Abstract
Genes are transferred between bacteria in dental plaque by transduction, conjugation, and transformation. Membrane vesicles can also provide a mechanism for horizontal gene transfer. DNA transfer is considered bacterial sex, but the transfer is not parallel to processes that we associate with sex in higher organisms. Several examples of bacterial gene transfer in the oral cavity are given in this review. How frequently this occurs in dental plaque is not clear, but evidence suggests that it affects a number of the major genera present. It has been estimated that new sequences in genomes established through horizontal gene transfer can constitute up to 30% of bacterial genomes. Gene transfer can be both inter- and intrageneric, and it can also affect transient organisms. The transferred DNA can be integrated or recombined in the recipient's chromosome or remain as an extrachromosomal inheritable element. This can make dental plaque a reservoir for antimicrobial resistance genes. The ability to transfer DNA is important for bacteria, making them better adapted to the harsh environment of the human mouth, and promoting their survival, virulence, and pathogenicity.
Keywords: bacteria, bacterial sex, DNA transfer, dental plaque
In an excellent review on the origins and evolution of antibiotic resistance, Davies and Davies (1) recently proposed that the intestinal tract of animals and humans is a bordello for microorganisms since bacterial transfer of resistance genes occurs here ad libitum. Indeed, recent studies have found diverse antibiotic resistance genes in the human gut microbiome, indicating such transfer (2). Bacterial conjugation has long been regarded as the equivalent of sexual reproduction or mating in bacteria since an exchange of genetic material is involved. Also, sex pheromones that induce mating responses in bacteria have been detected (e.g. in commensal oral streptococci) (3). They may promote intergeneric gene acquisition among bacteria in oral biofilms. The close proximity of cells in the dense and constricted bacterial populations of multispecies dental biofilm facilitates plasmid dispersal through conjugation (4, 5). In biofilm-grown Streptococcus mutans, cells were transformed at rates 10- to 600-fold higher than in planktonic S. mutans cells (6). Furthermore, DNA release and transformation seem to be parts of the biofilm-related life cycle, and released DNA has the capacity to stabilize biofilm structure and architecture (7). The source for this genetic material is probably extracellular DNA (eDNA) derived from biofilm cells or transitory bacterial cells reaching the mouth (7). Clearly, genes are transferred between bacterial cells in dental plaque with enhanced efficiency, but does this phenomenon imply that dental plaque can be considered a bordello for plaque bacteria? This review focuses on bacterial sex in dental plaque.
Bacterial sex
Bacteria are predominantly asexual and reproduce by binary fission. A single cell hereby replicates its genome and divides into two identical daughter cells. Variations in this process usually occur by mutation or duplication of existing intragenomic information (8). However, the genetic information of bacteria can be expanded and modified through mechanisms that introduce DNA from external sources. Bacterial sex is defined as the inheritance of DNA from any source aside from a bacterium's one parent cell (8).
Most bacterial species consist of a huge population of strains, which genetically are not very similar. Therefore, a bacterial species can be described by its pan-genome, which includes the core genome containing the genes that are present in all strains of the species, a dispensable genome containing genes that are present in two or more strains, and genes that are unique to single strains (9). Bacterial sex can take place between strains to recombine dispensable and unique genes into a new ‘offspring.’ In contrast to sex among eukaryotes, sex in bacteria is unidirectional and does not involve gamete fusion or reproduction. The acquirement of DNA through bacterial sex causes increased diversity through the alteration of existing genes and the introduction of new DNA sequences. Streptococcus is an example of a genus that is very diverse, with high levels of gene gain and loss (i.e. with extreme levels of evolutionary plasticity) (10). This genetic diversity is the result of mainly horizontal gene transfer in biofilms occurring as conjugation, transduction, and transformation (discussed later in this review).
The formation of new genotypes by homologous recombination is commonly referred to as ‘bacterial sex’ because the result is similar to that of higher eukaryotes: production of offspring with a mixture of traits from each parent. Bacterial sex can also introduce a limited amount of new sequences to bacteria through horizontal gene transfer of mobile elements, and increasing the gene repertoire of the recipient may change the virulence of the bacteria (e.g. through establishment of pathogenicity islands, new metabolic pathways, and plasmids or transposons encoding antibiotic resistance). Actually, sex and virulence are intimately integrated in a wide variety of microbes (11). The majority of genes in bacterial genomes seem to have been acquired by horizontal gene transfer at some time during the evolutionary history of the lineage, whereas the diversity of gene repertoires in eukaryotes is mainly due to gene duplication and loss (12).
DNA transfer between bacterial cells
DNA can be transferred between bacterial cells mainly by transduction, conjugation, or transformation. Transduction is caused by bacterial viruses, conjugation by a bacterial sex pilus, and transformation through DNA uptake by naturally competent bacteria (13). After transfer, DNA can be physically recombined into the chromosome by various cytoplasmic proteins or, in the case of a plasmid, maintained as a separate molecule. A number of the bacterial genomes that have been sequenced contain genes acquired through horizontal gene transfer.
In transduction, bacteria-infecting viruses (bacteriophages) introduce their own or sometimes alien DNA into the new host's genome. Transduction requires specific receptors on the cell surface of the recipient bacterium. The amount of DNA that can be packed within a phage can be about 100 kb but varies greatly. The proteins of the bacteriophage protect its double-stranded DNA from destruction by host endonucleases. Bacteriophages can become dormant after integration in the host chromosome (temperate), or they can cause lysis of the host (lytic). Bacterial viruses, together with eukaryote-infecting viruses, may be the most common sources of genetic material on the planet (8).
Conjugation requires cell-to-cell contact. In conjugation, DNA is transferred between two metabolically active cells by self-transmissible and mobilizable plasmids called F (sex factor) plasmids, integrative conjugation elements (plasmids integrating into a chromosome producing high-frequency recombinant (Hfr) strains), or conjugative transposons (which encode proteins for their excision and transposition into recipient strains) (8). Conjugation can result in the exchange of homologous or heterologous stretches of DNA between the mating pair. Conjugative transposons occur frequently in Gram-positive bacteria but have also made their way to Gram-negative organisms because they are highly evolved for transfer among a broad host range. Their omnipresence makes them important players in the dissemination of a large variety of antibiotic resistance determinants (14). The high density of cells in biofilms increases the spread of plasmids by conjugation, and conjugation itself may stimulate biofilm formation (15).
Transformation is the process driven by cells that display competence, which is the ability to take up DNA from the environment. The development of competence involves a quorum-sensing process encoded by different com genes. Cells that are competent are able to bind free double-stranded DNA from the environment to cell surfaces, transfer one strand of the DNA across the cell membrane, and then integrate the DNA into the chromosome through recombination with its homologous counterpart. Many bacterial genera in the mouth are naturally competent at all times (e.g. Haemophilus, Campylobacter, and Neisseria), while others such as Streptococcus are competent only during specific physiological states (16). A likely source of free DNA in the dental plaque is the matrix, or slime layer, that surrounds bacteria in a biofilm. DNA released by dead lyzed bacteria can be found in the matrix, as well as DNA secreted by living members of the community, to provide structural support for the slime. Transformation by eDNA has been shown to mediate transfer of antibiotic resistance in an oral biofilm model (16).
Gene transfer among Gram-negative bacteria via membrane vesicles
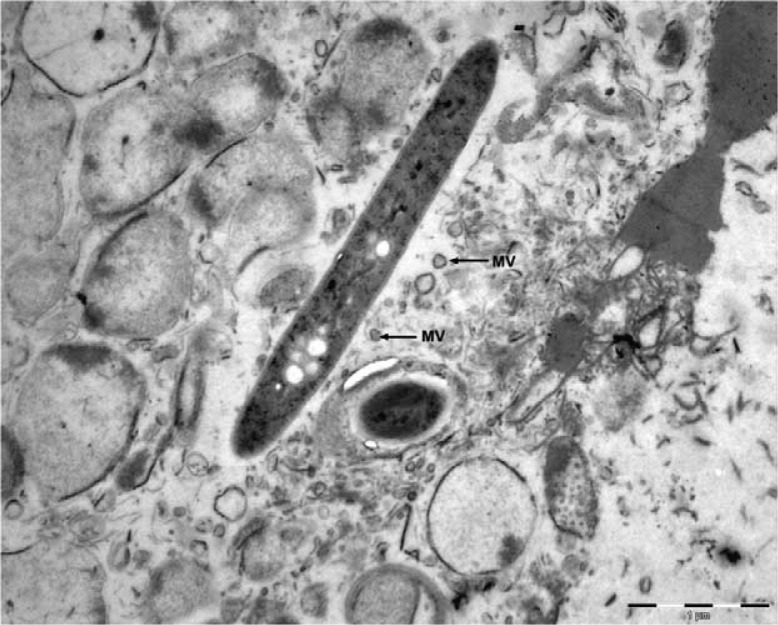
Many Gram-negative bacteria, including bacteria in dental plaque, form and release membrane vesicles (MVs) (Fig. 1) during growth (17–20). Using Pseudomonas aeruginosa as a model system, Beveridge et al. (21) demonstrated that Gram-negative bacteria can segregate and package periplasmic components into MVs. These MVs can protect the genetic content that is packaged inside from harsh environmental conditions such as DNase digestion (20, 22). MVs from Gram-negative bacteria can fuse into the surfaces of other species, indicating the possibility of MVs as delivery vehicles for transferring genetic materials and virulence factors from donor to recipient (10, 18). Indeed, it has been reported that MVs transfer virulence genes to recipient Gram-negative bacteria of the same or different species (17, 19, 20, 22). Because of their small dimensions (approximately 20–100 nm) (20), MVs could easily reach inaccessible areas such as the interior of biofilms and transport protected DNA to other bacteria, even when the donors and recipients are not in direct contact. These studies suggest that MV release by Gram-negative bacteria represents a novel mechanism for the transfer of genetic materials by these bacteria.
Fig. 1.
Transmission electron micrograph of a Leptotrichia buccalis cell with derived membrane vesicles (MVs) that provide an alternative mode of gene transfer. Bar=1 µm. Courtesy of Emenike R.K. Eribe.
Dental plaque and microbiome DNA exchange
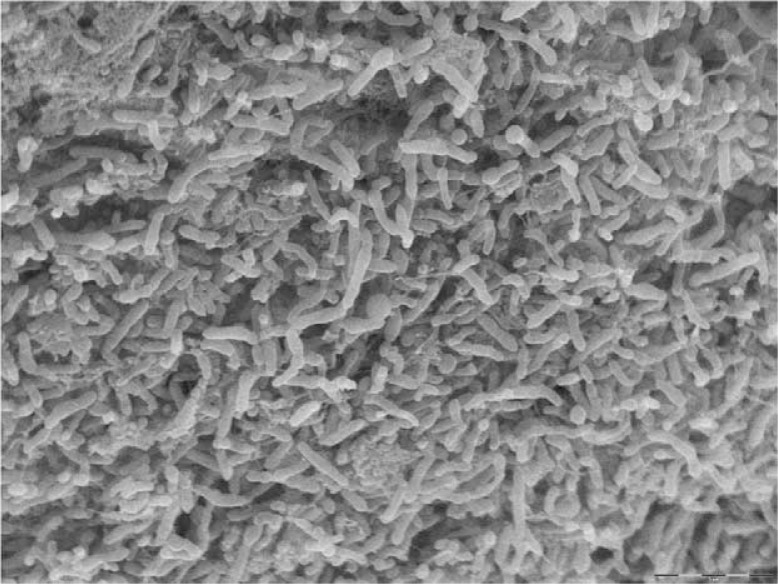
Dental plaque is a biofilm consisting of multiple bacterial species (Fig. 2). Since the tooth surface is nonshedding, this biofilm is actually the most diverse one in the human body in terms of microorganisms. With pyrosequencing, which detects even rare organisms at low concentrations, 1000 species have been found in plaque (23). This is in contrast to the previous estimate of more than 500 species in the dental biofilm (24). With these large numbers of bacteria in constant contact with each other, and with other bacteria transiently passing through the oral cavity, the potential for gene transfer is high. Recent metagenomic and bioinformatic studies have confirmed that horizontal DNA transfer is a common occurrence in human microbiota, and bacteria of the oral cavity are major players in this process (25, 26). In fact, horizontal DNA transfer was detected as commonly occurring between members of the oral microbiota and other bacteria in the body, especially those in the intestine such as the Bacteroides. As the oral cavity is the entry point for the entire digestive tract, oral bacteria will be in contact not only with other oral bacteria but also with intestinal bacteria that are transiently passing through the mouth and environmental bacteria that are present in food and drink products. As such, the oral microbiota could represent a reservoir for the exchange of virulence genes, such as genes for antibiotic resistance.
Fig. 2.
Scanning electron micrograph of a multispecies subgingival biofilm in vivo from adult periodontitis. Bar=5 µm. Courtesy of Steinar Stølen.
For example, DNA from a transient Phytoplasma species, which is normally associated with plants, has been detected in saliva (27). DNA is able to survive in a neutral pH environment, including saliva, for various time lengths (28, 29). Accordingly, the transfer of environmentally derived antibiotic resistance genes into oral bacteria could take place by transformation in addition to conjugation, and the subsequent spread of the newly acquired antibiotic resistance genes could then occur within the plaque population. Also, genetic material could then be transferred into intestinal genera that are only transient in the mouth. In this way, the normal microbiota in the mouth can potentially mediate the transfer of resistance genes between diverse sources and recipients (30). This is similar with findings from the colon, where extensive gene transfer has been found to occur within the genus Bacteroides and among Bacteroides species and Gram-positive bacteria (31). The colon is another body site that is considered highly conductive to horizontal gene transfer and to stable maintenance of transferred resistance.
DNA transfer in major dental plaque bacteria
By determining genome sequences of Porphyromonas gingivalis, a major pathogen in adult periodontitis, Naito et al. (32) detected extensive genomic rearrangement between the strains ATCC 33277T and W83. Both strains contained large numbers of mobile elements like insertion sequences, miniature inverted-repeat transposable elements, and conjugative transposons frequently connected with genomic rearrangements. Plasmid or chromosomal DNA can be transferred within and between strains of P. gingivalis (33). Strains W83 and ATCC 33277 can exchange chromosomally located antibiotic resistance markers. Natural competence was the major driving force behind DNA exchange in P. gingivalis, while conjugative DNA transfer played a minor role (33). Although DNA transfer in P. gingivalis is complex, comF-dependent transformation by uptake of eDNA appears to be the most important mechanism of DNA transfer in strain W83. Additionally, Tribble et al. (34) demonstrated the presence of eDNA in P. gingivalis biofilms. eDNA is predicted to be the source of the DNA exchanged between different strains during cohabitation in biofilms. comF is critical for DNA uptake from the environment. A transcriptome comparison analysis of planktonic and biofilm-cultured P. gingivalis showed that in the latter, comF expression increased twofold (35). Also, studies with other bacteria have indicated that eDNA is an important part of biofilm matrixes (36–39). The DNA exchange between strains may be important for the evolution of P. gingivalis from a commensal to a pathogenic state. It is also likely to be important for the short-term survival of P. gingivalis in the periodontal pocket, a stressful and highly competitive habitat. Changes in antigens obtained through genetic transfer may facilitate survival of the organism through protection against the adaptive immune responses.
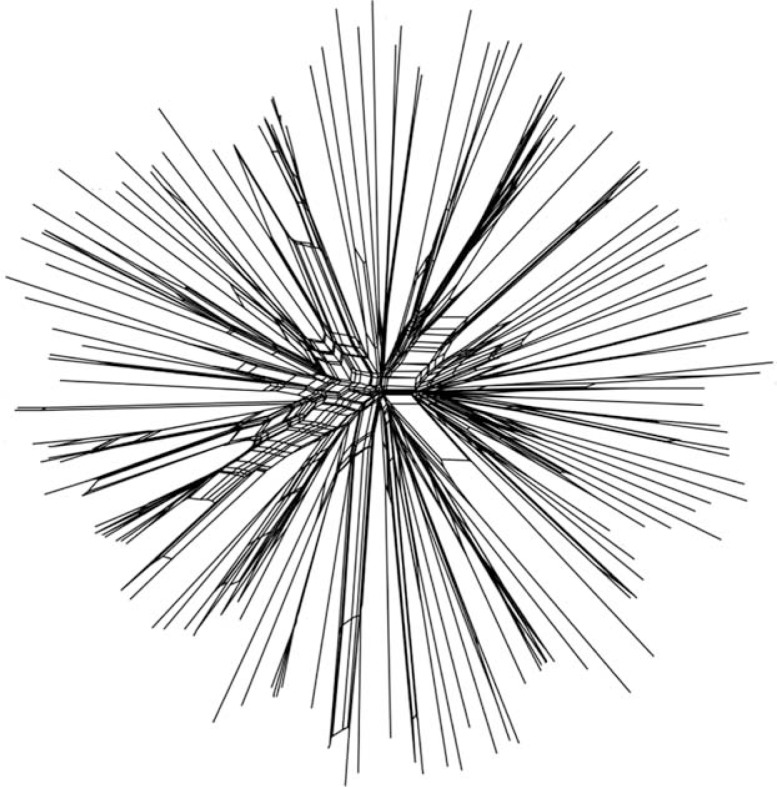
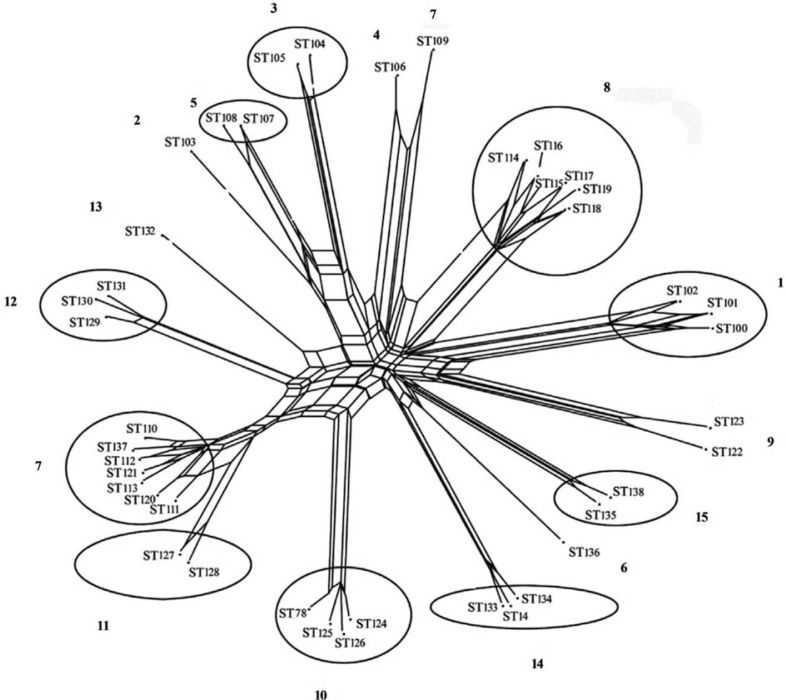
A multilocus sequence typing (MLST) scheme for P. gingivalis has been established (www.pubmlst.org/pgingivalis). It indicates a high degree of genetic diversity and a weakly clonal population structure comparable to that seen in Neisseria meningitidis (40) (Fig. 3). Multiple sequence types (STs) were detected in one site in several patients with ‘refractory’ periodontitis (Fig. 4). This reflected allelic variation in two housekeeping genes and recombination between different clones in vivo in subgingival plaque of the periodontal pocket (40). Accordingly, the genetic variation in P. gingivalis strains is considerable and reinforces the concept that exchange of genes occurs in subgingival plaque to improve the chances of P. gingivalis’ survival.
Fig. 3.
Split decomposition analysis graph constructed by Split Tree (v. 4.6) (www.splitstree.org) from allelic profiles of the 138 sequence types (STs) of P. gingivalis in the multilocus sequence typing (MLST) database. Recombination is illustrated by the net- or bushlike structures in the center. The star- or treelike structures indicate clonality. From Reference [40]. Courtesy of Morten Enersen.
Fig. 4.
The degree of recombination between 93 isolates of P. gingivalis sampled in vivo from 15 single sites of ‘refractory’ periodontitis patients is illustrated by a SplitsTree graph with superimposed results of the eBurst analysis to the graph. Patient and site numbers are marked in bold. Several STs detected in each periodontal site are located in groups (circles and ellipses) except for patients 2, 4, 6, 7, 9, and 13, for whom only one ST was found per site. See text in Reference [40]. Courtesy of Morten Enersen.
Plasmids may play an important role in horizontal gene transfer in fusobacteria. Genetic analyses of Fusobacterium nucleatum, which is another opportunistic pathogen implicated in periodontitis, demonstrate considerable diversity. Horizontal gene transfer is thought to play a prominent role in the evolution of this species. Several plasmids of F. nucleatum, including pFN1, have relaxase gene homologs that may operate in plasmid mobilization. Recently, Claypool et al. (41) found that the fusobacterial plasmid pFN1 encodes genes, enabling efficient mobilization between strains of Escherichia coli by the broad-range plasmid RP4. A role was suggested for plasmid mobilization in DNA transfer in F. nucleatum. Plasmid or plasmid-related sequences were identified in as many as 11.5% of the strains in a panel of clinical F. nucleatum isolates (41). Roberts and Lansciardi (42) showed that two strains of F. nucleatum were able to transfer tetracycline resistance by conjugation encoded by the tet(M) determinant to strains of F. nucleatum, Peptostreptococcus anaerobius, and Enterococcus faecalis. Another study showed that P. anaerobius could transfer tet(M) to recipients of P. anaerobius and F nucleatum (43).
Veillonella dispar was shown to transfer the conjugative transposon Tn916 to four different Streptococcus species in a multispecies oral biofilm established in a constant-depth biofilm fermenter (16). In the same system, purified V. dispar DNA conferred tetracycline resistance to S. mitis. None of the consortia members could develop tetracycline resistance when they were grown as monocultures. In a model oral biofilm established in another constant-depth film fermenter, the conjugative transposon Tn5397 was transferred from Bacillus subtilis to a Streptococcus species (44).
Transfer of conjugal elements in oral black-pigmented Prevotella species (P. denticola and P. intermedia) conferred tetracycline and penicillin resistance in the absence of plasmids (45). Transverse alternating-field electrophoresis of restricted chromosomal DNA from the transconjugants revealed rearrangements that indicated that the transfer occurred at strongly preferred sites in the recipient chromosome's genes. In another Prevotella study, a small conjugative transposon was identified that encoded a tet(Q) gene identical to genes found in the intestinal Bacteroides (46). Although the tet(Q) gene was the same, the Prevotella mobile element was an IS21 transposon not found in the Bacteroides, indicating that the Prevotella reassembled the resistance gene ‘module’ into its own transposon.
Transfer of a nonconjugative plasmid from Treponema denticola to Streptococcus gordonii was demonstrated in mixed-species biofilms (47). Transformation of S. gordonii occurred using the erythromycin resistance (Ermr) shuttle plasmid pKMR4PE as a donor source. Possibly there is a naturally occurring genetic transfer system within oral spirochetes, or they have the ability to take up and maintain mobilizable plasmids (48). Upregulation of various putative pro-phage genes and transposases was seen in a continuous-culture biofilm model of T. denticola ATCC 35405T (49). Also, a functional bacteriophage was isolated from this organism.
Transfer of genetic information in Actinomyces species was achieved by transfection with the genomic DNA of bacteriophages from human dental plaque (50). A study concerning a genetic transfer system for Actinomyces spp. has shown that multiple genes are involved in fimbriae synthesis and function (51). Furthermore, genetic analysis found various enzymes of strains of Actinomyces involved in the adherence of fimbrial receptors on host cells. These circumstances may determine the selection of Actinomyces spp. in certain ecological niches in the oral cavity.
In Aggregatibacter actinomycetemcomitans strains, Willi et al. (52) observed transduction of antibiotic resistance markers by temperate bacteriophages. Periodontal isolates of A. actinomycetemcomitans have been found to contain bacteriophages (53–55). The tet(B) determinant was transferable in vitro between isolates of A. actinomycetemcomitans and a Haemophilus influenza recipient and was probably associated with conjugative plasmids (56). Natural competence in some A. actinomycetemcomitans strains also takes place. UrpA is a novel gene that is required for natural competence in A. actinomycetemcomitans and is different from corresponding natural-competence genes in other bacterial species (57). A recent study demonstrated differences in full genome content and gene expression between A. actinomycetemcomitans strains of the JP2 and non-JP2 genotypes (58). This information might help us understand the pathogenic mechanisms of this important periodontopathogen. Also specific geographic distribution has been observed for the JP2 strain, suggesting limited genetic and species transfer between individuals from different subpopulations (59).
Several streptococcal species enter a physiological state of competence that allows them to bind, transport, and incorporate DNA into their chromosomes. Thereby, foreign DNA from the environment is acquired without cell–cell contact. Competence is linked to biofilm formation through generation of the quorum-sensing molecule competence-stimulating peptide (CSP) in S. pneumoniae, S. mutans, and S. gordonii. Biofilm communities may show peaks in competence over defined time periods or growth rates (60). The acquisition of exogenous DNA by naturally competent oral streptococci might enhance their survival under environmental stress. In S. gordonii, a genetic determinant encoded a peptide with activity similar to that of the sex pheromone cAM373 in E. faecalis, which promoted intergeneric DNA transfer (3). Interspecies genetic exchange in streptococci has also been demonstrated through the appearance of so-called mosaic genes, which are partially derived from donor DNA and the host chromosome. Additionally, it is clear from the genome of Streptococcus sanguinis that a 70 kb region-encoding pathway for vitamin B12, biosynthesis, and degradation of ethanolamine and propanediol has been acquired through horizontal gene transfer (61).
Roberts et al. (62) showed in microcosm dental plaque that conjugative transposon Tn916-like elements in four tetracycline-resistant streptococcal species could be transferred to other streptococci in filter-mating experiments and within biofilms in a fermenter. This was the first demonstration that a native conjugative transposon could be transferred within a model oral biofilm and that oral streptococci harbor and disseminate these mobile elements to other components of the oral microbiota. Particularly, S. pneumoniae and related streptococci are capable of exchanging genes. While oral streptococci can act as donors of DNA for pneumococci, the opposite can also occur, emphasizing the fact that streptococci are genetic reservoirs for transient organisms on their way through the mouth.
A transferable tetracycline resistance Tn916S element was characterized in S. intermedius (63), the genetic support of which was a conjugative transposon closely related to Tn916. Therefore, Tn916 is apparently involved in the dissemination not only of tet(M) but also of tet(S). Also, erythromycin resistance genes are prevalent in oral streptococci. Such isolates can be resistant to erythromycin and tetracycline simultaneously (64).
Warburton et al. (65) reported the transfer of antimicrobial resistance from one oral streptococcus species to another during systemic antibiotic treatment of periodontitis. In this case, an S. cristatus strain acquired a novel conjugative transposon CTn6002 that was derived in part from Tn916 during treatment, making it resistant to doxycycline and erythromycin.
S. mutans, which is hypertransformable when grown in biofilms, uses a peptide pheromone quorum-sensing signal transduction system for promoting the uptake and incorporation of foreign DNA (6). Seven ComC alleles encoding for three CSPs have been observed in S. mutans (66). Kuramitsu and Trapa (67) demonstrated that this organism could be a donor of plasmid DNA that gave rifampicin and erythromycin resistance to S. sanguinis and S. milleri. Also, transfer of genetic information from S. sanguinis to S. milleri was seen. Furthermore, fluoride resistance could be transferred between S. mutans strains (68) and chromosomally encoded genes providing resistance to rifampin, streptomycin, and spectinomycin as well as plasmids conferring resistance to tetracycline and erythromycin (69). The ability of streptococci to mediate interspecies DNA transfer seems to be common, as a filter-mating study showed that 4 of 12 isolates of oral streptococci were able to transfer genes encoding erythromycin resistance [erm(B)] and tetracycline resistance [tet(M)] to E. faecalis (70).
Mutant genotypes can also accumulate within dental biofilms. In S. mutans, natural selection but not increased mutation rate was found to be the primary driving force behind the accumulation of deletion mutants (71). The JP2 clone strains of A. actinomycetemcomitans constitute a unique clonal type that includes a 530 base pair deletion in the leukotoxin operon that is implicated in the enhanced leukotoxic activity of the clone (72).
Interference of gene transfer in dental plaque
Oral bacteria compete with each other for limited nutrients in the oral cavity. Therefore, it is not surprising that some products from different species can interfere with the gene transfer process of another species, since homologous recombination of foreign DNA into the host chromosome may give the recipient survival advantages. It has been demonstrated that proteases produced by both early and late colonizers of dental plaque can inactivate the S. mutans CSP, an essential factor to initiate competence in S. mutans’ natural transformation, which renders this naturally transformable species incompetent in horizontal genetic transfer (73, 74).
Another example of DNA transfer resistance and interference is found in Prevotella. Introduction of foreign DNA into Prevotella species is notoriously difficult, and some ruminal strains of Prevotella have been shown to produce extracellular endonucleases and high levels of cytoplasmic restriction and modification proteins (75, 76). The presence of secreted DNA nucleases could be predicted to interfere with DNA transfer within the local biofilm environment, via the degradation of eDNA. This could impact not only the strain secreting the DNase but also immediate neighbors that are competent and ‘in the market’ for eDNA. Many oral bacterial strains aside from the Prevotella ones have been found to secrete extracellular DNases (77).
Concluding remarks
There is no doubt that genes are transferred between bacteria in dental plaque, probably by all three major mechanisms of horizontal gene transfer: transduction, conjugation, and transformation. Several examples have been given in this review, which is not meant to be exhaustive. Of particular interest is that dental plaque can be a reservoir for antimicrobial resistance genes, which can be incorporated outside species’ boundaries. The foreign DNA can come from cells (even dead ones) of the same species or from distantly related organisms. The transmitted DNA can be integrated or recombined in the recipient's chromosome or remain as an extrachromosomal inheritable element. Although we do not know much about the frequency of such in vivo horizontal gene transfers, genomic studies reveal that they probably account for between 5 and 43% of genes in different oral species (78). Furthermore, species of oral genera (e.g. Actinomyces, Bifidobacterium, Fusobacterium, Haemophilus, Peptostreptococcus, Streptococcus, Porphyromonas, Prevotella, and Veillonella) contain transposons that promote transfer of DNA between bacteria through conjugation. We also know that, for example, tetracycline use in the community not only leads to selection for the maintenance of resistance genes [tet(Q)] but also causes them to be transferred in the first place. This transfer is not parallel to processes that we associate with sex in higher organisms as it does not cause reproduction. Bacterial sex is simply associated with the uptake of genetic material that can be transferred vertically or horizontally to other cells. It is hardly accompanied by pleasure for the bacteria (although who knows?), unlike the ultimate goal for activities in a bordello. Therefore, dental plaque can hardly be considered a bordello in the usual context of the word, even for bacteria. However, there is no doubt that bacterial sex occurs, which here means that genes are transferred between cells. This transfer is an important mechanism for the bacteria in terms of survival in a harsh environment, for their virulence and their pathogenicity.
Conflict of interest and funding
The authors declare no conflict of interest. Funding was from the Faculty of Dentistry, University of Oslo, Norway. P. gingivalis and Prevotella research was supported by funding from the NIH National Institute of Dental and Craniofacial Research DE019634, and past funding from DE16562 was awarded to G.D.T.
References
- 1.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Rev. 2010;74:417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vickerman MM, Flannagan SE, Jesionowski AM, Brossard KA, Clewell DB, Sedgley CM. A genetic determinant in Streptococcus gordonii Challis encodes a peptide with activity similar to that of enterococcal sex pheromone cAM373, which facilitates intergeneric DNA transfer. J Bacteriol. 2010;192:2535–45. doi: 10.1128/JB.01689-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 1910;8:471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 5.Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of biofilm structure. Curr Opin Biotechnol. 2003;14:255–61. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 6.Li Y-H, Lau PCY, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 8.Narra HP, Ochman H. Of what use is sex to bacteria? Current Biol. 2006;16:R705–10. doi: 10.1016/j.cub.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–94. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Lefébure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli; an evolutionary perspective. Mol Microbiol. 2006;60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLOS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redfield RJ. Do bacteria have sex? Perspectives. 2001;2:624–9. doi: 10.1038/35084593. [DOI] [PubMed] [Google Scholar]
- 14.Rice LB. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob Agents Chemother. 1998;42:1871–7. doi: 10.1128/aac.42.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burmølle M, Bahl MI, Jensen LB, Sørensen SJ, Hansen LH. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology. 2008;154:187–95. doi: 10.1099/mic.0.2007/010454-0. [DOI] [PubMed] [Google Scholar]
- 16.Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, Roberts AP. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol Med Microbiol. 2010;59:345–9. doi: 10.1111/j.1574-695X.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 17.Yaron S, Kolling GL, Simon L, Matthews KR. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol. 2000;66:4414–20. doi: 10.1128/aem.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadurugamuwa JL, Beveridge TJ. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology. 1999;145:2051–60. doi: 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- 19.Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae . J Bacteriol. 1989;171:2499–505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:1843–8. doi: 10.1128/aem.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beveridge TJ, Makin SA, Kadurugamuwa JL, Li ZJ. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20:291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 22.Kahn ME, Barany F, Smith HO. Transformasomes: specialized membranous structures that protect DNA during Haemophilus transformation. Proc Natl Acad Sci USA. 1983;80:6927–31. doi: 10.1073/pnas.80.22.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ten Cate JM. Biofilms, a new approach to the microbiology of dental plaque. Odontology. 2006;94:1–9. doi: 10.1007/s10266-006-0063-3. [DOI] [PubMed] [Google Scholar]
- 24.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontology 2000. 2005;38:135–87. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu L, Chen X, Skogerbø G, Zhang P, Chen R, He S, et al. The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics. 2012;100:265–70. doi: 10.1016/j.ygeno.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 27.Seville LA, Patterson AJ, Scott KP, Mullany P, Quail MA, Parkhill J, et al. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb Drug Resist. 2009;15:159–66. doi: 10.1089/mdr.2009.0916. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. Release and persistence of extracellular DNA in the environment. Environ Biosafety Res. 2007;6:37–53. doi: 10.1051/ebr:2007031. [DOI] [PubMed] [Google Scholar]
- 29.Mercer DK, Scott KP, Bruce-Johnson WA, Glover LA, Fint HJ. Fate of free DNA and transformation of the oral bacterium Streptococcus gordonii DL1 by plasmid DNA in human saliva. Appl Environ Microbiol. 1999;65:6–10. doi: 10.1128/aem.65.1.6-10.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti Infect Ther. 2010;8:1441–50. doi: 10.1586/eri.10.106. [DOI] [PubMed] [Google Scholar]
- 31.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environment Microbiol. 2001;67:561–8. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis . DNA Res. 2008;15:215–25. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tribble GD, Lamont GJ, Progulske-Fox A, Lamont RJ. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis . J Bacteriol. 2007;189:6382–8. doi: 10.1128/JB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tribble GD, Rigney TW, Dao D-HV, Wong CT, Kerr JE, Taylor BE, et al. Natural competence is a major mechanism for horizontal DNA transfer in the oral pathogen Porphyromonas gingivalis . MBio. 2012;3:e00231–11. doi: 10.1128/mBio.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo AW, Seers CA, Boyce JD, Dashper SG, Slakeski N, Lissel JP, et al. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 2009;9:18. doi: 10.1186/1471-2180-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harmsen M, Lappann M, Knøchel S, Molin S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes . Appl Environ Microbiol. 2010;76:2271–9. doi: 10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lappann M, Claus H, van Alen T, Harmsen M, Elias J, Molin S, et al. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis . Mol Microbiol. 2010;75:1355–71. doi: 10.1111/j.1365-2958.2010.07054.x. [DOI] [PubMed] [Google Scholar]
- 38.Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis . Mol Microbiol. 2009;72:1022–36. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilain S, Pretorius JM, Theron J, Brözel VS. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol. 2009;75:2861–8. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enersen M. Porphyromonas gingivalis: a clonal pathogen? Diversities in housekeeping genes and the major fimbriae gene. J Oral Microbiol. 2011;3:1–11. doi: 10.3402/jom.v3i0.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Claypool BM, Yoder SC, Citron DM, Finegold SM, Goldstein EJC, Haake SK. Mobilization and prevalence of a fusobacterial plasmid. Plasmid. 2010;63:11. doi: 10.1016/j.plasmid.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts MC, Lansciardi J. Transferable tetM in Fusobacterium nucleatum . Antimicrob Agents Chemother. 1990;34:1836–8. doi: 10.1128/aac.34.9.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts MC. Tetracycline resistance in Peptostreptococcus anaerobius . Antimicrob Agents Chemother. 1991;35:1682–4. doi: 10.1128/aac.35.8.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts AP, Pratten J, Wilson M, Mullany P. Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol Lett. 1999;177:63–6. doi: 10.1111/j.1574-6968.1999.tb13714.x. [DOI] [PubMed] [Google Scholar]
- 45.Guiney DG, Hasegawa P. Transfer of conjugal elements in oral black-pigmented Bacteroides (Prevotella) spp. involves DNA rearrangements. J Bacteriol. 1992;174:4853–5. doi: 10.1128/jb.174.14.4853-4855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tribble GD, Garza JJ, Yeung VL, Rigney TW, Dao D-HV, Rodrigues PH, et al. Genetic analysis of mobile tetQ elements in oral Prevotella species. Anaerobe. 2010;16:604–9. doi: 10.1016/j.anaerobe.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang BY, Chi B, Kuramitsu HK. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol Immunol. 2002;17:108–12. doi: 10.1046/j.0902-0055.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- 48.Chan EC, Klitorinos A, Gharbia S, Caudry SD, Rahal MD, Siboo R. Characterization of a 4.2-kb plasmid isolated from periodontopathic spirochetes. Oral Microbiol Immunol. 1996;11:365–8. doi: 10.1111/j.1399-302x.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- 49.Mitchell HL, Dashper SG, Catmull DV, Paolini RA, Cleal SM, Slakeski N, et al. Treponema denticola biofilm-induced expression of a bacteriophage, toxin-antitoxin systems and transposases. Microbiology. 2010;156:774–88. doi: 10.1099/mic.0.033654-0. [DOI] [PubMed] [Google Scholar]
- 50.Yeung MK, Kozelsky CS. Transfection of Actinomyces spp. by genomic DNA of bacteriophages from human dental plaque. Plasmid. 1997;37:141–53. doi: 10.1006/plas.1997.1285. [DOI] [PubMed] [Google Scholar]
- 51.Yeung MK. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med. 1999;10:120–38. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
- 52.Willi K, Sandmeier H, Kulik EM, Meyer J. Transduction of antibiotic resistance markers among Actinobacillus actinomycetemcomitans strains by temperate bacteriophages Aa phi 23. Cell Mol Life Sci. 1997;53:904–10. doi: 10.1007/s000180050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandmeier H, van Winkelhoff AJ, Bär K, Ankli E, Maeder M, Meyer J. Temperate bacteriophages are common among Actinobacillus actinomycetemcomitans isolates from periodontal pockets. J Periodontal Res. 1995;30:418–25. doi: 10.1111/j.1600-0765.1995.tb01296.x. [DOI] [PubMed] [Google Scholar]
- 54.Preus HR, Olsen I, Namork E. The presence of phage-infected Actinobacillus actinomycetemcomitans in localized juvenile periodontitis patients. J Clin Periodontol. 1987;14:605–9. doi: 10.1111/j.1600-051x.1987.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 55.Olsen I, Namork E, Myhrvold V. Electron microscopy of phages in serotypes of Actinobacillus actinomycetemcomitans . Oral Microbiol Immunol. 1993;8:383–5. doi: 10.1111/j.1399-302x.1993.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 56.Roe DE, Braham PH, Weinberg A, Roberts MC. Characterization of tetracycline resistance in Actinobacillus actinomycetemcomitans . Oral Microbiol Immunol. 1995;10:227–32. doi: 10.1111/j.1399-302x.1995.tb00147.x. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka A, Fujise O, Chen C, Mura M, Hamachi T, Maede K. A novel gene required for natural competence in Aggregatibacter actinomycetemcomitans . J Periodontal Res. 2012;47:129–34. doi: 10.1111/j.1600-0765.2011.01410.x. [DOI] [PubMed] [Google Scholar]
- 58.Huang Y, Kittichotirat W, Mayer MPA, Hall R, Bumgamer R, Chen C. Comparative genomic hybridization and transcriptome analysis with a pan-genome microarray reveal distinctions between JP2 and non-JP2 genotypes of Aggregatibacter actinomycetemcomitans . Mol Oral Microbiol. 2013;28:1–17. doi: 10.1111/omi.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Åberg CH, Kwamin F, Claesson R, Johansson A, Haubek D. Presence of JP2 and non-JP2 genotypes of Aggregatibacter actinomycetemcomitans and attachment loss in adolescents in Ghana. J Periodontol. 2012;83:1520–8. doi: 10.1902/jop.2012.110699. [DOI] [PubMed] [Google Scholar]
- 60.Cvitkovitch DG. Genetic competence and transformation in oral streptococci. Crit Rev Oral Biol Med. 2011;12:217–43. doi: 10.1177/10454411010120030201. [DOI] [PubMed] [Google Scholar]
- 61.Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, et al. Genome of the opportunistic pathogen Streptococcus sanguinis . J Bacteriol. 2007;189:3166–75. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts AP, Cheah G, Ready D, Pratten J, Wilson M, Mullany P. Transfer of Tn916-like elements in microcosm dental plaques. Antimicrob Agents Chemother. 2001;45:2943–6. doi: 10.1128/AAC.45.10.2943-2946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lancaster H, Roberts AP, Bedi R, Wilson M, Mullany P. Characterization of Tn916S, a Tn916-like element containing the tetracycline resistance determinant tet(S) J Bacteriol. 2004;186:4395–8. doi: 10.1128/JB.186.13.4395-4398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Villedieu A, Diaz-Torres ML, Hunt N, McNab R, Spratt DA, Wilson M, et al. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob Agents Chemother. 2003;47:878–82. doi: 10.1128/AAC.47.3.878-882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warburton PJ, Palmer RM, Munson MA, Wade WG. Demonstration of in vivo transfer of doxycycline resistance mediated by a novel transposon. J Antimicrob Chemother. 2007;60:973–80. doi: 10.1093/jac/dkm331. [DOI] [PubMed] [Google Scholar]
- 66.Allan E, Hussain HA, Crawford KR, Miah S, Ascott ZK, Khwaja MH, et al. Genetic variation in comC, the gene encoding competence-stimulating peptide (CSP) in Streptococcus mutans . FEMS Microbiol Lett. 2007;268:47–51. doi: 10.1111/j.1574-6968.2006.00593.x. [DOI] [PubMed] [Google Scholar]
- 67.Kuramitsu HK, Trapa V. Genetic exchange between oral streptococci during mixed growth. J Gen Microbiol. 1984;130:2497–500. doi: 10.1099/00221287-130-10-2497. [DOI] [PubMed] [Google Scholar]
- 68.Chansley PE, Kral TA. Transformation of fluoride resistance genes in Streptococcus mutans . Infect Immun. 1989;57:1968–70. doi: 10.1128/iai.57.7.1968-1970.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murchison HH, Barrett JF, Cardineau GA, Curtiss RD., III Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun. 1986;54:273–82. doi: 10.1128/iai.54.2.273-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Villedieu A, Diaz-Torres ML, Roberts AP, Hunt N, McNab R, Spratt DA, et al. Genetic basis of erythromycin resistance in oral bacteria. Antimicrob Agents Chemother. 2004;48:2298–301. doi: 10.1128/AAC.48.6.2298-2301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banas JA, Miller JD, Fuschino ME, Hazlett KRO, Toyofuko W, Porter KA, et al. Evidence that accumulation of mutants in a biofilm reflects natural selection rather than stress-induced adaptive mutation. Appl Environ Microbiol. 2007;73:357–61. doi: 10.1128/AEM.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haubek D. The highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans: evolutionary aspects, epidemiology and etiological role in aggressive periodontitis. APMIS Suppl. 2010;130:1–53. doi: 10.1111/j.1600-0463.2010.02665.x. [DOI] [PubMed] [Google Scholar]
- 73.Wang BY, Alvarez P, Hong J, Kuramitsu HK. Periodontal pathogens interfere with quorum-sensing-dependent virulence properties in Streptococcus mutans . J Periodontal Res. 2011;46:105–10. doi: 10.1111/j.1600-0765.2010.01319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuramitsu HK, Wang BY. The whole is greater than the sum of its parts: dental plaque bacterial interactions can affect the virulence properties of cariogenic Streptococcus mutans . Am J Dent. 2011;24:153–4. [PMC free article] [PubMed] [Google Scholar]
- 75.Accetto T, Peterka M, Avguštin G. Type II restriction modification systems of Prevotella bryantii TC1-1 and Prevotella ruminicola 23 strains and their effect on the efficiency of DNA introduction via electroporation. FEMS Microbiol Lett. 2005;247:177–83. doi: 10.1016/j.femsle.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 76.Accetto T, Avgustin G. Non-specific DNAases from the rumen bacterium Prevotella bryantii . Folia Microbiol (Praha) 2001;46:33–5. doi: 10.1007/BF02825880. [DOI] [PubMed] [Google Scholar]
- 77.Palmer LJ, Chapple ILC, Wright HJ, Roberts A, Cooper PR. Extracellular deoxyribonuclease production by periodontal bacteria. J Periodontal Res. 2012;47:439–45. doi: 10.1111/j.1600-0765.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 78.Mira A. Horizontal gene transfer in oral bacteria. In: Rogers AH, editor. Molecular oral microbiology. England: Academic Press; 2008. pp. 65–85. Chapter 3. Caister Norfolk. [Google Scholar]