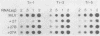Abstract
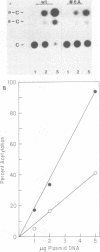
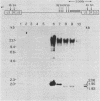
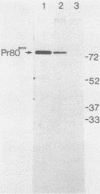
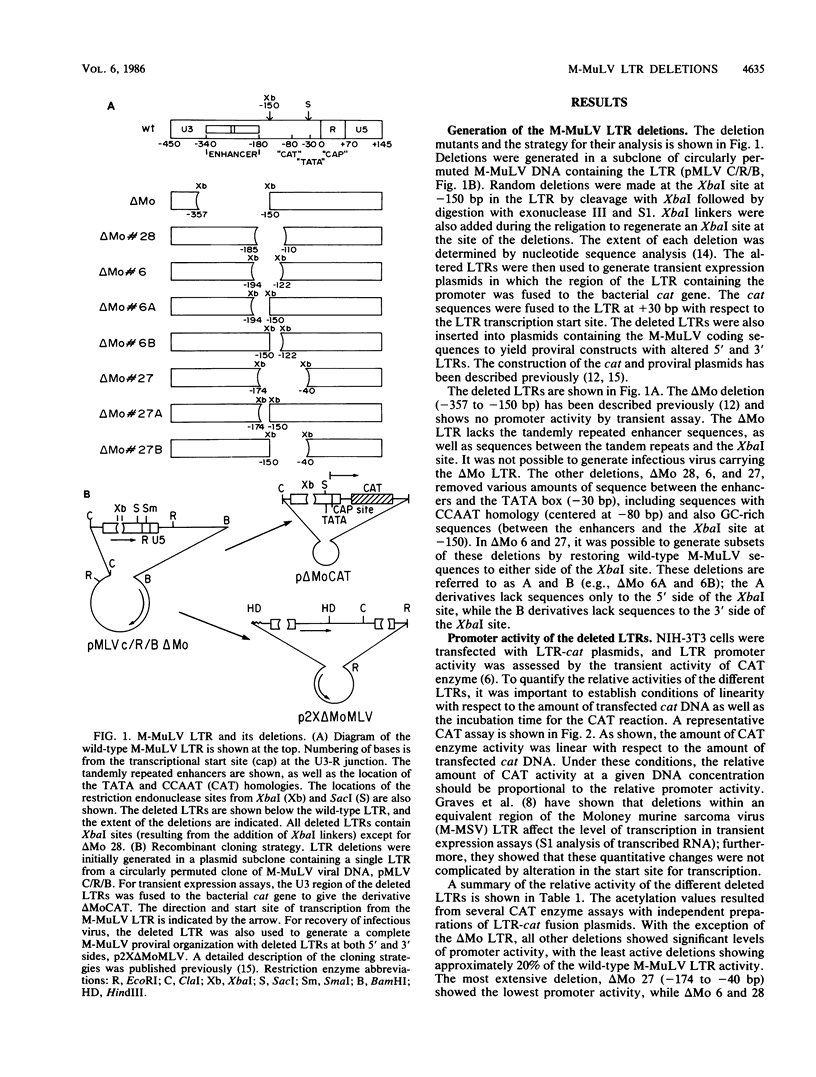
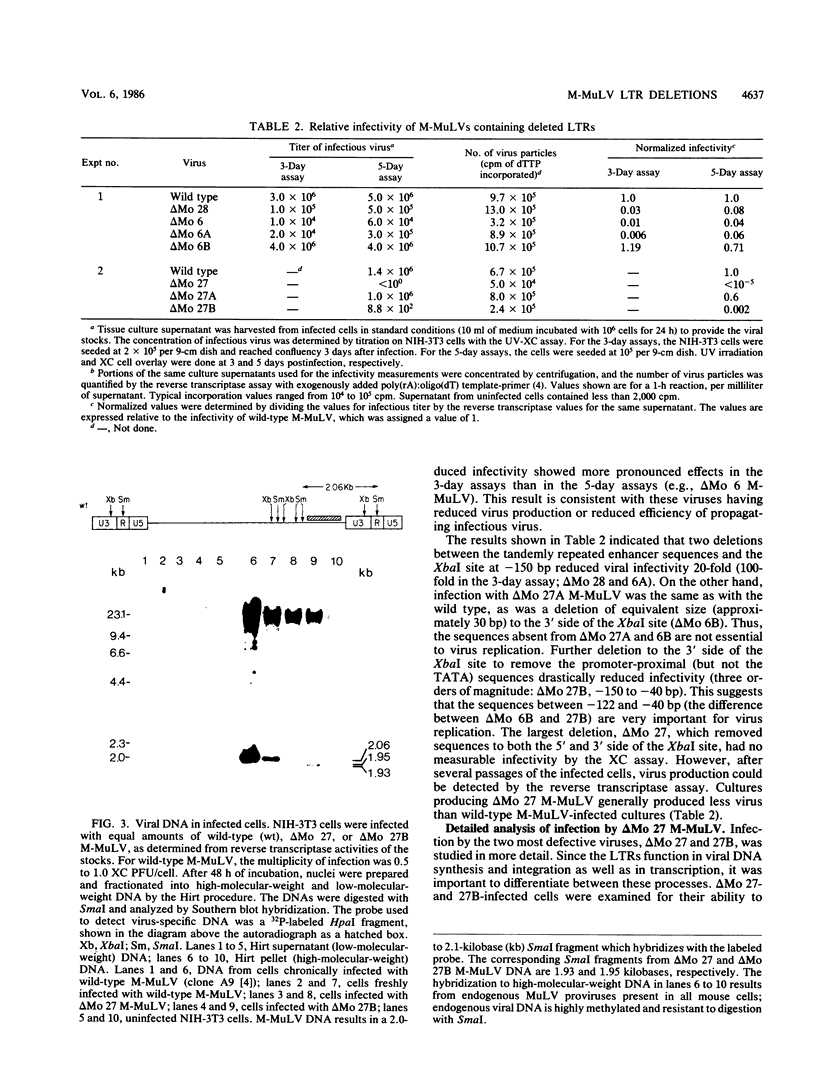
Deletional analysis within the long terminal repeat (LTR) of Moloney murine leukemia virus (M-MuLV) was performed. By molecular cloning, deletions were made in the vicinity of the XbaI site at -150 base pairs (bp) in the U3 region, between the tandemly repeated enhancers and the TATA box. The effects of the deletions on LTR function were measured in two ways. First, deleted LTRs were fused to the bacterial chloramphenicol acetyltransferase gene and used in transient expression assays. Second, infectious M-MuLVs were generated by transfection of M-MuLV proviruses containing the deleted LTRs, and the relative infectivity of the mutant viruses was assessed by XC-syncytial assay. Most of the deleted LTRs examined showed relatively high promoter activity in the transient chloramphenicol acetyltransferase assays, with values ranging from 20 to 50% of the wild-type M-MuLV LTR. Thus, the sequences between the enhancers and the TATA box were not absolutely required for transient expression. However, infectivity of viruses carrying the same deleted LTRs showed more pronounced effects. Deletion of sequences from -195 to -174 bp reduced infectivity 20- to 100-fold. Deletion of sequences within the region from -174 to -122 bp did not affect infectivity, indicating that this region is dispensable. On the other hand, deletion of sequences from -150 to -40 bp reduced infectivity from 5 to 6 logs, although the magnitude of the reduction partly may have reflected threshold envelope protein requirements for positive XC assays. The reduced infectivity did not appear to result from a failure of proviral DNA synthesis or integration by the mutant. Thus, the infectivity measurements identified three functional domains in the region between the enhancers and the TATA box.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Campos R., Villarreal L. P. An SV40 deletion mutant accumulates late transcripts in a paranuclear extract. Virology. 1982 May;119(1):1–11. doi: 10.1016/0042-6822(82)90059-9. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Edwards S. A., Fan H. gag-Related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J Virol. 1979 May;30(2):551–563. doi: 10.1128/jvi.30.2.551-563.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Jaenisch R., MacIsaac P. Low-multiplicity infection of Moloney murine leukemia virus in mouse cells: effect on number of viral DNA copies and virus production in producer cells. J Virol. 1978 Dec;28(3):802–809. doi: 10.1128/jvi.28.3.802-809.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallis B. M., Eisenman R. N., Diggelmann H. Synthesis of the precursor to avian RNA tumor virus internal structural proteins early after infection. Virology. 1976 Oct 15;74(2):302–313. doi: 10.1016/0042-6822(76)90337-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Rigby P. W., Lane D. P. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985 Sep;42(2):519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- Graves B. J., Eisenman R. N., McKnight S. L. Delineation of transcriptional control signals within the Moloney murine sarcoma virus long terminal repeat. Mol Cell Biol. 1985 Aug;5(8):1948–1958. doi: 10.1128/mcb.5.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E., Davis B., Overhauser J., Chao E., Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. 1984 Mar 29-Apr 4Nature. 308(5958):470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhauser J., Fan H. Generation of glucocorticoid-responsive Moloney murine leukemia virus by insertion of regulatory sequences from murine mammary tumor virus into the long terminal repeat. J Virol. 1985 Apr;54(1):133–144. doi: 10.1128/jvi.54.1.133-144.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Srinivasan A., Reddy E. P., Dunn C. Y., Aaronson S. A. Molecular dissection of transcriptional control elements within the long terminal repeat of the retrovirus. Science. 1984 Jan 20;223(4633):286–289. doi: 10.1126/science.6322296. [DOI] [PubMed] [Google Scholar]
- TODARO G. J., GREEN H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963 May;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T., Fan H. Mapping of DNase I-hypersensitive sites in the 5' and 3' long terminal repeats of integrated moloney murine leukemia virus proviral DNA. Mol Cell Biol. 1985 Apr;5(4):601–609. doi: 10.1128/mcb.5.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]