58.Hartig R, Shoeman RL, Janetzko A, Grüb S, Traub P. Active nuclear import of single-stranded oligonucleotides and their complexes with non-karyophilic macromolecules. Biol Cell. 1998;90:407–426. [PubMed] [Google Scholar]
- 59.Staufenbiel M, Deppert W. Intermediate filament systems are collapsed onto the nuclear surface after isolation of nuclei from tissue culture cells. Exp Cell Res. 1982;138:207–214. doi: 10.1016/0014-4827(82)90107-0. [DOI] [PubMed] [Google Scholar]
- 60.de Bruin K, Ruthardt N, von Gersdorff K, Bausinger R, Wagner E, Ogris M, Bräuchle C. Cellular dynamics of EGF receptor-targeted synthetic viruses. Mol Ther. 2007;15:1297–1305. doi: 10.1038/sj.mt.6300176. [DOI] [PubMed] [Google Scholar]
- 61.Grosse S, Aron Y, Thévenot G, Monsigny M, Fajac I. Cytoskeletal involvement in the cellular trafficking of plasmid/PEI derivative complexes. J Control Release. 2007;122:111–117. doi: 10.1016/j.jconrel.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 62.Akinc A, Langer R. Measuring the pH environment of DNA delivered using nonviral vectors: implications for lysosomal trafficking. Biotechnol Bioeng. 2002;78:503–508. doi: 10.1002/bit.20215. [DOI] [PubMed] [Google Scholar]
- 63.Gonçalves C, Pichon C, Guérin B, Midoux P. Intracellular processing and stability of DNA complexed with histidylated polylysine conjugates. J Gene Med. 2002;4:271–281. doi: 10.1002/jgm.277. [DOI] [PubMed] [Google Scholar]
- 64.Forrest ML, Pack DW. On the kinetics of polyplex endocytic trafficking: implications for gene delivery vector design. Mol Ther. 2002;6:57–66. doi: 10.1006/mthe.2002.0631. [DOI] [PubMed] [Google Scholar]
- 65.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. The possible “proton sponge” effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol Ther. 2013;21:149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kopatz I, Remy JS, Behr JP. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 67.Luzio JP, Parkinson MDJ, Gray SR, Bright NA. The delivery of endocytosed cargo to lysosomes. Biochem Soc Trans. 2009;37:1019–1021. doi: 10.1042/BST0371019. [DOI] [PubMed] [Google Scholar]
- 68.Wattiaux R, Laurent N, Wattiaux-De Coninck S, Jadot M. Endosomes, lysosomes: their implication in gene transfer. Adv Drug Deliv Rev. 2000;41:201–208. doi: 10.1016/s0169-409x(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 69.Lecocq M, Wattiaux-De Coninck S, Laurent N, Wattiaux R, Jadot M. Uptake and intracellular fate of polyethylenimine in vivo. Biochem Biophys Res Commun. 2000;278:414–418. doi: 10.1006/bbrc.2000.3809. [DOI] [PubMed] [Google Scholar]
- 70.Leroueil PR, Hong S, Mecke A, Baker JR, Orr BG, Banaszak Holl MM. Nanoparticle interaction with biological membranes: does nanotechnology present a Janus face? Acc Chem Res. 2007;40:335–342. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.