Abstract
Isocitrate dehydrogenase-1 (IDH1) mutations have been discovered to be frequent and highly conserved in secondary glioblastoma (GBM) and lower grade gliomas. Although IDH1 mutations confer a unique genotype that has been associated with a favorable prognosis, the role of the mutated IDH1 enzyme and its metabolites in tumor initiation and maintenance remains unresolved. However, given that IDH1 mutations are homogenously expressed and are limited solely to tumor tissue, targeting this mutation could potentially yield novel treatment strategies for patients with GBM.
Keywords: Isocitrate Dehydrogenase, IDH1, glioma, mutation, genetics
Introduction
Based on their clinical presentation and molecular phenotype, GBMs can be subdivided into two groups: primary versus secondary. GBMs are considered to be primary when they develop de novo and are often found in older patients. Primary GBMs develop with a relatively rapid clinical history and are associated with EGFR overexpression as well as alterations in PTEN, CDKN2A, and MDM2. By contrast, secondary GBMs typically develop in younger patients and arise due to transformation from prior lower grade tumors. Although they possess similar histological and morphological features to their primary counterparts, secondary GBMs differ significantly on a molecular basis due to their association with genetic alterations including mutations in TP53, IDH1/2 and ATRX.
A genome-wide analysis of somatic mutations occurring in GBMs revealed highly-conserved and tumor-specific mutations in the active-site-encoding portion of the IDH1 gene.31 Importantly, IDH1 mutations have been shown to be expressed at high frequencies (~75%) in almost all glioma subtypes, except primary GBM, as well as other tumors but at a significantly lower incidence. Because mutations in IDH1 are homogenously expressed in all tumor cells—even single infiltrating cells20—it has been hypothesized that mutations in IDH1 might represent some of the earliest genetic events that could drive the malignant transformation of lower-grade tumors. This review will address the function of IDH1 and its mutant form, the clinical role of IDH1 in gliomas, how to screen for mutated IDH1, therapeutic approaches to targeting mutated IDH1, and a novel engineering application of IDH1 mutational data in the synthesis of nylon.
What is IDH?
Isocitrate dehydrogenase 1 (IDH1) catalyzes the oxidative decarboxylation of isocitrate to 2-oxoglutarate. The most widely-cited tumor-specific mutation of IDH1 consists of a missense mutation at amino acid 132 that replaces an active-site arginine residue with histidine; this somatic heterozygous mutation was first discovered in a genome-wide analysis of central nervous system tumors.31 More specifically, IDH1 mutations have been described in WHO Grade IV secondary GBMs, WHO Grade II diffuse astrocytomas, WHO Grade II oligodendrogliomas, WHO Grade III anaplastic astrocytomas, WHO Grade III anaplastic oligodendroglioma, and WHO Grade III anaplastic oligoastrocytoma.4,5,8,31 IDH mutations were subsequently observed in several other cancers and syndromes, including acute myeloid leukemia, preleukemic clonal malignancies, central and periosteal cartilaginous tumors, colorectal cancer, prostate carcinoma, adult supratentorial primary neuroectodermal tumor (PNET), paraganglioma, intrahepatic origin cholangiocarcinomas, Ollier disease, and Maffucci syndrome).1,2,4,14,16,19,23 IDH1 mutations are rarely found in primary GBM (4.9%) and not found in pilocytic astrocytomas, ependymomas, or medulloblastomas (Table 1).
Table 1.
| Tumor Type | Mutations (%) and median (range) | Number of mutations/total number of tumors |
|---|---|---|
| Diffuse Astroctyoma | 74% (0–88%) | 307/407 |
| Anaplastic Astrocytoma | 59% (0–78%) | 294/457 |
| Secondary GBM | 83% (50–100%) | 94/121 |
| Oligodendroglioma | 76% (67–84%) | 283/366 |
| Anaplastic Oligodendroglioma | 67% (49–94%) | 237/355 |
| Oligoastrocytoma | 80% (50–100%) | 153/196 |
| Anaplastic Oligoastrocytoma | 75% (63–94%) | 245/345 |
| Ganglioglioma | 37.5% | 3/8 |
| Anaplastic Ganglioglioma | 60% | 3/5 |
| Sporadic Paraganglioma | 2% | 1/66 |
| Primitive Neuroectodermal Tumor | 17% (0–33%) | 3/17 |
| Acute Myelogenous Leukemia | 1% (0–9%) | 21/389 |
IDH1 mutants and their impact on metabolism and tumor initiation
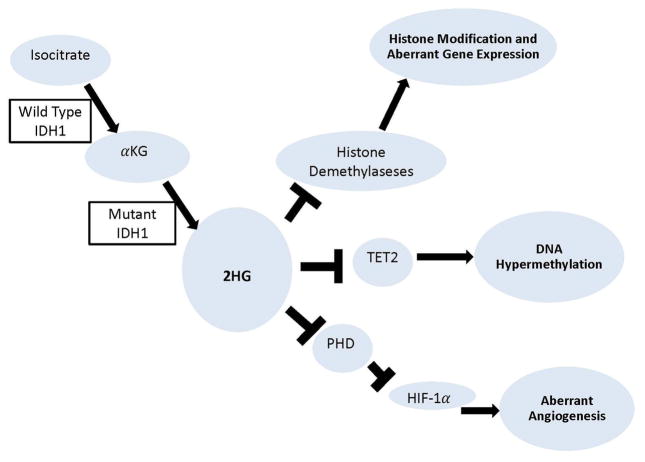
IDH1 is an enzyme located in the cytoplasm as well as in peroxisomes where it participates in lipid metabolism and glucose sensing. A number of potential hypotheses have been offered that implicate IDH1 mutations in malignant progression and oncogenesis. The altered IDH1 enzyme found in tumors is known to consist of a dimer between the wild-type and mutant proteins and thus possesses a function distinct from the normal enzyme. While normal IDH1 converts isocitrate into alpha-ketoglutarate (αKG), mutant IDH1 instead possesses the catalytic activity to convert αKG into 2-hydroxyglutarate (2HG).11 It has been suggested that 2HG may inhibit a variety of dioxygenases including PHD (prolyl hydroxylase), TET2, and histone demethylases,22 which in theory could trigger aberrant angiogenesis and aberrant gene expression (Figure 1). Mutations of IDH1 may also reduce the cellular level of glutathione synthase (GSH) by depleting NADPH and rendering the cells vulnerable to oxidative DNA damage, leading to the development of mutations in other genes.21 Moreover, IDH1 mutations have been proposed to facilitate “glycolytic flux”, where energy is produced predominately by aerobic glycolysis in the cytosol of cancer cells. During glioma progression, this allows the cancer cells to adopt these adaptive mechanisms in environmental conditions where nutrients and oxygen may otherwise be lacking.30
Figure 1.
Diagram of the molecular consequences of the IDH1 mutation.
Alternatively, 2HG produced by the mutant IDH1 expression is known to compete with αKG to inhibit degradation of HIF-1α, a transcription factor crucial to the cellular response to hypoxia. As a result, high levels of HIF-1α may promote the expression of VEGF-mediated angiogenesis, leading to greater tumor growth. Further implicating this pathway, mutant IDH1 is also known to HIF-1α inducible genes.32 However, a recent study showed that activation of EGLN, a HIF prolyl 4-hydroxylase, by the R enantiomer of 2HG (R-2HG), and subsequent downregulation of HIF-1α, contributes to the pathogenesis of IDH mutant gliomas. This data suggests that R-2HG quantitatively shifts the dose–response linking HIF-1 activation to hypoxia, leading to a blunted HIF-1α response for a given level of hypoxia. This controversial data suggests that modulation of the HIF-1α response over time, perhaps in conjunction with alterations in other enzymes affected by 2HG, results in epigenetic changes that are ultimately responsible for glioma transformation.
IDH1 as a biomarker
IDH1 mutations have been shown to be a prognostic indicator, but not predictive of response to therapy.12,15,26,28,29,31 It has been extensively shown that patients with mutated IDH1 have a better prognosis (5 year survival rate of 93%) than those without the mutation (5 year survival rate of 51%), in terms of overall and progression free survival.29 Even though patients with IDH1 mutation are generally younger, multivariate analysis has shown that IDH1 mutations are an independent prognostic factor after adjustment for age, grade, MGMT status, treatment, and genomic profile.26 The caveat of these studies arises from the comparison of genetically distinct tumors; gliomas that carry IDH1 mutations are biologically different and unique, and therefore comparing them with gliomas that do not carry the mutation can be misleading. This difference from other types of gliomas is supported by the fact that the IDH1 mutation is tightly associated with a distinct, homogenous DNA CpG island hypermethylated phenotype, resulting in reshaping of the epigenome.10,22,24 Recently, a pivotal genetic study in gliomas also supports the idea of refining the classification of malignant gliomas based on distinct genetic signatures.17
A few studies have evaluated the predictive value of IDH1 mutations in response to therapy. It has been shown that in patients with anaplastic oligodendroglial tumors treated with radiotherapy alone or radiotherapy with adjuvant PCV (procarbazine, lomustine, and vincristine), IDH1 mutations had no predictive value for outcome to PCV.29 In another study of patients with dedifferentiated low-grade astrocytomas progressive after radiotherapy, response to temozolomide (TMZ) did not differ between IDH1 mutant and wild-type tumors.12 However, in a recent study it was found that in secondary glioblasomas, IDH1 mutations, MGMT promoter methylation status (classically known to predict benefit from alkylating agents), and 1p19q codeletion were strongly correlated with increased overall survival. Three subgroups of different chemosensitivity were identified: patients with both IDH1 mutation and MGMT promoter methylation had the best response rate to TMZ (median progression-free survival of 13.4 months), patients with IDH1 mutation alone had the second best response rate to TMZ (10.2 months), and patients with none of these genetic alterations had the worst response rate to TMZ (6.1 months).28
Clinical screening for IDH1 mutations
There are several current methods that have proven useful for the diagnosis and biological evaluation of IDH1 mutation-bearing gliomas, including genotyping of the DNA extracted from the brain tumor specimen. It is a relatively straightforward method that many clinical molecular diagnostic laboratories use to perform IDH1 and IDH2 sequencing. The second method includes immunohistochemical analysis of the tumor specimen to detect IDH1 expression using specific monoclonal antibodies to target mutant IDH1. For these purposes, Imab-1, an anti-IDH1-R132H-specific antibody has been developed that reacts with mutated IDH1.7,8,20 Interestingly, in immunohistochemical studies using this antibody, every glioma cell in IDH1-positive samples stains positive for mutated IDH1, even along the infiltrating edge of the tumor into cortex.
Another modality for screening involves magnetic resonance spectrometry of the downstream metabolite of the mutant IDH1 pathway, 2HG.3,9,13 This noninvasive modality utilizes the presence of the L-enantiomer of 2HG and enables a detailed molecular characterization of gliomas bearing this mutation. This modality has been implicated in identifying 2HG “hot spots”, guiding biopsy procedures, assisting in diagnostic workup, and monitoring the response of targeted therapies. No current report exists of increased 2HG in blood, urine, or cerebrospinal fluid of glioma patients harboring IDH1 mutations.
Treatment strategies
Gain-of-function mutations in IDH1-associated glioma and the potential role of these mutations in the pathogenesis of malignant disease make IDH1 a promising, novel therapeutic target. The replacement of critical active-site arginine residues by alternative side chains offers an opportunity to develop selective inhibitors of mutant IDH1 that seek to limit its metabolic function. Certainly, a great deal of effort has been placed on the identification of small molecules that may directly inhibit mutant IDH1, which in theory would lead to the reduction of 2HG levels in gliomas, effectively modulating the impact of the metabolite on cancer-relevant processes. However, controversy remains regarding whether a therapeutically relevant impact can be achieved via mutant IDH1 inhibition alone.18 Additional strategies to reduce the production of 2HG in malignant cells have been suggested that focus on inhibition of enzymes that metabolize substrates up-stream of IDH1, including glutaminase and glutamate carboxypeptidase II (GCPII). Seltzer et al. targeted glutaminase, an enzyme upstream of the mutated IDH1 pathway that produces glutamate from glutamine, to determine if inhibiting this enzyme would perturb αKG homeostasis and yield an anti-tumor response.27 GCPII has the potential to reduce extracellular glutamate and represents an opportune target for treating cancers. GCPII is identical to a tumor marker, prostate-specific membrane antigen, and has drawn significant interest as a potential therapeutic target in cancers where excess glutamate is considered pathogenic. However because these enzymes also have normal functions in pathways of central metabolism, global inhibition may ultimately risk off-target toxicity in even normal, healthy tissues. In addition, given that the IDH1 mutation is tightly associated with a hypermethylated phenotype, DNA methyltransferase inhibitors and histone deacetylase inhibitors may also have therapeutic efficacy. Although these inhibitors have been studied in a phase 1/2 trial in a handful of patients in AML with limited efficacy6, they have yet to be pursued in glioma therapeutic targeting.
Neomorphic IDH mutant activity informs enzyme redesign
Generally, cancer can be thought of a selective biological process by which gain-of-function mutations, like those associated with IDH1, may actually lend a “competitive” advantage for tumor cells throughout their growth and development. Interestingly, knowledge of such neomorphic catalytic activities—as well as their associated functional active site alterations—may provide useful insight in the field of metabolic engineering, wherein the need for enzymes with novel catalytic activities is great. A recent advance toward this end is the use of human IDH1 mutational data to aid in the rapid design of enzymes that satisfy a significant industrial need in the large-scale production of adipic acid, a central substrate precursor in the synthesis of nylon.25 Studies such as these suggest that an intimate understanding of cancer-derived mutations may not only reveal underlying mechanisms of tumor pathogenesis, but may also have broad, far-reaching, applications in the fields of metabolic engineering and industrial enzyme redesign.
Conclusion
IDH1 is a key metabolic enzyme in the glycolytic pathway, in which mutations of this enzyme are consistent and frequent in low grade gliomas and secondary GBM. IDH1 mutations can currently be detected via immunohistochemistry, genetic analysis, and magnetic resonance spectrometry. Although IDH1 mutations denote a unique genotype with a favorable prognosis, the role of the mutated IDH1 enzyme and its metabolites in tumor initiation and maintenance remains largely unresolved. Since it is a driver mutation expressed early in tumor development and expressed in every tumor cell, the IDH1 mutation has the potential to be targeted and every tumor cell affected.
Acknowledgments
We would like to thank Tecca Wright for her assistance in the preparation of this manuscript.
Footnotes
Disclosure
The authors do not disclose any potential conflicts of interest.
References
- 1.Alderton GK. Genetics: IDH mosaicism in enchondromatosis syndromes. Nat Rev Cancer. 2012;12:6. doi: 10.1038/nrc3194. [DOI] [PubMed] [Google Scholar]
- 2.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 3.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4:116ra114. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 5.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 6.Borthakur G, Huang X, Kantarjian H, Faderl S, Ravandi F, Ferrajoli A, et al. Report of a phase 1/2 study of a combination of azacitidine and cytarabine in acute myelogenous leukemia and high-risk myelodysplastic syndromes. Leuk Lymphoma. 2010;51:73–78. doi: 10.3109/10428190903318329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capper D, Reuss D, Schittenhelm J, Hartmann C, Bremer J, Sahm F, et al. Mutation-specific IDH1 antibody differentiates oligodendrogliomas and oligoastrocytomas from other brain tumors with oligodendroglioma-like morphology. Acta Neuropathol. 2011;121:241–252. doi: 10.1007/s00401-010-0770-2. [DOI] [PubMed] [Google Scholar]
- 8.Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen BC, Smith AA, Zheng SC, Koestler DC, Houseman EA, Marsit CJ, et al. DNA Methylation, Isocitrate Dehydrogenase Mutation, and Survival in Glioma. Journal of the National Cancer Institute. 2011;103 doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubbink HJ, Taal W, van Marion R, Kros JM, van Heuvel I, Bromberg JE, et al. IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology. 2009;73:1792–1795. doi: 10.1212/WNL.0b013e3181c34ace. [DOI] [PubMed] [Google Scholar]
- 13.Elkhaled A, Jalbert LE, Phillips JJ, Yoshihara HA, Parvataneni R, Srinivasan R, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4:116ra115. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaal J, Burnichon N, Korpershoek E, Roncelin I, Bertherat J, Plouin PF, et al. Isocitrate dehydrogenase mutations are rare in pheochromocytomas and paragangliomas. J Clin Endocrinol Metab. 2010;95:1274–1278. doi: 10.1210/jc.2009-2170. [DOI] [PubMed] [Google Scholar]
- 15.Gravendeel LA, Kloosterhof NK, Bralten LB, van Marion R, Dubbink HJ, Dinjens W, et al. Segregation of non-p.R132H mutations in IDH1 in distinct molecular subtypes of glioma. Hum Mutat. 2010;31:E1186–1199. doi: 10.1002/humu.21201. [DOI] [PubMed] [Google Scholar]
- 16.Hayden JT, Fruhwald MC, Hasselblatt M, Ellison DW, Bailey S, Clifford SC. Frequent IDH1 mutations in supratentorial primitive neuroectodermal tumors (sPNET) of adults but not children. Cell Cycle. 2009;8:1806–1807. doi: 10.4161/cc.8.11.8594. [DOI] [PubMed] [Google Scholar]
- 17.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin G, Pirozzi CJ, Chen LH, Lopez GY, Duncan CG, Feng J, et al. Mutant IDH1 is required for IDH1 mutated tumor cell growth. Oncotarget. 2012;3:774–782. doi: 10.18632/oncotarget.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 20.Kato Y, Jin G, Kuan CT, McLendon RE, Yan H, Bigner DD. A monoclonal antibody IMab-1 specifically recognizes IDH1R132H, the most common glioma-derived mutation. Biochem Biophys Res Commun. 2009;390:547–551. doi: 10.1016/j.bbrc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32:1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 22.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitman ZJ, Choi BD, Spasojevic I, Bigner DD, Sampson JH, Yan H. Enzyme redesign guided by cancer-derived IDH1 mutations. Nat Chem Biol. 2012;8:887–889. doi: 10.1038/nchembio.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 27.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, et al. Inhibition of Glutaminase Preferentially Slows Growth of Glioma Cells with Mutant IDH1. Cancer Research. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103:269–273. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- 29.van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 30.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]