Transcriptional elongation control by RNA polymerase II and its associated factors is an essential process for the regulation of gene expression throughout development. Smith and Shilatifard review recent work identifying the factors functioning in the regulation of the transcriptional elongation checkpoint control (TECC) stage of gene expression and how the misregulation of these factors is associated with disease pathogenesis.
Keywords: cancer, chromatin, elongation, RNA polymerase II, transcription
Abstract
Transcriptional elongation control by RNA polymerase II and its associated factors has taken center stage as a process essential for the regulation of gene expression throughout development. In this review, we analyze recent findings on the identification of factors functioning in the regulation of the transcriptional elongation checkpoint control (TECC) stage of gene expression and how the factors' misregulation is associated with disease pathogenesis, including cancer.
It has long been recognized that gene expression can be regulated at the level of transcription elongation (Shilatifard 1998; Saunders et al. 2006). For example, the presence of transcriptionally engaged RNA polymerase (Pol II) without productive elongation (promoter-proximal paused Pol II) was observed for the Hsp70 gene in Drosophila and at the c-myc and c-fos genes in mammalian cells (Gilmour and Lis 1986; Fort et al. 1987; Rougvie and Lis 1988; Krumm et al. 1992). Subsequent genome-wide analyses of Pol II occupancy and nascent transcription have shown that proximal pausing of Pol II is a common feature of developmentally regulated genes (Guenther et al. 2007; Muse et al. 2007; Zeitlinger et al. 2007). Since the phenomenon of paused Pol II was initially described for the c-fos and Hsp70 genes, both of which are rapidly induced in response to environmental signals, it was considered that rapid transcriptional induction might be the major function of having Pol II present at the promoter prior to transcriptional induction. However, recent work has demonstrated that genes can be rapidly induced without having paused Pol II (Lin et al. 2011). Elegant in vivo studies by Boettiger and Levine (2009) have also demonstrated that developmental genes bearing paused Pol II on their promoters have a greater propensity to be induced synchronously. Based on these and additional studies demonstrating an essential role for the super elongation complex (SEC) in rapid transcriptional induction with or without paused Pol II, the emerging consensus is that genes with paused Pol II can be induced in an equitable and synchronous manner compared with genes without paused Pol II that are subject to potentially stochastic initiation events leading to more variable induction (Boettiger and Levine 2009; Levine 2011; Lin et al. 2011). This well-regulated release of paused Pol II is referred to as the transcriptional elongation checkpoint control (TECC) (Luo et al. 2012b).
Numerous factors have been identified that regulate the transcription elongation stage of gene expression, and many such factors were initially identified from in vitro biochemical studies (Shilatifard et al. 2003; Sims et al. 2004). In vitro assays typically use fractionated nuclear extracts and a runoff transcription template to measure either elongation rates or the ability to extend RNA chains by RNA Pol II (Shilatifard et al. 2003). The identified factors can be classified according to the step at which they are thought to be required. For example, TFIIF helps recruit Pol II to the promoter and has a role in the transcription of the first few nucleotides (Yan et al. 1999). The RNA Pol II elongation factor TFIIS is required whenever Pol II is irreversibly backtracked, in which case TFIIS stimulates Pol II to cleave the nascent RNA chain to allow realignment of the catalytic pocket with the 3′-OH of the nascent chain (Reines et al. 1996). The elongation factors Elongin A and ELL were identified by their ability to increase the rate (Vmax) of transcription elongation by Pol II in vitro, and even though they possess similar biochemical properties in vitro, they have been found to participate in distinct complexes and to have nonredundant functions in vivo (Aso et al. 1995; Shilatifard et al. 1996, 2003; Eissenberg et al. 2002; Gerber et al. 2004; Smith et al. 2008; Chopra et al. 2009; Yasukawa et al. 2012). The RNA Pol II elongation factors NELF and DSIF were purified as negative elongation factors that cooperate to promote the paused Pol II state, but DSIF, composed of SPT4 and SPT5, also has a distinct role in regulating the rate of elongation by Pol II (Wada et al. 1998; Yamaguchi et al. 1999).
Many of the Pol II elongation factors identified from these in vitro assays were independently identified from genetic studies as regulators of development in Drosophila, zebrafish, or mammals. ELL (eleven-nineteen lysine-rich leukemia) was identified as a Pol II elongation factor isolated from rat liver nuclear extracts and turned out to be the ortholog of the ELL gene product from humans (Thirman et al. 1994; Shilatifard et al. 1996). ELL is a frequent translocation partner of MLL (mixed-lineage leukemia), and even though MLL has a large number of translocation partners in leukemia, ELL was the first partner for which a biological function was proposed (Shilatifard 1998). Studies from our laboratory and others demonstrated that ELL is a component of the SEC that contains other ELL family members in addition to MLL translocation partners AFF1, AFF4, AF9, and ENL and Pol II elongation factor P-TEFb (He et al. 2010; Lin et al. 2010; Mohan et al. 2010; Sobhian et al. 2010). In vivo studies demonstrated that chromosomal translocations that result in the fusion of MLL with subunits of SEC can lead to the improper recruitment of SEC to MLL target genes, suggesting that misregulation of the TECC stage of gene expression contributes to leukemic pathogenesis (Lin et al. 2010; Mohan et al. 2010).
As indicated above, SEC contains the kinase module P-TEFb, itself a factor that was originally identified as an elongation factor from in vitro biochemical studies (Marshall and Price 1995). The first characterized in vivo role of P-TEFb was as a cellular cofactor for the viral Tat protein's ability to remove an elongation block of transcription of the HIV-1 provirus (Mancebo et al. 1997; Zhu et al. 1997). P-TEFb's kinase activity is required for the release of paused Pol II through the phosphorylations of the RNA Pol II C-terminal domain (CTD), the NELF-E subunit, and the CTD of the SPT5 subunit of DSIF (Fujinaga et al. 2004; Peterlin and Price 2006; Yamada et al. 2006). Phosphorylation of SPT5 results in the release of DSIF from NELF, and DSIF proceeds to travel with Pol II as a positive elongation factor. Foggy, the zebrafish ortholog of SPT5, was identified in a genetic screen for factors required for neuronal development (Guo et al. 1999). The mutant allele showed a single nucleotide change that resulted in an amino acid substitution of a conserved C-terminal residue. The mutant version of Foggy was able to stimulate elongation as well as the wild type but lacked the negative elongation activity, indicating the importance of Pol II pause control for development (Guo et al. 2000).
In this review, we discuss recent findings that provide insight into the nature of transcriptional regulation on genes at the level of paused Pol II and the TECC and the potential for diverse regulatory mechanisms for its regulation in normal developmental contexts as well as disease contexts.
Setup and release of RNA Pol II: Paused Pol II is not a prerequisite for rapid induction of gene expression
The Drosophila Hsp70 gene is characterized by the presence of transcriptionally engaged Pol II that is primarily producing 30- to 40-nucleotide (nt) transcripts in the unstressed state. Occasionally, about every 10 min, Pol II escapes and produces a full-length transcript, thus producing a low basal level of the Hsp70 protein. Upon heat shock, Pol II is released about every 4 sec, but the presence of Pol II and the production of shorter transcripts can still be detected even during a full heat-shock response (Nechaev and Adelman 2011). “Constitutively expressed” genes such as actin and tubulin are even known to have a “pileup” of Pol II at their 5′ ends and have engaged Pol II in producing the short transcripts while maintaining high levels of expression of the full-length transcripts (Boehm et al. 2003). These studies support a model in which genes bearing paused Pol II at their promoters are not in an off state, but rather in a preassembled state ready to respond rapidly to cellular signals. In addition to a possible role in rapid induction, one benefit of a preassembled system is to allow a synchronous response to developmental cues between cells in a developing organism, avoiding stochastic steps, including preinitiation complex (PIC) assembly and promoter melting (Boettiger and Levine 2009). Studies in mouse embryonic stem (ES) cells demonstrated that the most rapidly induced gene in response to retinoic acid, Cyp26a1, lacked paused Pol II in the undifferentiated state, demonstrating that paused Pol II is not a prerequisite for rapid transcriptional induction (Lin et al. 2011). Studies by Lin et al. (2011) also suggested that paused Pol II may function in the regulation of a uniform or equitable induction of gene expression in a given cell, similar to what was proposed on the role of paused Pol II in the regulation of uniform and synchronous transcriptional induction of a given gene in the cells of a developing embryo (Boettiger and Levine 2009).
Release of paused Pol II through CTD phosphorylation
A prominent distinction between paused and elongating Pol II is the phosphorylation state of the heptad repeat in the CTD. This repeat contains three serines, one threonine, and one tyrosine, all of which can be phosphorylated. Notably, the Ser 2 and Ser 5 phosphorylations have been the most widely investigated modification sites (for review, see Hsin and Manley 2012). Ser 5 phosphorylation is enriched in the promoter-proximal region, including genes such as Hsp70 that exhibit pausing. Ser 2 phosphorylation is enriched in the gene bodies and corresponds to the elongating form of Pol II. Therefore, the transition from paused Pol II to elongating Pol II through phosphorylation has been identified as a key regulatory step in gene expression (Mohan et al. 2010). Given the fact that the SEC family of Pol II elongation factors contains the most catalytically active versions of P-TEFb, their role in the regulation of pause release is of great interest (Luo et al. 2012a).
In addition to phosphorylating the Pol II CTD, P-TEFb also phosphorylates the CTD of SPT5 and the “E” subunit of the NELF complex. Both SPT5 and NELF are implicated in setting up the paused state of RNA Pol II. Phosphorylations of Pol II, Spt5, and NELF may all contribute to the release of the paused state. Phosphorylations of the CTDs of both SPT5 and Pol II also occur in the budding yeast Saccharomyces cerevisiae. As in metazoans, the Pol II CTD in yeast is phosphorylated on Ser 5 at the 5′ ends of genes, with a transitioning to Ser 2 phosphorylation toward the 3′ ends. However, yeast lacks NELF and may not regulate the pausing of RNA Pol II as seen in metazoans (Kim et al. 2010; Mayer et al. 2010; Churchman and Weissman 2011; Yearling et al. 2011).
An important difference between yeast and metazoan SPT5 is the addition of a conserved region that is C-terminal to the phosphorylated CTD, referred to as the extreme C terminus. This domain is implicated in the pausing of RNA Pol II and is the affected region in the zebrafish foggy mutant (Guo et al. 1999). It is known that phosphorylation of the CTDs of SPT5 and Pol II is required for the interaction of these proteins with a large number of partners mediating various cotranscriptional processes from mRNA capping, histone modifications, and transcription termination (Hsin and Manley 2012; Werner 2012; Zhou et al. 2012). Although the precise molecular details are not known, it is thought that a combination of interactions between Pol II, SPT5, and NELF mediates pausing of Pol II, with the P-TEFb-dependent phosphorylation of these factors disrupting these interactions (Hsin and Manley 2012; Zhou et al. 2012). The P-TEFb within SEC has emerged as the form of P-TEFb that is required for the release of Pol II at Hsp70, c-myc, c-fos, the HIV-LTR, and many of the developmentally regulated genes such as the Hox genes in ES cells (Fig. 1; Luo et al. 2012b). Much attention has also focused on the Brd4-associated form of P-TEFb (Zhou et al. 2012). Determining precisely how P-TEFb transitions between different complexes to regulate the various steps of the transcription cycle or the different classes of genes will be an important area of investigation (Bartholomeeusen et al. 2012; Zhou et al. 2012).
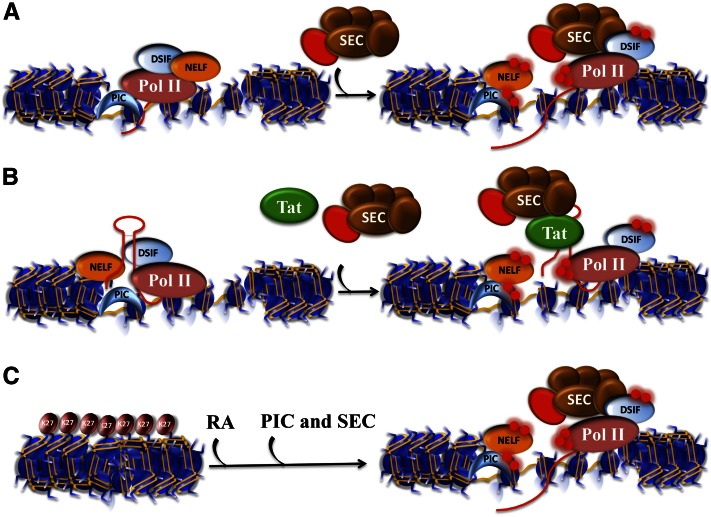
Figure 1.
Transcriptional activation with and without paused Pol II. (A) The classic example of transcriptional regulation by paused Pol II was demonstrated for the Hsp70 gene in Drosophila. Pol II is initially recruited to the gene promoter by the PIC (represented here with a TBP-shaped crescent). (Left panel) DSIF cooperates with NELF to keep Pol II within the first 40–70 nt of the transcription start site (TSS). (Right panel) Upon heat shock, the P-TEFb kinase (red sphere), as a component of the SEC, phosphorylates both DSIF and the Pol II CTD to allow the release from NELF, which allows productive elongation by Pol II, while DSIF becomes a positive factor for processivity. (B) Paused Pol II and its release by SEC have been extensively studied during transactivation of the HIV-1 provirus by the HIV-1 encoded Tat protein. Early in the viral life cycle, provirus transcription is inefficient until enough Tat protein has accumulated. Tat binds to a stem–loop structure early in the first 80 nt of the transcript from which it recruits SEC to cause the release of Pol II into productive transcription. (C) Transcriptional induction without paused Pol II. Recent studies in mouse ES cells revealed that one of the most rapidly induced genes in response to differentiation signals, Cyp26a1, lacks paused Pol II in the ES cell state and is covered by H3K27me3. In the presence of retinoic acid (RA), the PIC, Pol II can quickly initiate transcription, and in the presence of SEC, rapid transcriptional induction is ensued (see also Fig. 4A,B).
New regulators of pause–release
Recent yeast studies of nascent transcription suggest that a subset of genes may have some form of promoter-proximal pausing (Churchman and Weissman 2011; McKinlay et al. 2011), raising the possibility that alternate mechanisms for pausing may exist in yeast (Yearling et al. 2011). Furthermore, recent studies in mammalian cells have raised the possibility that there are multiple ways of regulating the release of paused Pol II, further increasing the complexity of transcriptional regulation at the level of TECC. One factor that was recently implicated in regulating paused Pol II is Gdown1, a protein associated with 30%–50% of Pol II (Hu et al. 2006). The Gdown1 form of Pol II was shown to require Mediator in activator-dependent in vitro transcription, while Pol II without Gdown1 did not require Mediator in these in vitro assays. Therefore, Gdown1 was seen as repressing transcription in a way that could be relieved by Mediator. Later studies indicated that Gdown1 and TFIIF compete with each other for association with Pol II (Cheng et al. 2012; Espinosa 2012; Jishage et al. 2012). Since TFIIF can act as both a preinitiation factor and an early elongation factor, Gdown1 could prevent transcription at either step. Genome-wide profiling of Gdown1 in HeLa cells shows that Gdown1 colocalizes extensively with Pol II and, despite being found on gene bodies, is particularly prominent at transcription start sites (Cheng et al. 2012). It was noted that on some genes, there were multiple peaks of Pol II and Gdown1, possibly due to alternative transcription start sites. There were many cases in which Gdown1 was strongly bound to both start sites, but Pol II was stronger on one site than the other. This could reflect differential pausing and release of Pol II at the alternate start sites, but differences in initiation cannot be ruled out by these assays.
While Gdown1 can antagonize TFIIF function (Cheng et al. 2012; Espinosa 2012; Jishage et al. 2012), Price and colleagues (Cheng et al. 2012) also found that Gdown1 can inhibit the activity of the Pol II termination factor TTF2. TTF2 is an SWI2/SNF2-related ATPase that has a prominent role in terminating transcription before chromosomes condense at mitosis (Jiang et al. 2004). Interestingly, TTF2 was recently found by the Bentley group (Brannan et al. 2012) to be associated with the XRN2 exonuclease. XRN2 degrades RNA in a 5′–3′ manner to facilitate termination of transcription using the 5′ end of the transcript created after cleavage by 3′ end processing enzymes. XRN2 is proposed to act as a torpedo, catching up to and helping to evict Pol I or Pol II, as the polymerases would otherwise continue transcribing well past the 3′ end cleavage site, which, in the case of Pol II, normally occurs just after the polyadenylation signal. Bentley and colleagues (Brannan et al. 2012) also found decapping enzymes in their XRN2 immunoprecipitates. Cytoplasmic decapping factors are used to destabilize transcripts by generating a 5′ phosphate for the 5′–3′ exonuclease XRN1. Chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) analysis to map the genome-wide distribution of XRN2, TTF2, and the decapping enzyme DCP1A demonstrated an association with Pol II at the 5′ end of genes. Furthermore, depletion of these factors by shRNA led to global changes in Pol II distribution. Therefore, it has been proposed that the decapping factors act to initiate the XRN2 torpedo in order to terminate Pol II at an early elongation stage (Fig. 2). Taking into account the work by Price and colleagues (Cheng et al. 2012), who suggested that Gdown1 might help to form a particularly stable form of paused Pol II, Gdown1 could regulate paused Pol II by preventing early termination by the TTF2/XRN2/DCP1A complex.
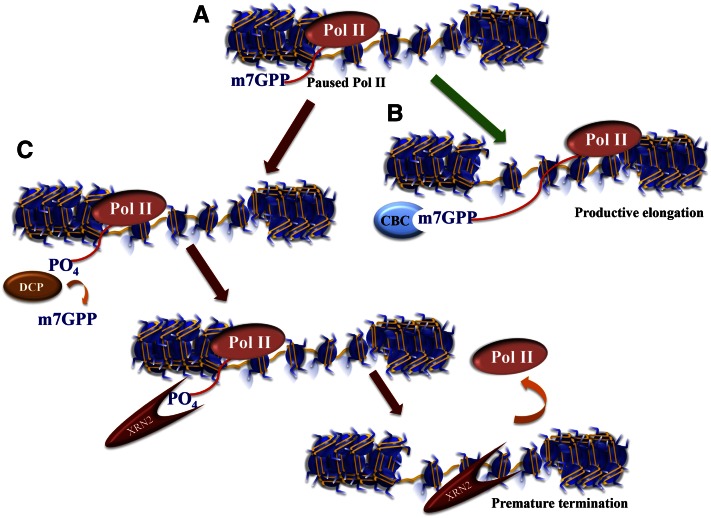
Figure 2.
Multiple ways to regulate paused Pol II. Recent studies have revealed other mechanisms to regulate paused Pol II in addition to the DSIF/NELF pause–release mechanism shown in Figure 1. (A) Paused Pol II is a nonproductive state producing short ∼50-nt transcripts that have already been modified with a methyl-7-guanosine cap (m7GPP) protecting the 5′ end of the mRNA from exonucleolytic cleavage. (B) Normally, the cap-binding protein complex (CBC) recognizes the m7G cap and stays with the transcript to facilitate RNA processing and the export of the RNA to the cytoplasm. (C) A decapping enzyme (DCP) can remove the m7G cap, thereby restoring the 5′ phosphate of the RNA, which allows the RNA to be degraded by the XRN2 exonuclease. The 5′ -to-3′ processivity of the XRN2 exonuclease helps push the stalling Pol II off the transcript, resulting in transcription termination. Note that an entry point for XRN2-dependent Pol II termination can also come about from any endonucleolytic cleavage event, such as when the miRNA processing factor Drosha cleaves the stem–loop structure of the TAR region of the HIV-1 transcript (also see Fig. 1).
XRN2 was also recently found to participate in premature termination of HIV transcription (Wagschal et al. 2012). Expression of the full-length RNA constituting the HIV genome requires the viral-encoded transcription activator Tat, which specifically recognizes a stem–loop structure (called the transcription activation response element [TAR]) that forms at the 5′ end of all HIV transcripts. Tat binding to TAR recruits P-TEFb within SEC, which can phosphorylate the Pol II CTD and promotes the production of full-length transcripts. Without Tat, HIV transcription is mostly unproductive, and most Pol II molecules transcribe a short distance and are considered to exist in a paused state. A small percentage of Pol II molecules are able to escape to make longer transcripts, which include those needed to express the Tat protein. Eventually, enough Tat protein is made to bind to TAR elements on nascent transcripts, and the full activation of HIV transcription begins. Wagschal et al. (2012) noticed that the TAR element resembled microRNAs (miRNAs) and asked whether the Drosha endonuclease required for miRNA cleavage regulated HIV transcription. They found that the knockdown of the Drosha levels led to activation of HIV transcription in the absence of Tat, indicating that Drosha was responsible for the nonproductive transcription of HIV. They proposed that Drosha stimulates the premature termination of HIV transcription by cleaving the TAR stem–loop structure. This cleavage generates a 5′ fragment containing the 7meG cap and a 3′ fragment that is associated with the engaged Pol II. The exposed 5′ phosphate of the 3′ fragment provides an entry point for the XRN2 torpedo, leading to premature termination of transcription. They proposed that in addition to the decapping/torpedo mechanism of termination observed by the Bentley group (Brannan et al. 2012), the Drosha-dependent cleavage is another path for the XRN2-dependent early termination of RNA Pol II.
A challenging area for the future will be to determine whether genes exhibiting engaged, nonproductive Pol II can be classified into being regulated by NELF/SPT5, XRN2/TTF2, or Gdown1 or to what extent combinations of these mechanisms can regulate the same gene. A subset of genes could have paused Pol II that is later released to elongate, while other genes exhibiting nonproductive Pol II might require termination of the proximally paused Pol II to allow a new productive form of Pol II to initiate a new transcript. However, both mechanisms appear to be used at the HIV LTR whose transcriptional regulation requires NELF for the pausing of Pol II (Zhang et al. 2007), SEC for releasing paused Pol II for productive elongation (He et al. 2010; Sobhian et al. 2010), and Drosha and XRN2 for controlling HIV-1 proviral transcription (Wagschal et al. 2012).
Regulating TECC in the context of chromatin
The in vitro transcription assays that have been used to identify elongation factors have traditionally used naked DNA templates. Attempts to use chromatin-containing templates for in vitro transcription were initially unsuccessful due to competition between nucleosomes and PIC formation (Lorch et al. 1987; Workman and Roeder 1987). The addition of nucleosome remodelers could restore the ability of Pol II to initiate on chromatin templates, but these transcripts were unable to elongate efficiently (Orphanides et al. 1998). Reinberg and colleagues (Orphanides et al. 1998, 1999) therefore assayed for factors that could facilitate transcription on chromatin templates and identified FACT, which is composed of two subunits, SPT16 and SSRP. SPT16 was identified in the same screen for factors affecting TY element transcription in budding yeast that identified SPT4 and SPT5, which were also independently identified biochemically as forming the elongation factor DSIF (Malone et al. 1991; Swanson et al. 1991; Hartzog et al. 1998; Wada et al. 1998; Kaplan et al. 2000, 2003). FACT directly interacts with H2A and H2B and facilitates both the disassembly and reassembly of nucleosomes associated with transcription by acting as a histone chaperone (Winkler and Luger 2011). SPT6, another hit in the TY transcription screen, also acts as a histone chaperone. In vivo, SPT6 associates with the Ser 2 phosphorylated CTD of Pol II (Yoh et al. 2007). Genome-wide studies in yeast have shown that SPT6 and SPT16 enter and leave the transcription unit at different places, indicating that they have nonredundant roles in vivo (Mayer et al. 2010).
Although FACT was identified as a factor that assisted RNA Pol II to transcribe chromatin templates, a function that seemingly would be required at all transcribed loci, FACT has also been implicated in the control of developmentally regulated genes. FACT subunits SPT16 and SSRP have been identified as interactors of TIF1γ, an E3 ubiquitin ligase that belongs to a family of coactivators and corepressors. TIF1γ is a major regulator of erythropoiesis from zebrafish to mammals (Ransom et al. 2004; He et al. 2006; Bai et al. 2010, 2013; Kusy et al. 2011) and is recruited to genes through its interactions with the SCL transcription factor complex (Schuh et al. 2005; Bai et al. 2010).
TIF1γ was first found to be involved in hematopoiesis when mutations in moonshine, shown to encode a TIF1γ ortholog, had severe defects in the differentiation of red blood cells (Ransom et al. 2004). Mutations in moonshine were used in the first genetic suppressor screen ever performed in a vertebrate system, which identified the zebrafish homolog of Cdc73 (named sunrise), a subunit of the PAF complex (Bai et al. 2010). The PAF complex has long been linked to transcription elongation through its association with Pol II, playing a role as a platform for the association of different factors mediating numerous cotranscriptional events, such as histone modifications and RNA processing events (for review, see Gerber and Shilatifard 2003; Sims et al. 2004). The foggy mutation, which lacks the negative elongation activity of SPT5, was also shown to suppress moonshine mutant phenotypes, suggesting a positive elongation activity dependent on TIF1γ. Defects in transcription elongation were demonstrated for several erythroid lineage genes in moonshine mutant zebrafish (Bai et al. 2010). Together with the biochemical association with the FACT complex, these molecular and genetic studies point to a role for TIF1γ in regulating FACT activity for the control of gene expression at the level of transcription elongation during development (Fig. 3).
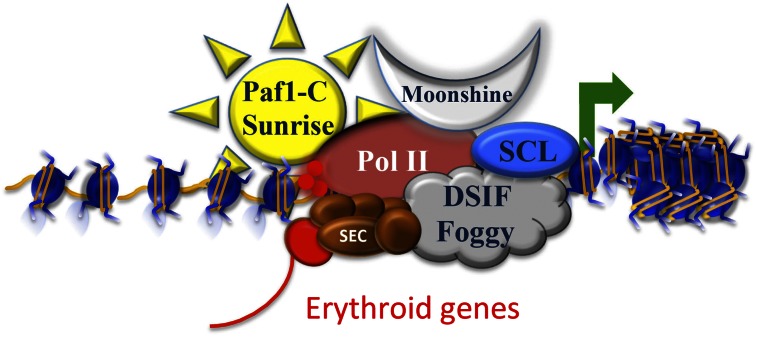
Figure 3.
Regulating paused Pol II in the context of chromatin. TIF1γ associates with histone chaperones and elongation factors to stimulate gene expression during erythropoiesis. Mutations in moonshine, encoding zebrafish TIF1γ, could be suppressed by mutations in sunrise, encoding the PAF-c subunit CDC73, and by mutations in foggy, which is an allele of the DSIF subunit SPT5, which loses its ability to maintain the paused Pol II state. The SCL complex, containing the basic helix–loop–helix (bHLH) SCL transcription factor, could recruit TIF1γ and the histone chaperones SPT6 and SPT16/FACT to remodel nucleosomes to facilitate Pol II release. The most active form of P-TEFb (red sphere) within SEC is involved in Pol II CTD phosphorylation and the release from the paused state. These findings implicate transcription elongation as a major regulatory step for the control of erythroid gene expression.
The FACT subunit SPT16 was also recently identified as physically interacting with JMJD3, a histone H3K27-demethylating enzyme in human cells (Chen et al. 2012). JMJD3 is associated with a second H3K27 demethylase, KIAA1718; the histone chaperone SPT6; the SPT6 interactor; and Pol II CTD-binding protein IWS1 as well as the H3K36 methyltransferase SET2, itself an interactor with the Pol II CTD. Using PMA induction for differentiation of HL60 cells as a model system, JMJD3 activity was shown to be required for the recruitment of SPT6 and SPT16 as well as for proper levels of the Ser 2 phosphorylated form of Pol II in the gene bodies of target genes. JMJD3 was found at many genes bearing paused Pol II and the bivalent H3K4me3/H3K27me3 mark, and its presence was required for their induction after TPA treatment.
While more work needs to be done in order to understand the mechanisms by which TIF1γ and JMJD3 regulate transcription elongation, both studies link the FACT subunits SPT16 and SPT6 to the regulation of genes through the release of paused Pol II. The copurification of SPT6 with SPT16 in the TIF1γ and JMJD3 purifications suggests that these factors may work in some coordinated manner to regulate the transcription elongation processes on chromatin. Although the histone chaperone activities of SPT6 and FACT may facilitate Pol II translocation through nucleosomes, both have been shown to have effects on transcription elongation outside the context of chromatin: SPT6 can stimulate transcription elongation by Pol II (Endoh et al. 2004), and FACT can relieve DSIF/NELF's transcription inhibitory activity, both on naked DNA templates in vitro (Wada et al. 2000). Therefore, interactions with Pol II and/or DSIF/NELF could also contribute to these factors' abilities to facilitate transcription elongation in vivo.
Establishment of the TECC in stem cells
Genome-wide studies in both Drosophila embryos and mouse ES cells have shown that proximal pausing of Pol II is found at many developmentally controlled genes well before their later activation in response to differentiation cues (Guenther et al. 2007; Muse et al. 2007; Zeitlinger et al. 2007). While several factors such as NELF are known to be required for the paused Pol II state, how this state is established in the early embryonic stage is largely unknown. Sequence-specific binding factors may help establish paused Pol II at specific promoters in the early embryo (Levine 2011). The Drosophila protein Zelda can recognize a specific motif, CAGGTAG, found at promoters and enhancers of many developmental genes, and Zelda is present at these genes before zygotic transcription has begun (Liang et al. 2008; Harrison et al. 2011; Levine 2011).
Although no Zelda-like protein has been found in mammals, early enhancer marking by a non-DNA-binding protein was shown for ELL3, which, like its homolog, ELL, can function as an elongation factor in vitro (Miller et al. 2000; Lin et al. 2013). ELL3 was found at some of the active, inactive, and poised enhancers in ES cells, being cobound with Mediator and cohesin. Mediator and cohesin were previously shown to be required for enhancer–promoter communication through chromatin looping (Kagey et al. 2010). ELL3's binding to some of the enhancers required cohesin. Additionally, loss of ELL3 led to reduced paused Pol II occupancy at some developmental genes (Lin et al. 2013). Induction of many lineage-specific genes requires the ELL2-containing SEC, and reductions in the levels of ELL3 can lead to defective induction of these genes. This observation suggests that there may be a handoff from ELL3, which helped establish the paused Pol II state and SEC that can mediate the release of paused Pol II during gene activation. In the absence of a Zelda-like protein in mammals, it is unclear at this time how Ell3, Mediator, and cohesin recognize enhancers (Lin et al. 2013).
ELL3 was originally described as a protein enriched in testes, but, recently, it was shown that ELL3 is also enriched in oocytes, with lower levels in ES cells and declining expression during embryoid body formation (Miller et al. 2000; Ahn et al. 2012). Immunogold electron microscopy showed that ELL3 and RNA Pol II are present in sperm nuclei, suggesting that transcription factors could be marking enhancers and promoters as early as the germ cell state (Lin et al. 2013). These studies raise the question of how early factors such as ELL3, Mediator, cohesin, and other transcription factors are brought to the enhancers and what the signals are that initiate enhancer–promoter communication establishing the paused Pol II state.
TECC and the biological significance of pausing
It has been widely assumed that the function of setting up paused Pol II at genes before their induction is to allow for a more rapid induction of the genes, since many of the steps associated with initiation have already taken place (Nechaev and Adelman 2011). This assumption was based in large part on the extraordinarily rapid induction of the Drosophila Hsp70 gene within seconds after heat shock (Boehm et al. 2003). However, human Hsp70—which, like its Drosophila counterpart, has paused Pol II that is released by SEC—instead takes hours to be induced, suggesting that paused Pol II is not just for rapid induction (Lin et al. 2010). Furthermore, among all of the genes induced in ES cells by retinoic acid, the most rapidly induced gene was identified as Cyp26a1, which lacks paused Pol II (Fig. 4A; Lin et al. 2011). Genes such as Hoxa1 that are rapidly induced, but whose levels need to be carefully regulated during development, have paused Pol II, suggesting that well-regulated expression is a major function of setting up paused Pol II at inducible genes. Collectively, the studies by Lin et al. (2011) suggest that genes bearing paused Pol II are equitably induced, while the presence of paused Pol II is not a prerequisite for rapid transcriptional induction (Fig. 4B).
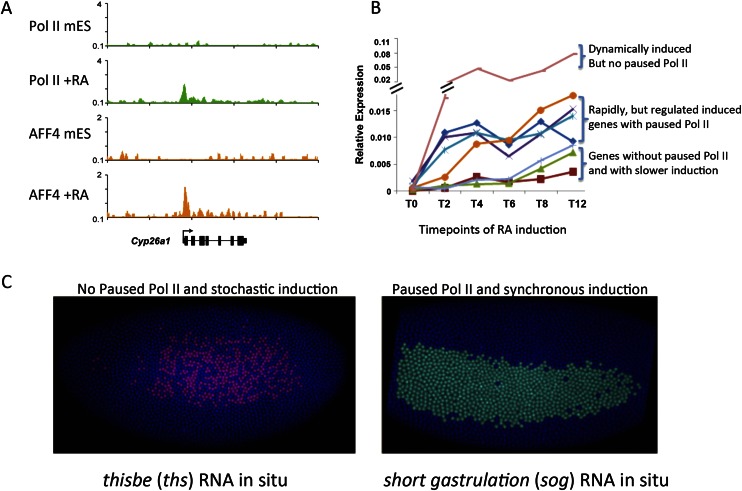
Figure 4.
Paused Pol II in equitable and synchronous transcriptional induction. (A) Rapid induction of gene expression can occur without prior paused Pol II. In mouse ES cells (mES), Pol II is absent from the promoter of Cyp26a1. Upon retinoic acid (RA) treatment, Pol II is recruited to Cyp26a1 along with the SEC component AFF4 (Lin et al. 2011). (B) Time course of transcriptional induction in retinoic acid-treated mouse ES cells. Cyp26a1 (pink line/top bracket), which A shows lacks paused Pol II in the ES state, is the most rapidly (dynamically) induced gene in response to retinoic acid treatment. Many genes that are rapidly induced by retinoic acid treatment had paused Pol II in mouse ES cells, although they are induced in a more regulated or equitable manner (middle bracket). This study demonstrated that paused Pol II is not a prerequisite for rapid transcriptional induction (Lin et al. 2011). (C) Paused Pol II correlates with the synchronous induction of genes in early Drosophila embryogenesis. Two genes, thisbe (ths) and short gastrulation (sog), can be distinguished by the absence of paused Pol II in very early embryos. (Left panel) ths in situ hybridization shows that ths, which lacked paused Pol II prior to induction, has a stochastic induction with a nonuniform pattern of expression. (Right panel) A gene with paused Pol II in the early embryo is sog, which, like ths, is induced in the neurogenic ectoderm but has a more uniform induction pattern. Therefore, Boettiger and Levine (2009) propose that the prevalence of paused Pol II on developmentally regulated genes is to ensure a well-regulated, less stochastic expression during embryogenesis. A and B are modified with permission from Genes & Development (Lin et al. 2011). C is modified from Levine 2011 with permission from Elsevier (© 2011).
Similarly, analysis of a Gro-seq study of estrogen response genes (Hah et al. 2011) showed that these genes could also be rapidly induced without prior paused Pol II (Gilchrist et al. 2012). Furthermore, a study of the Drosophila innate immune response pathway showed that rapidly induced anti-bacterial genes lack paused Pol II, while the master regulators of the immune pathway, whose levels need to be precisely controlled, have paused Pol II (Gilchrist et al. 2012). An elegant demonstration of the importance of paused Pol II in development comes from in situ hybridization studies of Drosophila embryos (Fig. 4C). Genes with paused Pol II were more synchronously induced compared with genes without Pol II that showed stochastic induction (Boettiger and Levine 2009), suggesting that genes without prior paused Pol II are subject to stochastic steps of preinitiation.
Summary and future directions
The past several years have seen a growing appreciation of transcription elongation as a critical aspect of the regulation of development. The number of elongation factors and the diverse ways in which they regulate post-initiation events will continue to expand at a rapid rate due to this heightened interest. Elegant genome-wide techniques that look at nascent transcription have provided an unprecedented view of transcribing Pol II (Core et al. 2008; Churchman and Weissman 2011; Khodor et al. 2011), and the power of these techniques for helping solve some of our remaining challenges will be even greater as they are used in combination with experimental perturbations such as the RNAi-mediated knockdown of elongation factors.
An emerging theme in the transcription field is that different elongation factors are required for different classes of genes. The ELL family has prominent roles in small nuclear RNA transcription as part of the little elongation complex (LEC) (Smith et al. 2011), in the rapid gene activation of developmental genes as part of SEC, and in enhancer–promoter communication for setting up the paused Pol II state and response to developmental signals (Lin et al. 2013). Even within the same complex, different factors have context-specific roles. For example, SPT4, which, with SPT5, forms DSIF, has a particular requirement for transcription of tracts of trinucleotide repeats (Liu et al. 2012). Like DSIF, which has both positive and negative elongation activities, the PAF complex, which is generally thought of as a positive elongation factor, is antagonistic to the productive elongation of erythroid genes, as evidenced by the suppression of the TIF1γ mutant in zebrafish (Schuh et al. 2005; Bai et al. 2010). Some of the diversity in elongation factors is matched by the multitude of challenges that Pol II encounters in different contexts. Discovering the diverse roles and regulation of P-TEFb as part of SEC and SEC-like complexes and within the Brd4 complex is an important challenge (Luo et al. 2012a; Zhou et al. 2012).
Recent studies of promoter-proximally paused Pol II suggest that there may be more than one way to regulate pausing, with Gdown1, TTF2, and XRN2 as new candidates of regulating the pause and release of Pol II (Brannan et al. 2012; Cheng et al. 2012; Wagschal et al. 2012). The paused Pol IIs observed at housekeeping genes, heat-shock loci, or developmental genes could each be controlled by different mechanisms and for different purposes, whether being dynamically or synchronously regulated. As with any step of transcription, knowing the underlying DNA features and their sequence-specific DNA-binding factors will help in understanding how specific elongation factors are recruited to specific classes of genes. Understanding the role of long-range communication between enhancer and promoter elements and the way that cohesin, Mediator, and elongation factors cooperate in this process will undoubtedly be part of the solution to understanding the intricate control of the pausing and release of Pol II in development and their perturbation in human disease.
Acknowledgments
We are grateful to Laura Shilatifard for editorial assistance. This work was supported in part by funds provided by the National Institute of Health (R01CA89455 and R01CA150265) to A.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.215137.113.
References
- Ahn HJ, Cha Y, Moon SH, Jung JE, Park KS 2012. Ell3 enhances differentiation of mouse embryonic stem cells by regulating epithelial–mesenchymal transition and apoptosis. PLoS ONE 7: e40293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC 1995. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science 269: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, et al. 2010. TIF1γ controls erythroid cell fate by regulating transcription elongation. Cell 142: 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Trowbridge JJ, Riley E, Lee JA, Dibiase A, Kaartinen VM, Orkin SH, Zon LI 2013. TiF1-γ plays an essential role in murine hematopoiesis and regulates transcriptional elongation of erythroid genes. Dev Biol 373: 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM 2012. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem 287: 36609–36616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol 23: 7628–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M 2009. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. 2012. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell 46: 311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W, Lv R, Li X, Villen J, Gygi SP, et al. 2012. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev 26: 1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, et al. 2012. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell 45: 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M 2009. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol 19: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A 2002. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci 99: 9894–9899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, et al. 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol 24: 3324–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM 2012. Get back TFIIF, don't let me Gdown1. Mol Cell 45: 3–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P, Rech J, Vie A, Piechaczyk M, Bonnieu A, Jeanteur P, Blanchard JM 1987. Regulation of c-fos gene expression in hamster fibroblasts: Initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res 15: 5657–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem 278: 26303–26306 [DOI] [PubMed] [Google Scholar]
- Gerber M, Eissenberg JC, Kong S, Tenney K, Conaway JW, Conaway RC, Shilatifard A 2004. In vivo requirement of the RNA polymerase II elongation factor elongin A for proper gene expression and development. Mol Cell Biol 24: 9911–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K 2012. Regulating the regulators: The pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev 26: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT 1986. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol 6: 3984–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Wilson SW, Cooke S, Chitnis AB, Driever W, Rosenthal A 1999. Mutations in the zebrafish unmask shared regulatory pathways controlling the development of catecholaminergic neurons. Dev Biol 208: 473–487 [DOI] [PubMed] [Google Scholar]
- Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A 2000. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408: 366–369 [DOI] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet 7: e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J 2006. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell 125: 929–941 [DOI] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Manley JL 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26: 2119–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt A 2006. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci 103: 9506–9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liu M, Spencer CA, Price DH 2004. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol Cell 14: 375–385 [DOI] [PubMed] [Google Scholar]
- Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, Chait BT, Gnatt A, Ren B, Roeder RG 2012. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol Cell 45: 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F 2000. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev 14: 2623–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT II, Rosbash M 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Liu CL, Yassour M, Holik J, Friedman N, Buratowski S, Rando OJ 2010. RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome Biol 11: R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev 6: 2201–2213 [DOI] [PubMed] [Google Scholar]
- Kusy S, Gault N, Ferri F, Lewandowski D, Barroca V, Jaracz-Ros A, Losson R, Romeo PH 2011. Adult hematopoiesis is regulated by TIF1γ, a repressor of TAL1 and PU.1 transcriptional activity. Cell Stem Cell 8: 412–425 [DOI] [PubMed] [Google Scholar]
- Levine M 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A 2010. AFF4, a component of the ELL/P–TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A 2011. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 25: 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A 2013. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CR, Chang CR, Chern Y, Wang TH, Hsieh WC, Shen WC, Chang CY, Chu IC, Deng N, Cohen SN, et al. 2012. Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell 148: 690–701 [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD 1987. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49: 203–210 [DOI] [PubMed] [Google Scholar]
- Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A 2012a. The super elongation complex family of RNA polymerase II elongation factors: Gene target specificity and transcriptional output. Mol Cell Biol 32: 2608–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A 2012b. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547 [DOI] [PubMed] [Google Scholar]
- Malone EA, Clark CD, Chiang A, Winston F 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol 11: 5710–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev 11: 2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270: 12335–12338 [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- McKinlay A, Araya CL, Fields S 2011. Genome-wide analysis of nascent transcription in Saccharomyces cerevisiae. G3 (Bethesda) 1: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Williams K, Johnstone RW, Shilatifard A 2000. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem 275: 32052–32056 [DOI] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A 2010. Licensed to elongate: A molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 10: 721–728 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K 2011. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288 [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Ransom DG, Bahary N, Niss K, Traver D, Burns C, Trede NS, Paffett-Lugassy N, Saganic WJ, Lim CA, Hersey C, et al. 2004. The zebrafish moonshine gene encodes transcriptional intermediary factor 1γ, an essential regulator of hematopoiesis. PLoS Biol 2: e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Conaway JW, Conaway RC 1996. The RNA polymerase II general elongation factors. Trends Biochem Sci 21: 351–355 [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT 2006. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7: 557–567 [DOI] [PubMed] [Google Scholar]
- Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, Rodriguez P, Strouboulis J, Enver T, Vyas P, et al. 2005. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol Cell Biol 25: 10235–10250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 1998. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J 12: 1437–1446 [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271: 1873–1876 [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW 2003. The RNA polymerase II elongation complex. Annu Rev Biochem 72: 693–715 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D 2004. Elongation by RNA polymerase II: The short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Smith ER, Winter B, Eissenberg JC, Shilatifard A 2008. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci 105: 8575–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, et al. 2011. The little elongation complex regulates small nuclear RNA transcription. Mol Cell 44: 954–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol 11: 4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD 1994. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci 91: 12110–12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Orphanides G, Hasegawa J, Kim DK, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, et al. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell 5: 1067–1072 [DOI] [PubMed] [Google Scholar]
- Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, et al. 2012. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell 150: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F 2012. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol 417: 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler DD, Luger K 2011. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J Biol Chem 286: 18369–18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Roeder RG 1987. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell 51: 613–622 [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21: 227–237 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51 [DOI] [PubMed] [Google Scholar]
- Yan Q, Moreland RJ, Conaway JW, Conaway RC 1999. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem 274: 35668–35675 [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Bhatt S, Takeuchi T, Kawauchi J, Takahashi H, Tsutsui A, Muraoka T, Inoue M, Tsuda M, Kitajima S, et al. 2012. Transcriptional elongation factor elongin A regulates retinoic acid-induced gene expression during neuronal differentiation. Cell Rep 2: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Yearling MN, Radebaugh CA, Stargell LA 2011. The transition of poised RNA polymerase II to an actively elongating state is a ‘complex’ affair. Genet Res Int 2011: 206290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA 2007. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev 21: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Gilmour DS, Henderson AJ 2007. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem 282: 16981–16988 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH 1997. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev 11: 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]