Programmed necrosis is induced in response to several forms of cellular stress. Fu et al. find that AlkB homolog 7 (ALKBH7) triggers the collapse of mitochondrial membrane potential and loss of mitochondrial function that lead to energy depletion and cell death. Depletion of ALKBH7 suppresses necrotic cell death induced by numerous alkylating and oxidizing agents by maintaining their mitochondrial membrane potential. These results uncover a role for a mammalian AlkB homolog in programmed necrosis, presenting a new target in cancer cells that are resistant to apoptotic cell death.
Keywords: AlkB, ALKBH7, death, DNA damage, necrosis
Abstract
Programmed necrosis has emerged as a crucial modulator of cell death in response to several forms of cellular stress. In one form of programmed necrotic cell death, induced by cytotoxic alkylating agents, hyperactivation of poly-ADP-ribose polymerase (PARP) leads to cellular NAD and ATP depletion, mitochondrial dysfunction, reactive oxygen species formation, and ensuing cell death. Here, we show that the protein encoded by the human AlkB homolog 7 (ALKBH7) gene plays a pivotal role in DNA-damaging agent-induced programmed necrosis by triggering the collapse of mitochondrial membrane potential and large-scale loss of mitochondrial function that lead to energy depletion and cellular demise. Depletion of ALKBH7 suppresses necrotic cell death induced by numerous alkylating and oxidizing agents while having no effect on apoptotic cell death. Like wild-type cells, ALKBH7-depleted cells undergo PARP hyperactivation and NAD depletion after severe DNA damage but, unlike wild-type cells, exhibit rapid recovery of intracellular NAD and ATP levels. Consistent with the recovery of cellular bioenergetics, ALKBH7-depleted cells maintain their mitochondrial membrane potential, plasma membrane integrity, and viability. Our results uncover a novel role for a mammalian AlkB homolog in programmed necrosis, presenting a new target for therapeutic intervention in cancer cells that are resistant to apoptotic cell death.
Programmed cell death can occur by a number of mechanisms that include classical apoptosis plus several recently described nonapoptotic pathways, such as lysosomal-mediated cell death, cell death with autophagy, and programmed necrosis (for review, see Kroemer and Jaattela 2005; Vandenabeele et al. 2010; Green et al. 2011; Vanlangenakker et al. 2012). One type of programmed necrosis, termed necroptosis, has emerged as a critical factor associated with a variety of pathological processes, including acute pancreatitis, inflammation, excitotoxicity, and cardiac cell death during ischemia–reperfusion (for review, see Vanlangenakker et al. 2012). In addition, necrosis is critical for an optimal immune response to bacterial and viral pathogens (for review, see Mocarski et al. 2012), underscoring the crucial role for programmed necrosis in maintaining organismal health.
Programmed necrosis can be induced by extensive DNA damage caused by oxidizing agents or cancer chemotherapeutic drugs such as alkylating agents (Yu et al. 2002; Du et al. 2003; Zong et al. 2004; Zong and Thompson 2006; Moubarak et al. 2007; Oka et al. 2008). DNA damage-induced necrosis is characterized by the hyperactivation of NAD+-dependent poly-ADP-ribose polymerase (PARP), which leads to intracellular NAD+ depletion, loss of ATP production, mitochondria depolarization, generation of reactive oxygen species (ROS), and eventual loss of total cellular bioenergetics (Ha and Snyder 1999; Cipriani et al. 2005; Alano et al. 2010; Tang et al. 2010; Goellner et al. 2011; Luo and Kraus 2012). In addition to mitochondrial dysfunction, severe DNA damage can induce the translocation of the apoptosis-inducing factor (AIF) from the mitochondria to the nucleus, where AIF forms a DNA-degrading complex with histone H2AX and cyclophilin D (Yu et al. 2002; Baritaud et al. 2010). This cascade of events contributes to the ultimate demise of the cell, characterized by cell swelling, large-scale chromosome fragmentation, and eventual plasma membrane disintegration (Vandenabeele et al. 2010).
Cells possess numerous mechanisms to repair DNA damage and counteract the cytotoxic and mutagenic effects of DNA damage. One of the most recently discovered repair mechanisms for alkylated DNA bases is direct DNA damage reversal catalyzed by the bacterial AlkB dioxygenase (for review, see Sedgwick et al. 2007; Yi and He 2013). The Escherichia coli AlkB protein has been characterized as a DNA repair enzyme that catalyzes oxidative demethylation of the cytotoxic alkyl DNA base lesions 1-methyladenine (1meA) and 3-methylcytosine (3meC) (Falnes et al. 2002; Trewick et al. 2002). The mammalian genome encodes nine AlkB homologs (ALKBH1 through ALKBH8 plus the FTO gene product). Of the nine ALKBH proteins, ALKBH1, ALKBH2, ALKBH3, and FTO display DNA repair activity on methylated DNA substrates in vitro (Duncan et al. 2002; Aas et al. 2003; Gerken et al. 2007; Jia et al. 2008; Westbye et al. 2008). Mouse knockout studies have shown that ALKBH2 is the major repair protein for 1meA in DNA (Ringvoll et al. 2006), while ALKBH3 has been shown to play a role in the repair of 3meC lesions in human cells (Dango et al. 2011).
Besides the repair of alkylated DNA base substrates, bacterial AlkB, ALKBH1, ALKBH2, ALKBH3, and FTO can repair methylated RNA bases in vitro and potentially in vivo using the same oxidative demethylation reaction as for DNA (Aas et al. 2003; Ougland et al. 2004; Lee et al. 2005; Jia et al. 2008; Westbye et al. 2008; Vagbo et al. 2013). Moreover, mammalian ALKBH8 has been shown to modify specific tRNAs, which in turn modulate the DNA damage response and enhance survival (Fu et al. 2010; Songe-Moller et al. 2010), thus expanding the role of mammalian ALKBH enzymes beyond nucleic acid repair to include RNA modification. Even more recently, ALKBH5 has been shown to catalyze the demethylation of N(6)-methyladenosine in mRNA to affect mRNA export and RNA metabolism (Zheng et al. 2013). The large number of AlkB homologs in mammalian cells, combined with their emerging functions in diverse cellular processes, suggests additional biological roles for the remaining ALKBH proteins.
Here, we show that one of the human AlkB homologs, ALKBH7, is a mitochondrial protein required for programmed cell necrosis induced by alkylating agents or oxidative stress. Loss of ALKBH7 suppresses such necrosis and prevents the loss of mitochondrial function and cellular bioenergetics induced by DNA-damaging agents that lead to necrotic cell death. The findings in this study identify a novel cellular role for an ALKBH dioxygenase protein in the execution of programmed necrotic cell death induced by genotoxic stress.
Results
ALKBH7 is a nuclear-encoded protein that is imported into mitochondria
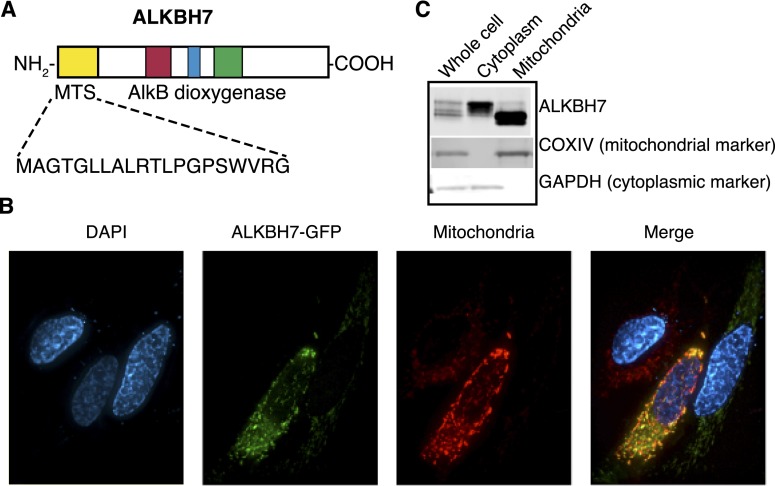
Previous studies by our laboratory and others have shown that mammalian ALKBH proteins localize to different subcellular compartments; namely, the nucleus, cytoplasm, and mitochondria (Aas et al. 2003; Tsujikawa et al. 2007; Westbye et al. 2008; Fu et al. 2010; Thalhammer et al. 2011). Based on multiple subcellular localization prediction algorithms, it appears that ALKBH7 contains a mitochondrial targeting sequence (MTS) at the N terminus (Fig. 1A). Through protein tagging with a C-terminal green fluorescent protein (GFP), we found that ALKBH7 exhibits a punctate-like pattern of localization that does indeed coincide with mitochondria (Fig. 1B). In addition, we found that transiently expressed ALKBH7 fractionates into two distinct pools consisting of a larger isoform found in the cytoplasm and a faster-migrating species found only in purified mitochondria (Fig. 1C). The presence of a truncated form of ALKBH7 in the mitochondria is most likely due to processing of the N-terminal mitochondrial localization signal after import from the cytoplasm, a common feature of many nuclear-encoded proteins that exhibit mitochondrial localization. The localization of human ALKBH7 in mitochondria is consistent with the mitochondrial localization of endogenous mouse ALKBH7 (Solberg et al. 2013). Finally, comprehensive proteomic analysis from two independent studies has detected endogenous ALKBH7 in the purified mitochondria of both human and mouse tissues (Pagliarini et al. 2008; Rhee et al. 2013). These criteria establish ALKBH7 as a nuclear-encoded protein that is translated in the cytoplasm and imported into mitochondria, suggesting that ALKBH7 plays a biological role in the mitochondria.
Figure 1.
ALKBH7 is targeted to mitochondria. (A) ALKBH7 contains a MTS at the N terminus, as predicted through primary amino acid sequence analysis and subcellular localization prediction. (B) Transiently expressed ALKBH7-GFP colocalizes with mitochondria in HeLa human cervical carcinoma cells. (C) ALKBH7 fractionates into a higher-molecular-weight species in the cytoplasm and a lower-molecular-weight form in purified mitochondrial fractions, indicative of mitochondrial import and processing of ALKBH7.
In addition to ALKBH7, another human AlkB homolog protein, ALKBH1, has been shown to be located in the mitochondria even though it lacks a detectable mitochondrial localization signal (Westbye et al. 2008). ALKBH1 catalyzes the direct repair of 3meC lesions in ssDNA in vitro (Westbye et al. 2008), suggesting that mitochondrial AlkB enzymes play a role in mitochondrial DNA repair. However, we found that purified human ALKBH7 lacked dioxygenase-mediated repair activity for both 1meA and 3meC lesions in either ssDNA or dsDNA oligonucleotides (Supplemental Fig. S1). The lack of oxidative demethylation activity exhibited by human ALKBH7 has also been observed with the mouse Alkbh7 protein (Lee et al. 2005). While ALKBH7 did not display DNA demethylase activity, we detected robust DNA repair activity for the ALKBH2 and ALKBH3 enzymes on both 1meA and 3meC DNA adducts (Supplemental Fig. S1), as previously described (Aas et al. 2003; Lee et al. 2005). Our studies indicate that ALKBH7 could play a unique cellular role in mitochondria distinct from known ALKBH functions in nucleic acid repair.
Depletion of ALKBH7 confers resistance rather than sensitivity to DNA-damaging agents
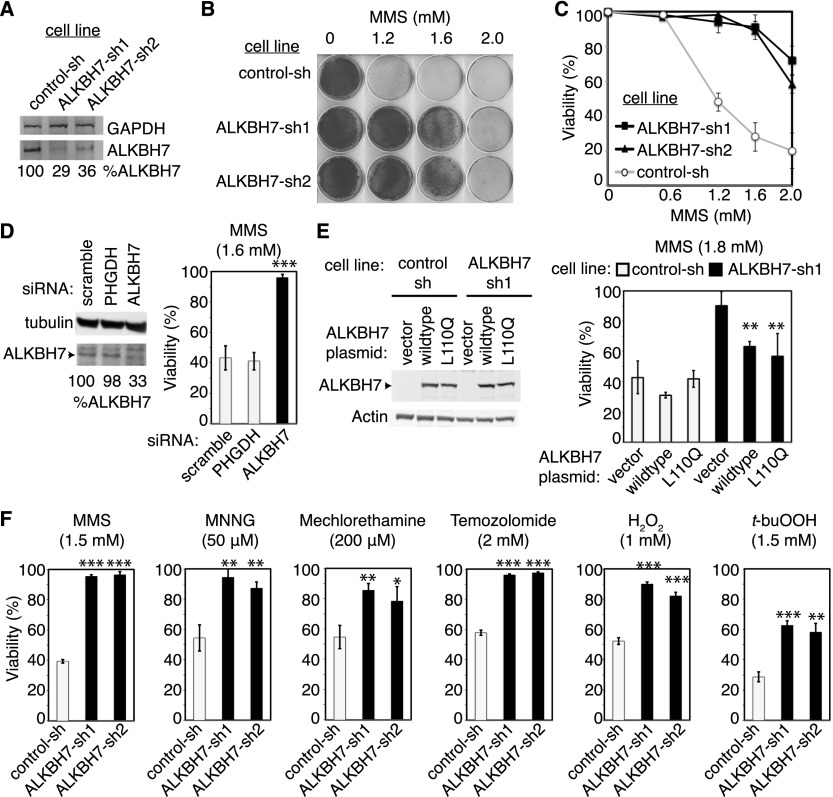
We showed previously that the mammalian AlkB homolog ALKBH8 confers cellular resistance to DNA-damaging agents by catalyzing tRNA modification, which in turn modulates the DNA damage response (Fu et al. 2010). Thus, AlkB homologs can influence cellular sensitivity to DNA damage through mechanisms other than direct DNA repair. To determine whether ALKBH7 plays a role in modulating sensitivity to DNA-damaging agents, we generated 293T human embryonic kidney fibroblast cell lines that were stably depleted of the ALKBH7 protein using lentiviral shRNA expression constructs targeting different regions of the ALKBH7 mRNA transcript (Fig. 2A). To our surprise, we found that ALKBH7-depleted cell lines became resistant to cell death induced by the alkylating agent methyl methanesulfonate (MMS) (Fig. 2B,C). Cell death was significantly reduced in ALKBH7-depleted cells even at doses of MMS that reduced viability to ∼20% in control 293T cells. Transient knockdown of ALKBH7 in 293T cells using siRNAs also conferred resistance to alkylating agents, recapitulating the results using stable shRNA vectors (Fig. 2D).
Figure 2.
Depletion of ALKBH7 in 293T human embryonic kidney cells confers resistance to alkylating and oxidizing agents. (A) Immunoblot analysis of ALKBH7 levels in 293T cells expressing a nonsilencing shRNA (control-sh) or either of two shRNAs targeting different regions of the ALKBH7 mRNA transcript (ALKBH7-sh1 and ALKBH7-sh2). The amount of ALKBH7 remaining in each cell type (%ALKBH7) is expressed relative to the control-sh cell line after normalization to the levels of GAPDH. (B) Representative growth of control-sh versus ALKBH7-depleted 293T cell lines after treatment with the indicated amount of MMS. (C) Viability of control-sh and ALKBH7-depleted 293T cell lines after treatment with the indicated concentration of MMS, as measured by trypan blue dye exclusion. (D) Immunoblot analysis and post-MMS viability of 293T cells transfected with nonsilencing siRNA (scramble) or pooled siRNAs targeting a non-ALKBH protein-coding transcript (PHGDH) or ALKBH7 mRNA. (E) Confirmation of ALKBH7 expression in 293T cell lines transfected with plasmids expressing the indicated ALKBH7 protein and viability of the transfected cells after treatment with MMS. (F) Viability of control-sh and ALKBH7-depleted cell lines after exposure to the indicated alkylating or oxidizing agent. Cells were treated with MMS, MNNG, H2O2, and t-buOOH at the indicated concentration for 1 h in serum-free medium, while mechlorethamine and temozolomide were left in culture medium until analysis. Error bars represent the standard deviation of at least three independent experiments (C,D,F) or three technical replicates (E). (*) P < 0.05 (***); (**) P < 0.01; P < 0.001.
To confirm that ALKBH7 is indeed required for cell death in response to MMS, we tested whether re-expression of ALKBH7 in an ALKBH7-depleted cell line could suppress the MMS-resistant phenotype exhibited by ALKBH7-depleted cells. We transiently transfected either an empty vector plasmid or an ALKBH7-expressing construct into the control-sh or ALKBH7-sh cell lines, followed by treatment with MMS (Fig. 2E). As expected, control shRNA cell lines transfected with the empty vector remained sensitive to MMS, while the ALKBH7-depleted cell line transfected with the empty vector remained resistant (Fig. 2E). In contrast, transfection of the ALKBH7 expression construct into ALKBH7-depleted cells significantly decreased their resistance to MMS (Fig. 2E). Due to incomplete transfection, not all cells were transfected with the ALKBH7 plasmid; hence, sensitivity to MMS was increased but not completely restored to the level of the control-sh cell line. Interestingly, expression of an ALKBH7 protein containing a mutation in the putative 2-oxoglutarate-binding site (L110Q) could also partially suppress MMS resistance in ALKBH7-depleted cells, suggesting that 2-oxoglutarate binding is dispensable for the prodeath function of ALKBH7.
To determine whether the resistant phenotype of ALKBH7-depleted cells was specific to MMS, we tested other alkylating agents that differ from MMS in their chemical reactivity; these include N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and the cancer chemotherapeutic alkylating drugs mechlorethamine (nitrogen mustard) and temozolomide. Importantly, MNNG, mechlorethamine, and temozolomide induce a different spectrum of DNA alkyl base lesions than MMS, since MNNG, mechlorethamine, and temozolomide are SN1 alkylating agents, while MMS is an SN2 alkylating agent. We found that ALKBH7-depleted cell lines were resistant to cell death induced by all three compounds (Fig. 2F), indicating that ALKBH7 plays a role in the induction of cell death in response to different types of cytotoxic alkylating agents.
In addition to alkylating agents, we tested the same cell lines for their sensitivity to the oxidizing agents hydrogen peroxide (H2O2) and tert-butyl-hydroperoxide (t-BuOOH). Notably, we found that ALKBH7 depletion conferred significant resistance to both H2O2 and t-BuOOH (Fig. 2F), indicating that ALKBH7 is involved in both alkylation and oxidation-induced cell death. Importantly, the resistance phenotype associated with ALKBH7 depletion was not limited to 293T human cells since knockdown of ALKBH7 in HeLa cervical carcinoma cells also confers resistance to either MMS or t-BuOOH (Supplemental Fig. S2). To our knowledge, this is the first ALKBH protein for which cellular depletion leads to resistance to cytotoxic damaging agents.
ALKBH7 is required for necrosis induced by DNA-damaging agents
The fact that ALKBH7 depletion confers resistance to alkylating and oxidizing agents suggests that ALKBH7 may play a novel role in promoting cell death in response to DNA damage. To explore the biological role of ALKBH7, we examined DNA damage stress signaling pathways in control and ALKBH7-depleted cells upon MMS exposure. Both control and ALKBH7-depleted cells displayed normal activation of canonical DNA damage stress signaling proteins, including phosphorylation of p53 and Chk1 (Supplemental Fig. S3A). These results suggest that ALKBH7-depleted cells undergo DNA damage to the same extent as the parental cells and that they elicit a normal DNA damage signaling response, at least initially.
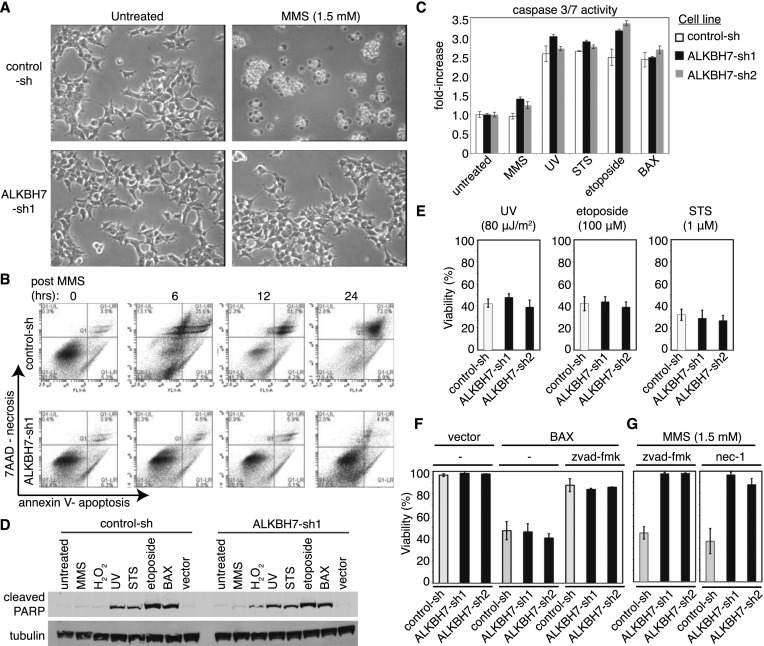
Previous studies have shown that severe DNA damage caused by alkylating or oxidizing agents can trigger a caspase-independent programmed necrotic cell death pathway that displays cellular and molecular features distinct from apoptotic cell death (Zong and Thompson 2006; Eisenberg-Lerner and Kimchi 2007; Trivedi et al. 2008). Moreover, a recent study has shown that 293T cells undergo necrosis after treatment with MNNG alkylating agent (Ethier et al. 2012). Indeed, we observed the typical hallmarks of necrotic cell death in 293T cells treated with either MMS or H2O2, including increased cell volume, translucent cytoplasm, aggregation of intact nuclei, and plasma membrane disintegration (Fig. 3A). The loss of plasma membrane integrity in MMS-treated cells was confirmed by 7-aminoactinomycin D (7AAD) staining of nuclear DNA, which revealed cell permeabilization starting at 6 h post-MMS treatment, leading to complete loss of cell integrity at 24 h (Fig. 3B). The initial loss of viability and plasma membrane integrity in MMS-treated cells occurred in the absence of typical apoptotic markers such as decreased cell volume, membrane blebbing, early phosphatidylserine exposure, caspase activation, or PARP cleavage (Fig. 3A–D). We thus infer that 293T cells undergo necrosis rather than apoptosis after exposure to MMS. In contrast to control cells, ALKBH7-depleted cells remained adherent to the plate and maintained plasma membrane integrity even at doses of alkylating agents that induce severe DNA damage, consistent with the maintenance of cell viability and resistance to alkylation-induced cell death (Fig. 3A,B).
Figure 3.
ALKBH7 depletion inhibits necrotic cell death but not apoptosis. (A) Phase-contrast images of control-sh and ALKBH7-depleted 293T cells that were left untreated or treated with the indicated concentration of MMS. Images were taken 24 h after initiation of treatment. (B) Scatter plots of 7AAD positivity for plasma membrane permeabilization (necrosis) and annexin V staining (apoptosis), as measured simultaneously through flow cytometry in single cells at the indicated time points post-MMS treatment. (C) Caspase 3/7 activation in 293T cell lines after treatment with the indicated agents. Bars represent the standard deviation of three independent experiments. (D) Immunoblot analysis of cleaved PARP in control-sh and ALKBH7-depleted cell lines after exposure to the indicated agent or transient expression of the proapoptotic protein BAX. (E–G) Viability of control-sh and ALKBH7-depleted cell lines after exposure to the indicated agent or after transient expression of the apoptosis-inducing protein BAX. Cells were treated with MMS (1.5 mM) or H2O2 (1 mM) for 1 h, while STS (1 μM) or etoposide (100 μM) were left in the culture medium until analysis. UV irradiation was delivered at 80 μJ/m2. In some experiments, the caspase inhibitor z-vad-fmk (100 μM) or the RIP kinase inhibitor nec-1 (50 μM) was added to the medium after transfection or treatment. Bars represent the standard deviation of at least three independent experiments.
Since ALKBH7 depletion can prevent necrotic cell death, we tested whether ALKBH7 could also protect against apoptosis. We treated control and ALKBH7-depleted cell lines with DNA-damaging agents known to induce apoptosis in 293T cells; namely, UV and etoposide (Mourtada-Maarabouni et al. 2009; Zalckvar et al. 2010). In addition, we tested staurosporine (STS), a well-characterized inducer of apoptosis. In contrast to being resistant to alkylation- and oxidation-induced necrosis, ALKBH7-depleted cell lines underwent levels of apoptosis similar to those of control cells after treatment with either UV, etoposide, or STS, as judged by caspase activation, PARP cleavage, and eventual cell death (Fig. 3C–E). In addition, we found that transient expression of the apoptosis-inducing protein BAX induced apoptosis and cell death in control and ALKBH7-depleted cells to a similar extent (Fig. 3F). Finally, while the pan-caspase inhibitor z-vad-fmk abrogated BAX-induced apoptotic cell death, there was no effect of z-vad-fmk on cell death induced by MMS (Fig. 3G), further indicating that alkylating agents induce a necrotic cell death pathway in 293T cells. Taken together, our results show that ALKBH7 depletion specifically protects against necrotic cell death without affecting apoptotic cell death pathways.
Recent studies have shown that the RIP kinases play a role in programmed necroptosis induced by death receptors and some forms of DNA damage (Biton and Ashkenazi 2011; Feoktistova et al. 2011; Tenev et al. 2011; Yang et al. 2011). However, programmed necrotic cell death induced by either alkylating or oxidizing agents has been shown to be independent of the RIP kinase-dependent necroptotic pathway (Tang et al. 2010; Wang et al. 2012). Indeed, we found that addition of the RIP kinase inhibitor necrostatin-1 (nec-1) was unable to prevent necrotic cell death induced by MMS (Fig. 3G). However, nec-1 was able to block programmed necrotic cell death induced by TNF-α in L929 mouse cells, indicating that the nec-1 inhibitor was active (data not shown). Thus, ALKBH7 is required for DNA damage-induced necrosis induced by alkylating or oxidizing agents that is independent of nec-1 inhibition.
ALKBH7 prevents the recovery of cellular bioenergetics after DNA damage-induced PARP hyperactivation
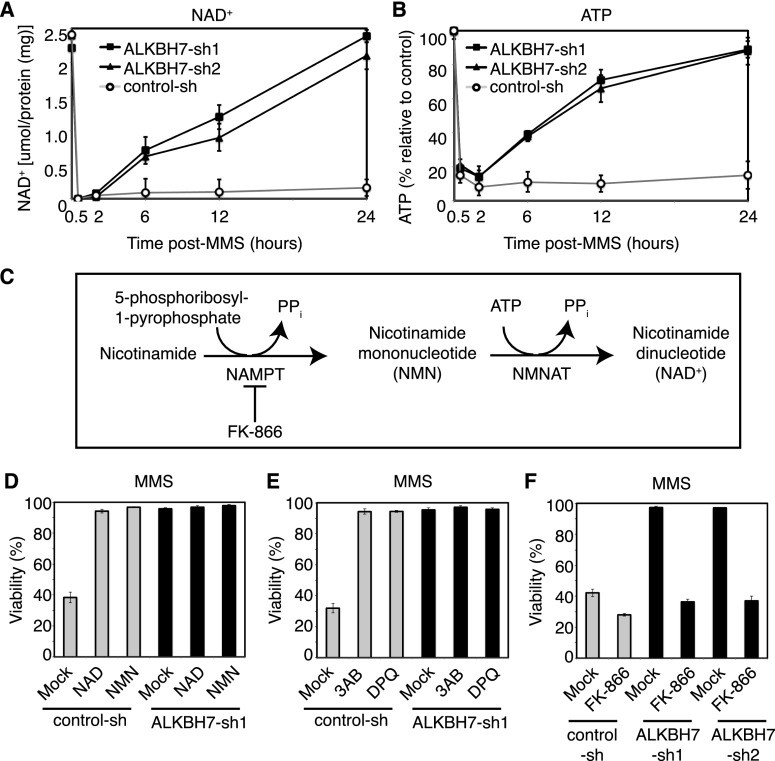
DNA damage-induced programmed necrosis has been linked to the hyperactivation of the NAD+-consuming enzyme PARP; PARP hyperactivation leads to NAD+ depletion, loss of ATP, and eventual bioenergetic collapse. We therefore tested whether PARP hyperactivation and NAD+ depletion are involved in the MMS-induced death of 293T cells and whether ALKBH7 plays a role in this process. Upon MMS treatment, we found that PARP was hyperactivated to the same extent in both control and ALKBH7-depleted cells, as indicated by the formation of poly(ADP ribose) (PAR)-modified proteins (Supplemental Fig. S3B). Moreover, we found that both control and ALKBH7-depleted cells suffered a dramatic decrease in both NAD+ and ATP starting as early as 30 min after MMS treatment (Fig. 4A,B), consistent with the hyperactivation of PARP in response to DNA damage. However, in notable contrast to control cells, the ALKBH7-depleted cells recovered their NAD+ and ATP levels starting 6 h post-MMS treatment, returning to basal levels by 24 h. Thus, both control and ALKBH7-depleted cells undergo DNA damage-induced PARP hyperactivation that leads to massive depletion of total cellular NAD+ and ATP, but ALKBH7-depleted cells regained NAD+/ATP homeostasis and maintained cell viability. Our results suggest that ALKBH7 functions to prevent wild-type cells from regaining NAD+/ATP homeostasis after acute exposure to genotoxic compounds.
Figure 4.
ALKBH7 prevents the recovery of NAD+ and ATP levels after MMS treatment that is necessary for cellular survival after exposure to MMS (1.5 mM for 1 h). (A,B) Measurement of NAD+ or ATP levels in the indicated 293T human embryonic kidney cell lines after MMS treatment. (C) Enzymes and metabolites of the NAD+ salvage pathway. (NAMPT) Nicotinamide phosphoribosyltransferase; (NMNAT) nicotinamide mononucleotide adenylyltransferase; (PPi) inorganic pyrophosphate. (D) Viability of control-sh or ALKBH7-depleted cell lines after treatment with MMS, followed by the addition of either mock buffer, NAD+ (10 mM), or NMN (10 mM). (E) Viability of control-sh or ALKBH7-depleted cell lines after treatment with MMS in the absence or presence of either of the PARP inhibitors: 3AB (2 mM) or DPQ (20 μM). (F) Viability of control-sh or ALKBH7-depleted cell lines after treatment with MMS, followed by the absence or presence of the NAMPT inhibitor FK-866 (10 nM). Bars for all graphs represent the mean of at least three independent experiments.
The rapid recovery of [NAD+] and [ATP] levels in ALKBH7-depleted cells, which coincides with the maintenance of cell viability, indicates that the recovery of cellular bioenergetic metabolites is a crucial step in preventing the execution of cellular necrosis. The recovery of cellular NAD+ could occur through the import of exogenous NAD+, if present at high enough levels; through the NAD+ salvage pathway from NAD+ precursors (Fig. 4C); or both. Indeed, the addition of exogenous NAD+ or the NAD+ precursor nicotinamide mononucleotide (NMN) to the culture medium suppresses MMS sensitivity in control 293T cells (Fig. 4D). As expected, ALKBH7-depleted cells were not affected by the addition of NAD+, since they were already MMS-resistant. As a corollary experiment, we tested whether PARP inhibition could suppress MMS sensitivity in control 293T cells after MMS treatment by preventing the massive decrease in [NAD+] and [ATP]. We tested the PARP inhibitors 3-aminobenzimide (3AB) and 3,4-dihydro-5[4-(1-piperindinyl)butoxy]-1(2H)-isoquinoline (DPQ) by coincubation during MMS treatment. Similar to NAD+ repletion, the blockade of PARP activation completely suppresses MMS-induced cell death, again indicating that NAD+ levels are crucial for cell survival after alkylating agent treatment (Fig. 4E).
The recovery of total cellular [NAD+] in ALKBH7-depleted cells suggests that, when present, ALKBH7 prevents NAD+ resynthesis after PARP hyperactivation, potentially by inhibiting NAD+-forming pathways that are required to regenerate the NAD+ consumed during PARP hyperactivation. To test this idea, we determined whether disruption of NAD+ salvage pathways in ALKBH7-depleted cells could perturb their MMS-resistant phenotype. For these analyses, we used the inhibitor FK-866, which inhibits the nicotinamide phosphoribosyltransferase (NAMPT) enzyme within the NAD+ salvage pathway (Fig. 4C; Yang et al. 2007; Pittelli et al. 2010). Previous studies have shown that FK-866 can dramatically enhance MMS-induced cell death in numerous mammalian cell types (Yang et al. 2007; Goellner et al. 2011). We treated control and ALKBH7-depleted cell lines with MMS, followed by the addition of FK-866 at a concentration that does not itself result in cell death within the time span of the experiment. As expected for the important role of maintaining NAD+ homeostasis, control cells treated with MMS underwent cell death that was further increased by NAMPT inhibition using FK-866 (Fig. 4F). More importantly, we found that the MMS-resistant phenotype exhibited by ALKBH7-depleted cells could be completely suppressed by treatment with FK-866 (Fig. 4F). Thus, inhibition of the NAD+ salvage pathway nullifies the MMS-resistant phenotype of ALKBH7-depleted cells. Together, these results indicate that ALKBH7 plays a role in suppressing the recovery of cellular bioenergetics and, as a consequence, promotes the onset of cellular necrosis. The recovery of physiological NAD+ and ATP levels in ALKBH7-depleted cells indicates that in wild-type cells, the initial loss of total cellular NAD+ precedes an ALKBH7-dependent step that prevents recovery of NAD+ pools, thus promoting necrotic cell death.
ALKBH7 plays a key role in mitochondrial dysfunction leading to programmed necrosis
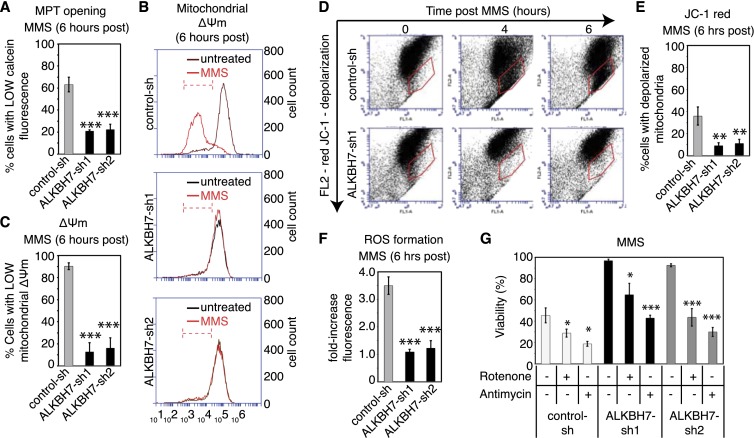
The regeneration of [NAD+] and [ATP] after PARP hyperactivation in ALKBH7-depleted cells indicates that ALKBH7 modulates a step after NAD+ depletion that inhibits recovery of cellular bioenergetic status. The opening of the mitochondrial permeability transition (MPT) pore, dissolution of the mitochondria membrane potential (ΔΨm), and formation of ROS are closely associated with programmed necrotic cell death, with each cellular event contributing to the eventual demise of the cell. Since ALKBH7 is required for necrotic cell death after PARP hyperactivation and NAD+/ATP depletion, we determined whether ALKBH7 plays a role in the initiation of mitochondrial dysfunction that occurs after severe DNA damage.
To probe for MPT in cells, we assayed mitochondrial retention of calcein-AM, a fluorescent probe that can only exit through an open MPT pore. In addition, we monitored mitochondrial ΔΨm using two different types of membrane potential indicators: tetramethylrhodamine (TMRE) and JC-1. Following MMS treatment, we found that control 293T cells lose mitochondrial calcein-AM staining as early as 6 h post-treatment (Fig. 5A), indicating that MPT has occurred. Furthermore, we detected an overall decrease in both TMRE staining and JC-1 red fluorescence in MMS-treated cells, denoting the loss of mitochondrial ΔΨm after MMS treatment (Fig. 5B–E). Finally, MMS treatment in control cells leads to a significant induction of mitochondrial ROS, as measured using MitoSox Red, a mitochondrial targeted form of the superoxide indicator dihydroethidium (Fig. 5F). Previous studies have shown that ROS formation is required for programmed necrotic cell death induced by alkylating agents or death receptor activation (Vandenabeele et al. 2010; Ethier et al. 2012; Vanlangenakker et al. 2012). Indeed, we found that preincubation of control cells with the antioxidant N-acetylcysteine can rescue control cells from necrotic cell death induced by MMS (Supplemental Fig. S4A).
Figure 5.
ALKBH7 depletion prevents the MPT, loss of mitochondria membrane potential, and formation of ROS that accompany MMS-induced cell death. For all experiments, cell lines were treated with 1.5 mM MMS for 1 h. (A) Measurement of MPT in control-sh or ALKBH7-depleted cell lines after exposure to MMS. Cells were loaded with calcein-FM, followed by CoCl2 quenching. (B) Representative flow cytometry plot of untreated and MMS-treated cell lines stained with TMRE to monitor mitochondria membrane potential (Δψm). (C) Percentage of cells with low TMRE staining, indicative of reduced mitochondrial Δψm after treatment with MMS. Bars represent the mean of three independent experiments. (D) Scatter plots of JC-1 staining for mitochondria membrane potential as measured through JC-1 staining. (E) Quantification of cells with depolarized mitochondria, as measured by the loss of JC-1 red fluorescence, which is indicative of mitochondria membrane depolarization. (F) Quantification of ROS formation. After treatment with MMS, the indicated cell lines were loaded with the mitochondrial superoxide indicator MitoSox Red (2 μM), which increases in fluorescence intensity after oxidation. The fold increase in fluorescence signal relative to untreated cells represents the mean of three independent experiments. (G) Viability of control-sh or ALKBH7-depleted cell lines after treatment with MMS in the absence or presence of the ETC inhibitors antimycin (5 μM) or rotenone (25 μM). Bars for all graphs represent the mean of at least three independent experiments. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
In vivid contrast to control cells, we found that ALKBH7-depleted cells treated with MMS maintained mitochondrial integrity and function, as measured by each of the aforementioned criteria (Fig. 5A–F). ALKBH7-depleted cells maintained a closed MPT pore as well as robust ΔΨm after MMS treatment, as evidenced by the retention of mitochondrial calcein fluorescence along with JC-1 red and TMRE staining (Fig. 5A–E). Consistent with the maintenance of mitochondrial function and homeostasis, ALKBH7-depleted cells did not exhibit any increase in ROS after MMS treatment (Fig. 5F), even though DNA damage and PARP hyperactivation had occurred, accompanied by [NAD+] and [ATP] depletion. The maintenance of mitochondrial function most likely explains the recovery of NAD+ and ATP levels in ALKBH7-depleted cells after MMS treatment, thus allowing cells to survive and prevent necrotic cell death.
Based on the ALKBH7-dependent loss of mitochondrial homeostasis after MMS treatment, we hypothesize that ALKBH7 inhibits essential mitochondrial functions to induce mitochondrial perturbation and subsequent programmed necrosis. Since the loss of respiration has previously been linked to caspase-independent mitochondrial cell death (Lartigue et al. 2009), we focused on the respiratory electron transport chain (ETC) as a potential mitochondrial function modulated by ALKBH7. Strikingly, we found that chemical inhibition of either complex I or III of the mitochondrial ETC via rotenone or antimycin, respectively, can suppress the MMS-resistant phenotype of ALKBH7-depleted cells (Fig. 5G). Importantly, treatment with rotenone or antimycin in the absence of MMS did not induce cell death within the time span of the experiment (Supplemental Fig. S4B). Moreover, we found that the presence or absence of glucose had no effect on cell survival after MMS treatment (Supplemental Fig. S4C), suggesting that blockade of respiration but not glycolysis plays a role in ALKBH7-dependent necrosis. We conclude that ALKBH7 plays a pivotal role in triggering the cascade of mitochondrial events leading to eventual programmed necrosis by inhibiting key aspects of mitochondrial function, such as respiration.
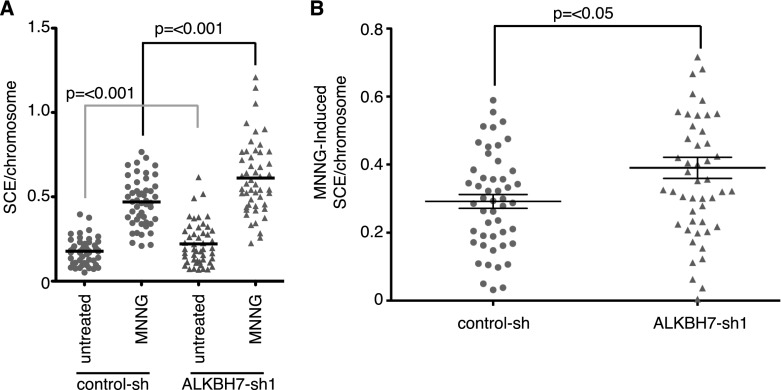
ALKBH7-mediated necrosis prevents the accumulation of cells with DNA damage
To evaluate the possible downstream biological effects of programmed necrosis modulated by ALKBH7, we compared alkylation-induced sister chromatid exchange (SCE) in control and ALKBH7-depleted. As expected, we detected a significant increase in the number of SCEs in MNNG-treated versus untreated cells for both the control and ALKBH7-depleted cell line (Fig. 6A; Supplemental Fig. S5). However, both the spontaneous frequency and MNNG-induced frequency of SCEs were significantly higher in ALKBH7-depleted cells versus control cells (Fig. 6). We infer that the induction of programmed necrosis by severe DNA damage may serve as a mechanism to eliminate cells harboring excessive genomic damage, thus preventing the accumulation of cells with chromosomal aberrations. Depletion of ALKBH7 allows these cells to survive even in the presence of recombinogenic and potentially mutagenic DNA damage.
Figure 6.
Programmed necrosis inhibits the accumulation of cells with genomic DNA damage. (A) SCEs per chromosome of the indicated cell lines with or without treatment with MNNG (20 μM). Scatter plots represent the aggregate of two independent experiments. (B) MNNG-induced frequency of SCE in the indicated cell lines.
Discussion
Programmed necrosis induced by excessive DNA damage can be initiated by PARP hyperactivation, leading to NAD+/ATP depletion, mitochondrial depolarization, ROS production, and eventual plasma membrane permeability. While many of the cellular aspects of programmed necrosis have been characterized, the factors responsible for initiating and carrying out programmed necrotic cell death are only beginning to be found. Here, we show that a mammalian ALKBH protein, ALKBH7, represents a crucial link in the necrotic cellular cascade induced after PARP hyperactivation by promoting the large-scale loss of mitochondrial function required for programmed necrosis. Loss of ALKBH7 expression facilitates the recovery of cellular metabolites after severe DNA damage, thus allowing cells to remain viable.
All previously characterized mammalian ALKBH dioxygenases have been shown to play a protective role upon exposure to DNA-damaging agents through either the repair of cytotoxic DNA damage (ALKBH2 and ALKBH3) or the modulation of DNA damage survival pathways via tRNA modification (ALKBH8). However, we found that another ALKBH protein, ALKBH7, actually facilitates cell death after exposure to alkylating or oxidizing agents, thus playing a novel prodeath function. These results expand the functions of ALKBH proteins beyond that of nucleic acid repair and modification and uncover a novel role for ALKBH7 in programmed necrosis.
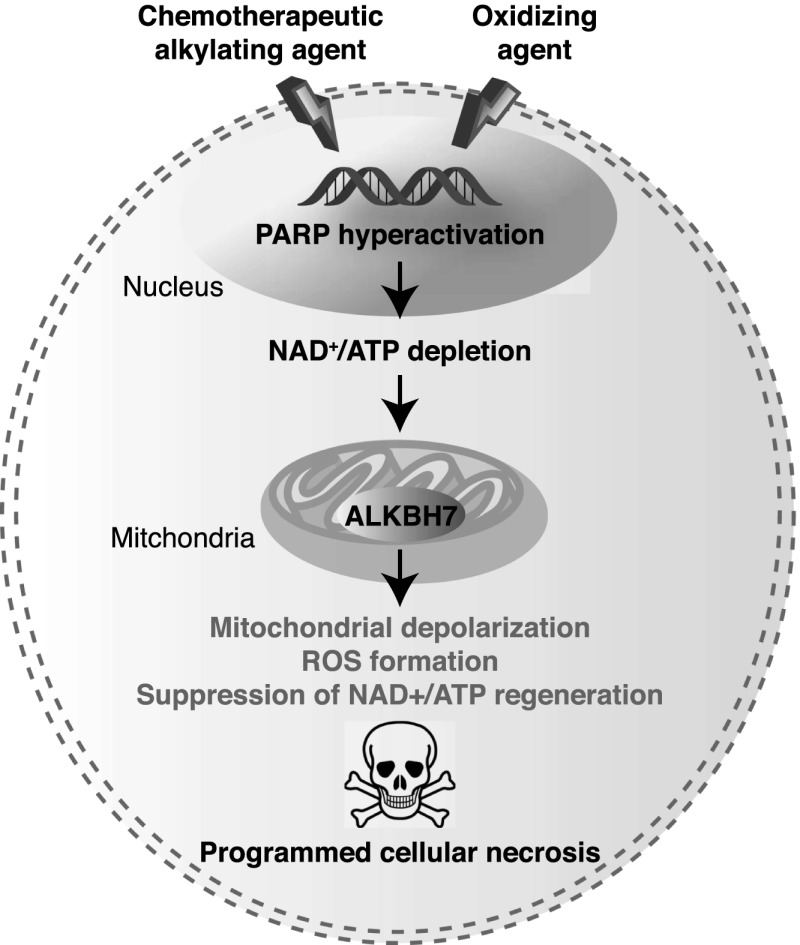
How does ALKBH7 suppress the recovery of biological metabolites to promote necrosis? We propose that ALKBH7 inhibits mitochondrial function after PARP hyperactivation to prevent the regeneration of vital bioenergetic metabolites such as NAD+ and ATP (Fig. 7). Based on our results, ALKBH7 is required for the massive loss of mitochondrial homeostasis, which leads to MPT pore opening and the efflux of NAD+ into the cytoplasm, allowing further consumption by nuclear PARP. NAD+ is particularly important in mitochondria, since numerous oxidative phosphorylation enzymes require NAD+ for function. Moreover, the maintenance of mitochondrial [NAD+] by the NAD salvage pathway is essential for cellular survival after DNA damage (Yang et al. 2007). Massive NAD+ consumption by PARP, coupled with NAD+ efflux, can lead to loss of all energy production and abrogation of the proton gradient as cells try to regenerate NAD+ using ATP; this vicious cycle eventually results in mitochondrial dysfunction and formation of toxic ROS. All of these features are observed in cells after MMS treatment and are abrogated in ALKBH7-depleted cells. Since the maintenance of NAD+ levels through the NAD+ salvage pathway after DNA damage requires ALKBH7, this protein is critical in determining cell fate by preventing NAD+ regeneration after cellular stress, thus promoting the loss of mitochondrial function and cell viability.
Figure 7.
Model for alkylating and oxidizing agent-induced necrotic cell death modulated by ALKBH7. See the Discussion for details.
Recently, the mitochondrial protein phosphatase PGAM5 and p53 were shown to be downstream effectors for the execution of programmed necroptotic cell death by inducing changes in mitochondrial permeability and dynamics (Vaseva et al. 2012; Wang et al. 2012). Like ALKBH7, both PGAM5 and p53 are necessary for programmed necrosis induced by oxidizing agents even though it remains to be seen whether PGAM5 or p53 is required for programmed necrosis induced by alkylating agents. It will be interesting to explore the potential interplay between ALKBH7, p53, and PGAM5 in the execution of mitochondrial dysfunction during necroptotic cell death.
Since many cancer cell types can evade apoptotic cell death through deficiencies in key apoptotic proteins or by inhibition of apoptotic pathways, necrosis presents an attractive target for activation in apoptosis-resistant cancer cells (Kreuzaler and Watson 2012). Indeed, recent studies showed that apoptosis is dispensable for complete tumor regression after alkylating agent chemotherapy because other forms of cell death can be activated, including programmed necrosis (Guerriero et al. 2008). Moreover, necrosis promotes the clearance of apoptosis-deficient tumors by stimulating the innate immune response, a process that is dependent on the necrosis-dependent release of the HMGB1 protein (Guerriero et al. 2011). Interestingly, 293T human cells are highly resistant to some forms of apoptosis induced by viral infection, TNF-α, or the anti-Fas antibody due to the expression of SV-40 T antigen and the adenoviral E1b19K and E1b55K oncoproteins, which inhibit p53-dependent and p53-independent apoptosis (Grimm et al. 1996; Fearnhead et al. 1997; Cuconati and White 2002). In contrast to apoptosis, however, ALKBH7-dependent necrotic cell death is readily triggered by alkylating or oxidizing agents. These studies suggest that activation of ALKBH7 in apoptosis-resistant cancer cells could serve to enhance the toxic effects of chemotherapeutic alkylating agents in tumors. Our discovery of a role for ALKBH7 in programmed necrosis highlights a potential alternative route to tumor eradication that will be the subject of future investigation. Intriguingly, protein profiling of cancer tissues has revealed that a majority of malignant gliomas and skin cancers exhibit low or negative expression of ALKBH7 (Uhlen et al. 2010).
All previously known substrates of AlkB and ALKBH dioxygenases consist of nucleic acid substrates such as DNA or RNA bases. However, we found that ALKBH7 lacks dioxygenase repair activity on canonical AlkB DNA substrates even though ALKBH7 contains all of the active site motifs and residues found in all known active AlkB dioxygenases. Interestingly, mutation of the putative 2-oxoglutarate-binding site in ALKBH7 that is critical for dioxygenase activity in other AlkB enzymes does not affect its ability to promote necrosis. Based on this result, ALKBH7 may display a novel enzymatic activity that does not require 2-oxoglutarate binding. Nondioxygenase activities for AlkB enzymes have been discovered previously, including tRNA methylation by the ALKBH8 enzyme (Fu et al. 2010; Songe-Moller et al. 2010) and DNA lyase activity by the human ALKBH1 and Schizosaccharomyces pombe Abh1 enzymes (Muller et al. 2010; Korvald et al. 2012). It is also possible that ALKBH7 binds nonenzymatically to protein or nucleic acid substrates to initiate the cascade of mitochondrial events that lead to necrosis. Future studies will be devoted to identifying the targets and binding partners of ALKBH7 that regulate the induction of programmed necrosis.
Materials and methods
Cell culture, plasmids, and shRNA
293T human embryonic kidney, HeLa S3 cervical carcinoma, and derivative stable cell lines were cultured in Dulbecco's minimal essential medium (Gibco Life Technologies) containing 10% fetal bovine serum (FBS) and 2 mM L-glutamine. For stable RNAi, lentiviral RNAi constructs (Open Biosystems) expressing a nonsilencing shRNA (RHS4459) or one of two shRNAs targeting the ALKBH7 transcript (TRCN0000072764 or TRCN0000072765) were cotransfected with packaging plasmids (psPAX2 and pMD2.G) into 293T cells for lentivirus production. As an additional shRNA set, we used the GIPZ RNAi constructs expressing either scramble nonsilencing shRNA (RHS4346) or shRNA targeting ALKBH7 (RHS4430-98514376). The 293T or HeLa cell lines were subsequently infected with lentivirus in the presence of polybrene, followed by stable clone selection using puromycin.
For colony-forming analysis, cells were seeded into 60-mm plates, allowed to attach for 24 h, and exposed to serum-free medium alone or containing MMS for 1 h, followed by replacement with complete medium containing serum. Surviving colonies were scored 6–10 d later by methylene blue staining. For short-term viability assays, cells were exposed to MMS, MNNG, mechlorethamine, temozolomide, H2O2, t-buOOH, etoposide, or STS (all from Sigma). MMS, MNNG, H2O2, and t-buOOH were incubated in serum-free medium for 1 h, followed by replacement with complete medium containing serum. Mechlorethamine, temozolomide, etoposide, and STS were left in the medium for the duration of the experiment until analysis. UV irradiation (80 J/m2) was performed using a UV cross-linker (Stratagene) set at a 254-nm wavelength. Viability for MMS, MNNG, mechlorethamine, temozolomide, H2O2, t-buOOH, UV, and BAX overexpression was determined using a Coulter counter coupled with trypan blue staining (total number of viable treated cells/total number of viable untreated cells), while viability for etoposide and STS was measured by the WST-1 cell proliferation reagent (Roche) according to the manufacturer's instructions. Additional reagents used (all from Sigma) were FK-866, NAD+, NMN, 3-aminobenzamide, DPQ, z-vad-fmk, nec-1, and 7AAD.
NAD+ and ATP levels were measured using a NADH/NAD Quantification kit (Abcam, ab65348) or an ATP Colorimetric/Fluorometric Assay kit (Abcam, ab83355). MPT pore status was determined via calcein staining/CoCl2 quenching via the MitoProbe Transition Pore Assay kit (Molecular Probes, Life Technologies). Mitochondrial membrane potential was measured with either the mitochondrial dye JC-1 or TMRE ethyl ester, while mitochondrial superoxide levels were detected using MitoSox Red (all from Molecular Probes, Life Technologies). Cells were analyzed on an Accuri C6 flow cytometer (BD Biosciences). Additional information on siRNA knockdown, mammalian expression constructs, and SCE analysis can be found in the Supplemental Material.
Subcellular predication, localization, and fractionation of ALKBH7
Subcellular localization predication was performed using three independent online algorithms: SherLoc2 (http://abi.inf.uni-tuebingen.de/Services/SherLoc2), MitoProtII (http://ihg.gsf.de/ihg/mitoprot.html), and TargetP1.1 (http://www.cbs.dtu.dk/services/TargetP).
Expression constructs encoding ALKBH7-GFP were transiently transfected into 293T cells via calcium phosphate-mediated DNA transfer. For mitochondria colocalization, cells were infected with virus expressing red fluorescent protein targeted to mitochondria (CellLight Mitochondria-RFP, BacMam 2.0, Molecular Probes, Life Technologies). Cells were collected at 48 h post-transfection for formaldehyde fixation and mounting in ProLong anti-fade reagent with DAPI (Molecular Probes, Life Technologies), followed by microscopy.
Subcellular fractionation of 293T cells was performed by differential centrifugation according to Graham (2001).
Protein analysis
Cellular extract preparation, antibodies, protein purification, and enzymatic activity assays were performed essentially as described (Fu et al. 2010) and are described in the Supplemental Material.
Acknowledgments
We thank Kyle Atmore and Peter Svensson for initial production of the ALKBH7 knockdown cell lines, Jennifer A. Calvo for manuscript comments, and Arne Klungland for sharing unpublished results. This work was funded by NIH grants R01-CA055042, P30-ES002109, and P30-CA014051. D.F. and J.J.J. are funded by American Cancer Society Post-doctoral Fellowships 116155-PF-08-255-01-GMC and PF-12-060-01-DMC, respectively.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.215533.113.
References
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. 2003. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 [DOI] [PubMed] [Google Scholar]
- Alano CC, Garnier P, Ying W, Higashi Y, Kauppinen TM, Swanson RA 2010. NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J Neurosci 30: 2967–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baritaud M, Boujrad H, Lorenzo HK, Krantic S, Susin SA 2010. Histone H2AX: The missing link in AIF-mediated caspase-independent programmed necrosis. Cell Cycle 9: 3166–3173 [DOI] [PubMed] [Google Scholar]
- Biton S, Ashkenazi A 2011. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-α feedforward signaling. Cell 145: 92–103 [DOI] [PubMed] [Google Scholar]
- Cipriani G, Rapizzi E, Vannacci A, Rizzuto R, Moroni F, Chiarugi A 2005. Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J Biol Chem 280: 17227–17234 [DOI] [PubMed] [Google Scholar]
- Cuconati A, White E 2002. Viral homologs of BCL-2: Role of apoptosis in the regulation of virus infection. Genes Dev 16: 2465–2478 [DOI] [PubMed] [Google Scholar]
- Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, Rubin M, Gygi S, Harper JW, Shi Y 2011. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell 44: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Zhang X, Han YY, Burke NA, Kochanek PM, Watkins SC, Graham SH, Carcillo JA, Szabo C, Clark RS 2003. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J Biol Chem 278: 18426–18433 [DOI] [PubMed] [Google Scholar]
- Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B 2002. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci 99: 16660–16665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Kimchi A 2007. DAP kinase regulates JNK signaling by binding and activating protein kinase D under oxidative stress. Cell Death Differ 14: 1908–1915 [DOI] [PubMed] [Google Scholar]
- Ethier C, Tardif M, Arul L, Poirier GG 2012. PARP-1 modulation of mTOR signaling in response to a DNA alkylating agent. PLoS ONE 7: e47978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Johansen RF, Seeberg E 2002. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419: 178–182 [DOI] [PubMed] [Google Scholar]
- Fearnhead HO, McCurrach ME, O'Neill J, Zhang K, Lowe SW, Lazebnik YA 1997. Oncogene-dependent apoptosis in extracts from drug-resistant cells. Genes Dev 11: 1266–1276 [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M 2011. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 43: 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD 2010. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 30: 2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. 2007. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318: 1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goellner EM, Grimme B, Brown AR, Lin Y-C, Wang X-H, Sugrue KF, Mitchell L, Trivedi RN, Tang J-b, Sobol RW 2011. Overcoming temozolomide resistance in glioblastoma via dual inhibition of NAD+ biosynthesis and base excision repair. Cancer Res 71: 2308–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JP 2001. Isolation of mitochondria from tissues and cells by differential centrifugation. Curr Protoc Cell Biol 3.3.1–3.3.15 doi: 10.1002/0471143030.cb0303s04 [DOI] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G 2011. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333: 1109–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S, Stanger BZ, Leder P 1996. RIP and FADD: Two ‘death domain’-containing proteins can induce apoptosis by convergent, but dissociable, pathways. Proc Natl Acad Sci 93: 10923–10927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero JL, Ditsworth D, Fan Y, Zhao F, Crawford HC, Zong WX 2008. Chemotherapy induces tumor clearance independent of apoptosis. Cancer Res 68: 9595–9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero JL, Ditsworth D, Catanzaro JM, Sabino G, Furie MB, Kew RR, Crawford HC, Zong WX 2011. DNA alkylating therapy induces tumor regression through an HMGB1-mediated activation of innate immunity. J Immunol 186: 3517–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HC, Snyder SH 1999. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci 96: 13978–13982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C 2008. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett 582: 3313–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korvald H, Falnes PO, Laerdahl JK, Bjoras M, Alseth I 2012. The Schizosaccharomyces pombe AlkB homolog Abh1 exhibits AP lyase activity but no demethylase activity. DNA Repair (Amst) 11: 453–462 [DOI] [PubMed] [Google Scholar]
- Kreuzaler P, Watson CJ 2012. Killing a cancer: What are the alternatives? Nat Rev Cancer 12: 411–424 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M 2005. Lysosomes and autophagy in cell death control. Nat Rev Cancer 5: 886–897 [DOI] [PubMed] [Google Scholar]
- Lartigue L, Kushnareva Y, Seong Y, Lin H, Faustin B, Newmeyer DD 2009. Caspase-independent mitochondrial cell death results from loss of respiration, not cytotoxic protein release. Mol Biol Cell 20: 4871–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O'Connor TR 2005. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem 280: 39448–39459 [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL 2012. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev 26: 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Upton JW, Kaiser WJ 2012. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nat Rev Immunol 12: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubarak RS, Yuste VJ, Artus C, Bouharrour A, Greer PA, Menissier-de Murcia J, Susin SA 2007. Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Mol Cell Biol 27: 4844–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT 2009. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28: 195–208 [DOI] [PubMed] [Google Scholar]
- Muller TA, Meek K, Hausinger RP 2010. Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites. DNA Repair (Amst) 9: 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y 2008. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J 27: 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO 2004. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell 16: 107–116 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A 2010. Inhibition of nicotinamide phosphoribosyltransferase: Cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem 285: 34106–34114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339: 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, et al. 2006. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J 25: 2189–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T 2007. Repair of alkylated DNA: Recent advances. DNA Repair (Amst) 6: 429–442 [DOI] [PubMed] [Google Scholar]
- Solberg A, Robertson AB, Aronsen JM, Rognmo O, Sjaastad I, Wisloff U, Klungland A 2013. Deletion of mouse Alkbh7 leads to obesity. J Mol Cell Biol doi 10.1093/jmcb/mjt012 [DOI] [PubMed] [Google Scholar]
- Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A 2010. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol 30: 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J-B, Goellner EM, Wang X-H, Trivedi RN, St Croix CM, Jelezcova E, Svilar D, Brown AR, Sobol RW 2010. Bioenergetic metabolites regulate base excision repair–dependent cell death in response to DNA damage. Mol Cancer Res 8: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, et al. 2011. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 43: 432–448 [DOI] [PubMed] [Google Scholar]
- Thalhammer A, Bencokova Z, Poole R, Loenarz C, Adam J, O'Flaherty L, Schodel J, Mole D, Giaslakiotis K, Schofield CJ, et al. 2011. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α (HIF-1α). PLoS ONE 6: e16210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419: 174–178 [DOI] [PubMed] [Google Scholar]
- Trivedi RN, Wang X-H, Jelezcova E, Goellner EM, Tang J-B, Sobol RW 2008. Human methyl purine DNA glycosylase and DNA polymerase β expression collectively predict sensitivity to temozolomide. Mol Pharmacol 74: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa K, Koike K, Kitae K, Shinkawa A, Arima H, Suzuki T, Tsuchiya M, Makino Y, Furukawa T, Konishi N, et al. 2007. Expression and sub-cellular localization of human ABH family molecules. J Cell Mol Med 11: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. 2010. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250 [DOI] [PubMed] [Google Scholar]
- Vagbo CB, Svaasand EK, Aas PA, Krokan HE 2013. Methylation damage to RNA induced in vivo in Escherichia coli is repaired by endogenous AlkB as part of the adaptive response. DNA Repair (Amst) 12: 188–195 [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G 2010. Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat Rev Mol Cell Biol 11: 700–714 [DOI] [PubMed] [Google Scholar]
- Vanlangenakker N, Vanden Berghe T, Vandenabeele P 2012. Many stimuli pull the necrotic trigger, an overview. Cell Death Differ 19: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM 2012. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149: 1536–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiang H, Chen S, Du F, Wang X 2012. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148: 228–243 [DOI] [PubMed] [Google Scholar]
- Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, et al. 2008. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem 283: 25046–25056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, et al. 2007. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S, Gandhi P, Munson M, Miyamoto S, Kelliher MA 2011. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-κB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol Cell Biol 31: 2774–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, He C 2013. DNA repair by reversal of DNA damage. Cold Spring Harb Perspect Biol 5: a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL 2002. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297: 259–263 [DOI] [PubMed] [Google Scholar]
- Zalckvar E, Yosef N, Reef S, Ber Y, Rubinstein AD, Mor I, Sharan R, Ruppin E, Kimchi A 2010. A systems level strategy for analyzing the cell death network: Implication in exploring the apoptosis/autophagy connection. Cell Death Differ 17: 1244–1253 [DOI] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Thompson CB 2006. Necrotic death as a cell fate. Genes Dev 20: 1–15 [DOI] [PubMed] [Google Scholar]
- Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB 2004. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev 18: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]