Many gene products involved in cellular processes are substrates of the E3 ubiquitin ligase Mule/Huwe1/Arf-BP1 (Mule). Inoue et al. show that Muleflox/flox;K14-Cre mice subjected to DMBA/PMA-induced skin carcinogenesis exhibit increased number and severity of skin tumors. In the absence of Mule, c-Myc/Miz1 transcriptional complexes accumulated, and levels of p21CDKN1A (p21) and p15INK4B (p15) were down-regulated. This study suggests that Mule suppresses Ras-mediated tumorigenesis by preventing the accumulation of c-Myc/Miz1 complexes that mediate p21 and p15 down-regulation.
Keywords: Mule, Huwe1, Miz1, c-Myc, p21, Ras
Abstract
Tumorigenesis results from dysregulation of oncogenes and tumor suppressors that influence cellular proliferation, differentiation, apoptosis, and/or senescence. Many gene products involved in these processes are substrates of the E3 ubiquitin ligase Mule/Huwe1/Arf-BP1 (Mule), but whether Mule acts as an oncogene or tumor suppressor in vivo remains controversial. We generated K14Cre;Muleflox/flox(y) (Mule kKO) mice and subjected them to DMBA/PMA-induced skin carcinogenesis, which depends on oncogenic Ras signaling. Mule deficiency resulted in increased penetrance, number, and severity of skin tumors, which could be reversed by concomitant genetic knockout of c-Myc but not by knockout of p53 or p19Arf. Notably, in the absence of Mule, c-Myc/Miz1 transcriptional complexes accumulated, and levels of p21CDKN1A (p21) and p15INK4B (p15) were down-regulated. In vitro, Mule-deficient primary keratinocytes exhibited increased proliferation that could be reversed by Miz1 knockdown. Transfer of Mule-deficient transformed cells to nude mice resulted in enhanced tumor growth that again could be abrogated by Miz1 knockdown. Our data demonstrate in vivo that Mule suppresses Ras-mediated tumorigenesis by preventing an accumulation of c-Myc/Miz1 complexes that mediates p21 and p15 down-regulation.
Oncogenes drive tumor development by dysregulating various cellular processes, including proliferation, apoptosis, differentiation, senescence, and responses to DNA damage. Oncogenic mutations of Ras occur in ∼30% of human cancers (Bos 1989) and activate Raf/MEF/ERK and PI3K/Akt signaling pathways that lead to uncontrolled proliferation and transformation (Karnoub and Weinberg 2008; DeNicola and Tuveson 2009). However, oncogenic Ras also activates signaling by tumor suppressor gene (TSG) pathways mediated by p53/p21, p16Ink4A, and/or p19Arf. These TSG pathways attempt to induce the cell cycle arrest and/or premature senescence of cells with DNA damage to prevent their malignant transformation (Bringold and Serrano 2000). Indeed, oncogenic Ras stimulation alone does not transform wild-type fibroblasts but can transform primary cells deficient in TSGs (Bringold and Serrano 2000; DeNicola and Tuveson 2009). Thus, oncogenic Ras must cooperate with other oncogenes and/or loss of TSGs to overcome the barrier of premature senescence and drive neoplastic transformation.
Myc is another oncogene frequently amplified or overexpressed in human tumors. Myc regulates cellular proliferation, apoptosis, differentiation, and transformation via transcription-dependent and -independent mechanisms (Eilers and Eisenman 2008; Meyer and Penn 2008). At a molecular level, Myc partners with various proteins in distinct heterodimeric complexes to activate or repress the transcription of two sets of target genes. The Myc/Max complex binds directly to the promoters of Myc target genes to stimulate cell cycle progression and proliferation (Levens 2002). Myc/Max also up-regulates p19Arf, which stabilizes p53 by inactivating Mdm2 (an E3 ligase for p53); the cell then undergoes cell cycle arrest or apoptosis. An alternative complex contains Myc and Miz1, a zinc finger transcription factor. The c-Myc/Miz1 complex represses transcription through direct binding of Miz1 to target gene promoters. Prominent c-Myc/Miz1 targets are the cyclin-dependent kinase (CDK) inhibitors p21, p15, and p57Kip2 (p57) (Wanzel et al. 2003; Herkert and Eilers 2010).
In the absence of Myc, Miz1 is free to activate the transcription of Myc targets such as integrins and cell adhesion proteins (Gebhardt et al. 2006). Studies of total Miz1 knockout mice have shown that Miz1 is essential for early embryogenesis (Adhikary et al. 2003). Using conditional knockout mice, roles for Miz1 in B- and T-cell differentiation (Kosan et al. 2010; Saba et al. 2011), keratinocyte adhesion and differentiation (Gebhardt et al. 2006), hair follicle orientation, keratinocyte proliferation and differentiation (Gebhardt et al. 2007), and skin carcinogenesis (Honnemann et al. 2012) have been demonstrated.
Mule/Huwe1/Arf-BP1 (Mule) is a large, multidomain member of the HECT (homologous to E5-AP C terminus) domain family of E3 ubiquitin ligases (Khoronenkova and Dianov 2011). Various Mule substrates are involved in apoptosis (Mcl-1) (Zhong et al. 2005), DNA replication, DNA damage responses, DNA repair (TopBP1, Cdc6, and DNA polymerases β and λ) (Hall et al. 2007; Herold et al. 2008; Parsons et al. 2009; Markkanen et al. 2012), and transcriptional regulation (p53, c-Myc, N-Myc, Miz1, HDAC2, and MyoD) (Adhikary et al. 2005; Chen et al. 2005; Zhao et al. 2008; Yang et al. 2010; Zhang et al. 2011; Noy et al. 2012). Mule attaches Lys 48 (K48)-linked polyubiquitin chains to most of its substrates, promoting their proteasomal degradation. However, Mule also attaches K63-linked polyubiquitin chains to c-Myc, suggesting a signaling function (Adhikary et al. 2005). Mule-mediated K63-linked ubiquitination of c-Myc is inhibited by Miz1 (Adhikary et al. 2005), implying that this c-Myc modification likely occurs when c-Myc is free or participating in a c-Myc/Max complex but not in a c-Myc/Miz1 complex. In addition to c-Myc, Mule interacts with p19Arf and ATM. Binding to p19Arf inhibits Mule E3 ligase activity (Chen et al. 2005), and Mule regulates the ATM–p53 axis either positively or negatively depending on whether DNA damage is present (Hao et al. 2012).
Total Mule knockout in mice is embryonic-lethal (Hao et al. 2012; Kon et al. 2012). Conditional Mule knockout in neuronal or glial cells results in accumulations of N-Myc and c-Myc and is also lethal (Zhao et al. 2009; D'Arca et al. 2010). Conditional Mule knockout in B cells leads to p53 accumulation and impaired B-cell development and homeostasis that can be rescued by concomitant p53 knockout (Hao et al. 2012). Similarly, conditional Mule knockout in pancreatic β cells causes p53 accumulation, loss of β cells, and severe diabetes preventable by concomitant p53 knockout (Kon et al. 2012). However, while these reports confirm that products of both oncogenes (N-Myc and c-Myc) and a TSG (p53) are Mule substrates in vivo, a role for Mule in tumorigenesis remains to be substantiated. Here we provide the first in vivo evidence that (1) Mule is a tumor suppressor, (2) the c-Myc/Miz1 complex is a Mule substrate, and (3) Mule suppresses Ras-induced tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15.
Results
Loss of Mule in keratinocytes promotes 7,12-dimethylbenz-(a)-anthracene (DMBA)/12-O-tetradecanoylphorbol-13-acetate (PMA)-induced tumorigenesis in a manner that does not depend on p53 or p19Arf but does involve c-Myc
To ascertain the role of Mule in tumorigenesis in vivo, we used the two-step model of skin carcinogenesis induced by oncogenic Ras. DMBA is applied to the skin as a tumor initiator, and PMA is subsequently applied as a tumor promoter (Kemp et al. 1993). Several major Mule substrates (p53, c-Myc, and Miz1) and Mule-associated proteins (p19Arf) are important in DMBA/PMA carcinogenesis (Kemp et al. 1993; Kelly-Spratt et al. 2004; Oskarsson et al. 2006; Honeycutt et al. 2010; Honnemann et al. 2012). In this model, p53 and p19Arf act as TSGs, while c-Myc and Miz1 act as oncogenes.
To generate mutants in which Mule was selectively deleted in skin epithelium, we crossed Mulefl/fl(y) (male or female) mice to K14Cre mice in which Cre recombinase is under the control of the human keratin 14 (K14) promoter. Cre-mediated deletion of floxed genes in these animals can be detected in basal epidermis and hair follicles in skin from embryonic day 13.5 (E13.5) (Jonkers et al. 2001; Kurek et al. 2007). K14Cre;Mulefl/lf(y) (Mule kKO) mice were born at the expected Mendelian ratio (1:1) (Supplemental Table I), did not spontaneously develop tumors, and had normal life spans (Supplemental Fig. 1A).
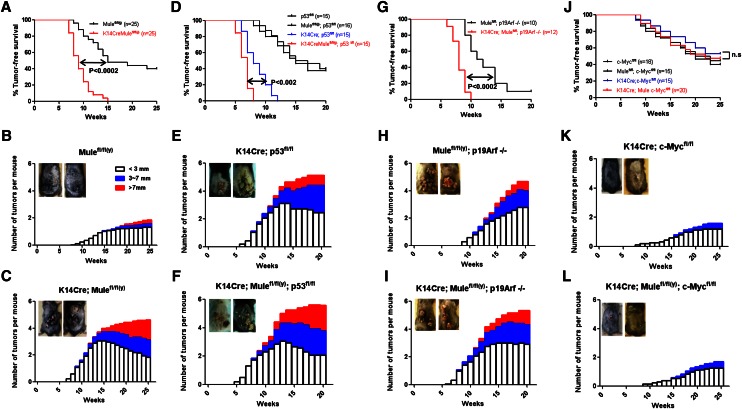
To examine induced tumorigenesis in Mule kKO mice, we exposed their dorsal skin to DMBA/PMA and monitored skin tumor development. Tumors were first observed in Mule kKO and control littermates at 7–8 wk post-DMBA exposure (Fig. 1A), indicating that initial tumor formation was not altered by Mule deficiency. However, by 15 wk post-DMBA, all Mule kKO mice had developed tumors, whereas only half of control mice had done so. Moreover, tumors in Mule kKO mice were more numerous and larger in size than in controls (Fig. 1B,C). Histologically, ∼70% of tumors in Mule kKO mice were keratoacanthomas (KAs), which are benign but highly proliferative, whereas all growths in control mice were benign papillomas typical of the DMBA/PMA model (Supplemental Table II). Thus, Mule plays a tumor-suppressive role in vivo, at least in skin.
Figure 1.
Skin tumorigenesis promoted by Mule deficiency can be reversed by c-Myc deficiency. (A,D,G,J) Tumor-free survival of mice of the indicated genotypes following exposure to DMBA/PMA. P-values, log-rank test. (n.s.) Not significant. (B,C,E,F,H,I,K,L) Numbers of tumors that were small (∼3 mm), medium (3∼7 mm) or large (>7 mm) that developed on the dorsal skin of the mice in A, D, G, and J. (Insets) Representative examples of dorsal skin tumors.
Next, we investigated whether p53 and p19Arf were involved in Mule’s tumor-suppressive function by subjecting K14Cre;p53fl/fl (p53 kKO) and p19Arf−/− mice to DMBA/PMA-induced carcinogenesis. In the absence of such induction, Mule deficiency did not increase spontaneous tumorigenesis (Supplemental Fig. 1B,C). However, all p53 kKO mice exposed to DMBA/PMA developed more numerous and larger tumors than control p53fl/fl littermates (Fig. 1D,E; Supplemental Fig. 2A). We then generated K14Cre;p53fl/fl;Mulefl/fl(y) (Mule/p53 kDKO) double mutants and observed that tumor development was accelerated (latency 8 wk post-DMBA) compared with p53 kKO mice (latency 12 wk post-DMBA) (Fig. 1D). Notably, tumors in Mule/p53 kDKO mice were even larger than those in p53 kKO mice (Fig. 1E,F). Similarly, K14Cre;Mulefl/fl(y);p19Arf−/− (Mule/p19Arf kDKO) double mutants showed more rapid tumor growth than p19Arf−/− mice (Fig. 1G). All Mule/p19Arf kDKO mutants had developed tumors by 10 wk post-DMBA, whereas by 20 wk, only ∼90% of p19Arf knockout mice had done so. Tumors in Mule/p19Arf kDKO mice were also larger than those in p19Arf−/− mice (Fig. 1H,I). Thus, neither p53 nor p19Arf is absolutely required for Mule’s tumor-suppressive function in this skin tumor model.
K14Cre;c-Mycfl/fl (c-Myc kKO) mice are known to be resistant to DMBA/PMA-induced skin carcinogenesis (Oskarsson et al. 2006). We therefore evaluated whether Mule deficiency would render these mutants vulnerable to such tumorigenesis. Confirming the study by Oskarsson et al. (2006), our c-Myc kKO mice showed no spontaneous tumors (Supplemental Fig. 1D) and were relatively resistant to DMBA/PMA-induced tumor formation (Fig. 1J). We generated K14Cre;Mulefl/fl(y);c-Mycfl/fl (Mule/c-Myc kDKO) double mutants and observed that tumors developed with kinetics similar to those in c-Myc kKO mice (Fig. 1J). Tumors in Mule/c-Myc kDKO mice were comparable in size with those in controls and only slightly larger than those in c-Myc kKO mice (Fig. 1K,L; Supplemental Fig. 2C,D). Thus, concomitant knockout of c-Myc largely “rescued” the Mule-deficient phenotype in the DMBA/PMA model, suggesting that c-Myc signaling might be involved in Mule’s tumor-suppressive function.
Mule deficiency increases tumor cell proliferation due to an accumulation of the c-Myc/Miz1 complex that down-regulates p21CDKN1A and p15INK4b
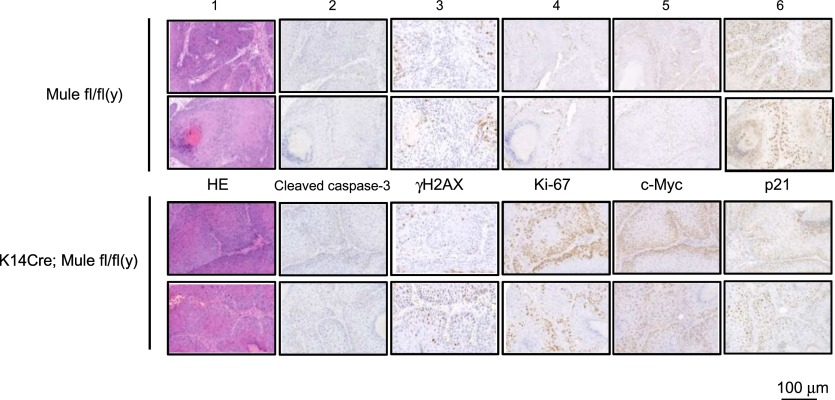
To investigate how Mule deficiency affects cancer growth, we subjected tumor samples from Mule kKO and control littermates to histological analysis. Equally low levels of cleaved caspase-3 (marker of apoptosis), senescence-associated β-galactosidase (SA-β-gal) staining (marker of cellular senescence), and γH2AX (marker of DNA damage) were found in tumors from mice of both genotypes (Fig. 2; Supplemental Fig. 3A), suggesting that Mule deficiency does not lead to significant changes in apoptosis, senescence, or responses to spontaneous DNA damage (at least in this skin tumor model). Interestingly, Ki67 staining (marker of proliferation) was significantly elevated in tumors of Mule kKO mice, and levels of CD31 (marker of angiogenesis) were slightly enhanced (Fig. 2; Supplemental Fig. 3B). These data suggest that the larger skin tumor size in DMBA/PMA-exposed Mule-deficient mice is due to a major increase in tumor cell proliferation and a minor increase in tumor angiogenesis. When we used immunohistochemistry (IHC) to examine key factors driving cancer cell growth in the absence of Mule, we found that Mule deficiency did not affect protein levels of p16Ink4a, p19Arf, or p53 in skin tumors but that c-Myc was markedly increased and p21 was decreased (Fig. 2; Supplemental Fig. 3B). These results raised the possibility that the tumor-suppressive function of Mule might be related to the regulation of proliferation exerted by c-Myc and p21.
Figure 2.
Skin tumors in Mule-deficient mice show enhanced cellular proliferation, increased c-Myc protein, and decreased p21 protein. Representative tumor samples from two Mulefl/fl(y) and two K14Cre;Mulefl/fl(y) (Mule kKO) mice were subjected to hematoxylin–eosin (HE) staining (panel 1) as well as IHC analysis to detect the indicated proteins (panels 2–6).
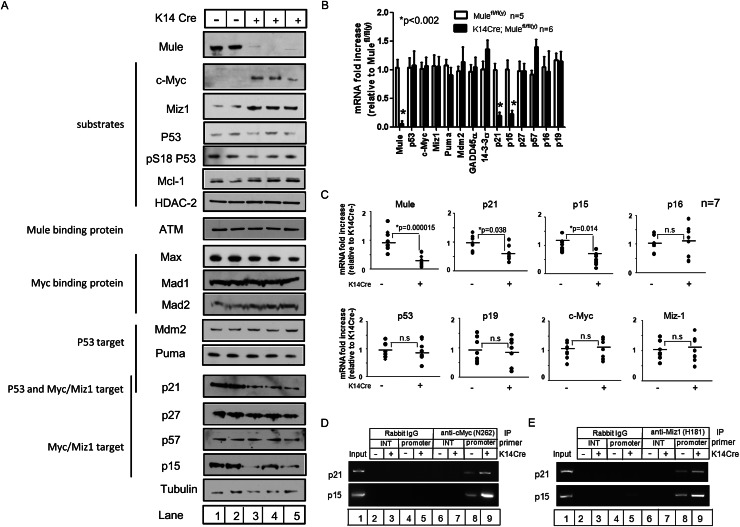
We next examined the effects of Mule deficiency on the protein expression of several Mule substrates and downstream targets in primary keratinocytes from both Mule kKO mice and Mulefl/fl(y) mice infected in vitro with Cre-expressing adenovirus (Ad-Cre). Immunoblotting confirmed that Mule protein was efficiently decreased in both cases (Fig. 3A; Supplemental Fig. 4). There were no detectable differences in Mcl-1, HDAC2, ATM, or p53 protein levels in the absence of Mule, and there were no changes in protein levels of the p53 targets Mdm2 and Puma or the c-Myc-binding partners Max, Mad1, and Mad2. In contrast, both c-Myc and Miz1 were consistently increased in Mule-deficient cells (Fig. 3A; Supplemental Fig. 4). As the c-Myc/Miz1 complex represses the transcription of several CDK inhibitors in a tissue context-dependent fashion (Seoane et al. 2001; Herold et al. 2002; Adhikary et al. 2003; Wu et al. 2003; Basu et al. 2009), we examined protein levels of p21, p27, p57Kip2 (p57), and p15 in Mule-deficient cells. Levels of p21 and p15, but not p27 and p57, were consistently reduced in Mule-deficient primary keratinocytes (Fig. 3A; Supplemental Fig. 4).
Figure 3.
Mule deficiency increases c-Myc and Miz1 and decreases p21 and p15. (A) Immunoblotting to detect the indicated proteins in lysates of primary keratinocytes from Mulefl/fl(y) (lanes 1,2) and K14Cre;Mulefl/fl(y) (lanes 3–5) mice. Tubulin was used as a loading control. (B) Quantitative RT–PCR determination of the indicated mRNAs in primary keratinocytes from Mulefl/fl(y) and K14Cre;Mulefl/fl(y) mice. Values were normalized to 18S rRNA. Data are the mean fold increase ± SD relative to levels in control keratinocytes. P-values are from unpaired Student’s t-test. (C) Quantitative RT–PCR of the indicated mRNAs in skin tumors from Mulefl/fl(y) (−) and K14Cre;Mulefl/fl(y) (+) mice, determined as for B. Data points represent tumor samples from individual mice (n = 7 per group). P-values are from unpaired Student’s t-test. (D,E) ChIP assays of primary keratinocytes from Mulefl/fl(y) (lanes 2,4,6,8) and K14Cre;Mulefl/fl(y) (lanes 3,5,7,9) mice. Lysates were immunoprecipitated (IP) with rabbit anti-IgG (control), rabbit anti-c-Myc (N262) Ab (D), or rabbit anti-Miz1 (H190) Ab (E). The coimmunoprecipitated promoter regions of the indicated genes were amplified by PCR. (Input) Positive control (lane 1); (INT) internal control.
We then determined mRNA levels of the above factors using real-time RT–PCR. Mule mRNA levels were significantly reduced in Mule-deficient primary keratinocytes (Fig. 3B) and skin tumor samples (Fig. 3C), as expected. The mRNA levels of p53, c-Myc, and Miz1 were not altered by Mule deficiency, also expected, since Mule acts on these substrates at the protein rather than the mRNA level. In addition, no significant differences in mRNA levels of most p53 target genes (Puma, Mdm2, and GADD45α) or in a c-Myc/Miz1 target gene (p27) were detected. For unknown reasons, levels of 14-3-3σ and p57 mRNAs were slightly up-regulated in Mule-deficient cells. Consistent with their protein profiles, p21 and p15 mRNAs were markedly down-regulated in Mule-deficient primary keratinocytes and skin tumor samples (Fig. 3B,C). Thus, rather than affecting the stability of the p21 and p15 proteins, Mule deficiency results in transcriptional down-regulation of these genes.
The transcription of p21 and p15 mRNAs can be repressed directly by the c-Myc/Miz1 complex (Seoane et al. 2001; Staller et al. 2001; Herold et al. 2002; Wu et al. 2003). To determine whether Mule deficiency enhances the recruitment c-Myc/Miz1 to the p21 and p15 promoters, we performed chromatin immunoprecipitation (ChIP) experiments using anti-c-Myc and anti-Miz1 antibodies (Abs). c-Myc and Miz1 were each able to bind to the p21 and p15 promoters, and this binding was enhanced in Mule-deficient cells compared with controls (Fig. 3D,E). Thus, Mule deficiency in keratinocytes results in accumulations of c-Myc and Miz1 that are directly associated with repression of p21 and p15 transcription.
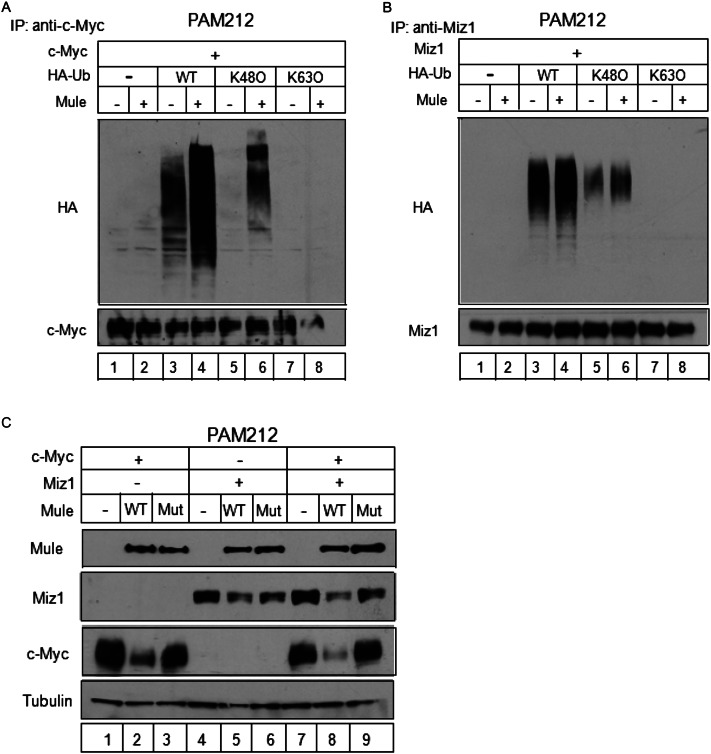
To investigate whether Mule prevents the accumulation of c-Myc/Miz1 complexes by mediating polyubiquitination and subsequent degradation of these proteins, we initially attempted to perform ubiquitination assays using primary keratinocytes from Mule kKO mice. Unfortunately, we could not obtain sufficient gene transduction efficiency without incurring lethal toxicity. As an alternative, we engineered PAM212 cells (a transformed murine keratinocyte cell line) (Yuspa et al.1980) to overexpress c-Myc (Fig. 4A) or Miz1 (Fig. 4B). When these cells also overexpressed Mule, we observed enhanced linkage of wild-type HA-ubiquitin to both c-Myc and Miz1. The same result was observed for all members of a series of HA-ubiquitin proteins mutated at individual lysines except K48 (K48O) (Fig. 4A,B). In contrast, Mule overexpression did not enhance the linkage of HA-ubiquitin mutated except at K63 (K63O) to either c-Myc or Miz1 (Fig. 4A,B). Interestingly, Mule deficiency had a more dramatic impact on the polyubiquitination of c-Myc than on that of Miz1 (Fig. 4A,B). These data suggest that Mule regulates c-Myc and Miz1 protein stability primarily through K48-linked polyubiquitination.
Figure 4.
Mule controls the stability of c-Myc and Miz1 by regulating their ubiquitin-mediated proteasomal degradation. (A,B) In vivo ubiquitination of c-Myc and Miz1 by Mule. PAM212 cells were cotransfected with vectors expressing Mule and c-Myc (A) or Miz1 (B), plus wild-type (WT) HA-ubiquitin (lanes 3,4), or HA-ubiquitin in which all lysine mutated except for K48 (K48O; lanes 5,6) or K63 (K63O; lanes 7,8). After MG132 exposure for 2 h, cell lysates were immunoprecipitated (IP) using rabbit anti-c-Myc (N262) Ab (A) or rabbit anti-Miz1 (H-190) Ab (B). The indicated proteins in the immunoprecipitates were identified by immunoblotting. (C) PAM212 cells were cotransfected with vectors expressing c-Myc (lanes 1–3), Miz1 (lanes 4–6), or both (lanes 7–9), plus wild-type (WT) Mule (lanes 2,5,8) or the enzymatically inactive C4341A Mule mutant (Mut; lanes 3,6,9). The indicated proteins were identified by immunoblotting.
As both c-Myc and Miz1 were increased in Mule-deficient cells (Fig. 3A; Supplemental Fig. 4), we investigated whether c-Myc or Miz1 was a direct target of Mule activity. PAM212 cells engineered to ectopically overexpress both wild-type Mule and c-Myc showed a marked reduction in c-Myc compared with controls, and this reduction was not observed when enzymatically inactive Mule was co-overexpressed with c-Myc (Fig. 4C). Overexpression of wild-type Mule also decreased ectopically expressed Miz1 but more modestly (Fig. 4C). The decrease in Miz1 mediated by wild-type Mule was slightly enhanced by co-overexpression of c-Myc (Fig. 4C). These findings collectively suggest that (1) Mule targets c-Myc more effectively than Miz1 and (2) Miz1 may be targeted more effectively by Mule when it participates in the c-Myc/Miz1 complex, at least in keratinocytes.
Mule deficiency increases keratinocyte proliferation via effects on Miz1
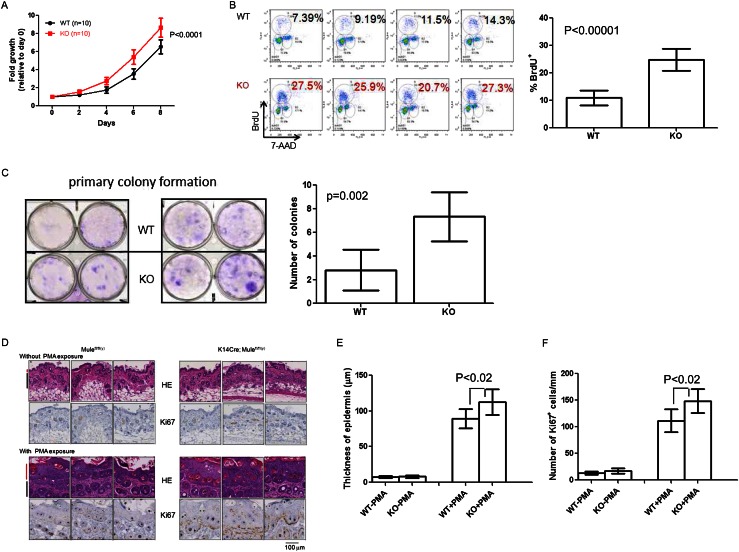
As our results showed that Mule controls Miz1 stability and Miz1 is known to regulate keratinocyte proliferation through p21 (Honnemann et al. 2012), we assessed the proliferation of control and Mule-deficient keratinocytes derived by infecting primary keratinocytes from Mulewt/wt and Mulefl/fl(y) mice with Ad-Cre. Cultures of Mule-deficient keratinocytes grew more rapidly than controls (Fig. 5A) and showed an approximately twofold elevation in BrdU incorporation (Fig. 5B), suggesting that the increased proliferation was due to cell cycle regulation rather than decreased apoptosis. In addition, although total numbers were very low, Mule-deficient keratinocytes formed more colonies than controls (Fig. 5C). Thus, keratinocytes acquire an enhanced potential for transformation in the absence of Mule.
Figure 5.
Mule deficiency increases keratinocyte proliferation. (A) Primary keratinocytes from Mulewt/wt (WT) and Mulefl/fl(y) (KO) mice (n = 10 per group) were exposed to Ad-Cre for 24 h. Cell numbers were counted on days 2, 4, 6, and 8 post-exposure. Data are the mean ± SD and are expressed as fold increase relative to cell numbers at day 0. P-value is from two-way ANOVA. (B) Flow cytometric analysis of BrdU incorporation by primary keratinocytes from the mice in A. (Left) Representative histograms for four mice per group. Numbers are percentage of BrdU+ cells. (Right) Statistical analysis of percentage of BrdU+ cells for all mice in A. Data are the mean ± SD (n = 10 per group). P-value is from unpaired Student’s t-test. (C) Colony formation assay of Mulefl/fl(y) (WT) and K14Cre;Mulefl/fl(y) (KO) keratinocytes that were seeded in six-well plates (2 × 104 per well) and cultured for 2 wk. Cells were stained with 0.5% crystal violet. (Left) Representative staining. (Right) Quantitation of number of colonies per well. Data are the mean ± SD (n = 6 per group). P-value is from unpaired Student’s t-test. (D) HE staining (top) and Ki67 staining (bottom) of dorsal skin from three Mulefl/fl(y) and three K14Cre;Mulefl/fl(y) mice that were either left untreated or treated with PMA every day for 5 d, as indicated. (Red line) Epidermis; (black line) dermis. (E,F) Quantitation of thickness of basal epidermis (E) and number of Ki67+ cells per millimeter (F) in dorsal skin samples from the mice in D. Data were acquired from 40 different images derived from four mice per group. Data are the mean ± SD. P-values are from unpaired Student’s t-test.
Next, we determined the effect of Mule deficiency on epidermal keratinocyte proliferation in vivo. First, we examined the dorsal skin morphology of untreated Mule kKO and control littermates and observed no significant differences in either epidermal thickness or number of Ki67+ cells per millimeter (Fig. 5D [top], E,F [left]). We then exposed the dorsal skin of Mule kKO and control littermates to PMA to induce keratinocyte hyperplasia (Oskarsson et al. 2006). In PMA-treated animals of both genotypes, epidermal thickness and number of Ki67+ cells per millimeter were dramatically increased (Fig. 5D [bottom], E,F [right]). However, both of these parameters were enhanced to a greater degree in PMA-treated Mule kKO mice compared with controls. Thus, Mule deficiency accelerates PMA-induced hyperplasia.
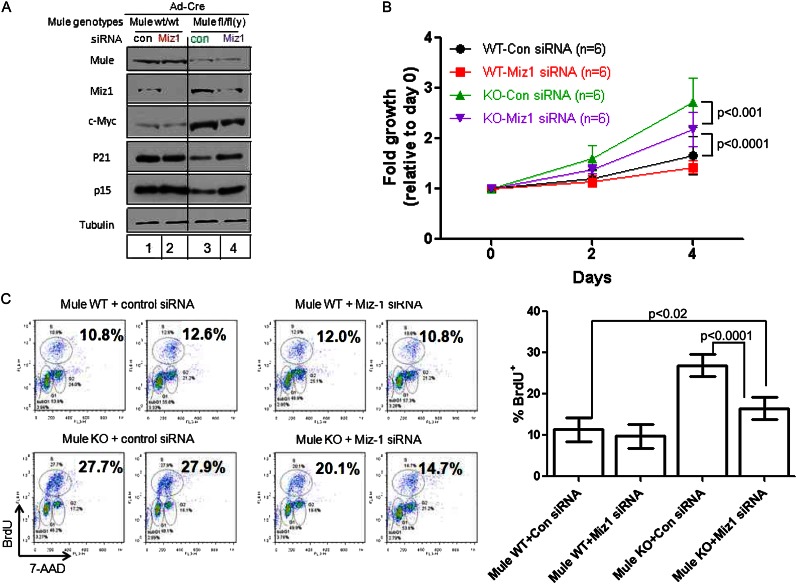
To investigate whether Miz1 is involved in Mule’s regulation of keratinocyte proliferation, we examined the effect of siRNA-mediated Miz1 knockdown on Mulewt/wt and Mulefl/fl(y) primary keratinocytes infected with Ad-Cre. Exposure to Miz1 siRNA for 48 h efficiently reduced Miz1 protein in Mule kKO and control cells (Fig. 6A). Levels of c-Myc protein were slightly decreased in Mule kKO cells by Miz1 knockdown compared with controls (Fig. 6A). Importantly, the decreased protein levels of p21 and p15 present in Mule kKO cells transfected with scrambled siRNA were restored by Miz1 depletion (Fig. 6A), demonstrating that Miz1 is crucial for the p21 and p15 down-regulation induced by Mule deficiency. Moreover, the enhancements of proliferation (Fig. 6B) and BrdU incorporation (Fig. 6C) in Mule kKO cells were partially impaired by Miz1 knockdown. Thus, the effects of Mule deficiency on keratinocyte proliferation are mediated primarily through Miz1.
Figure 6.
Miz1 knockdown reverses the enhanced proliferation associated with Mule deficiency. (A) Immunoblotting to detect the indicated proteins in Ad-Cre-infected Mulewt/wt and Mulefl/fl(y) primary keratinocytes transfected with control siRNA (Con) or Miz1 siRNA (Miz1). (B) The proliferation of the siRNA-expressing keratinocytes in A was analyzed by counting cell numbers at 2 and 4 d post-seeding. Data are the mean ± SD of six cultures per group. P-values are from two-way ANOVA. (C) The proliferation of the siRNA-transfected keratinocytes in A was analyzed by BrdU incorporation as for Figure 5B. (Left) Representative flow cytometric histograms for two cultures per group. Numbers are percentage of BrdU+ cells. (Right) Statistical analysis of percentage of BrdU+ cells for all cultures in A. Data are the mean ± SD of six cultures per group. P-values are from unpaired Student’s t-test.
Mule deficiency promotes Ras-induced tumorigenesis in vivo through a mechanism involving Miz1
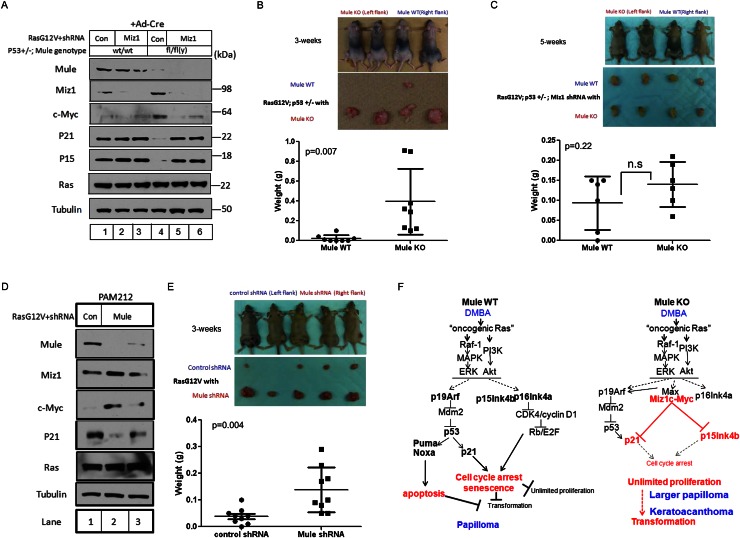
To confirm the effects of Mule deficiency on tumor growth in vivo, we initially planned to monitor the growth of transformed keratinocytes derived from Mule kKO mutants in athymic nude mice. Unfortunately, we could not obtain such transformed keratinocytes due to poor gene transduction efficiency. We therefore turned to the use of primary mouse embryonic fibroblasts (MEFs) in allograft experiments. Primary MEFs derived from Mulewt/wt;p53+/− (Mule WT;p53+/−) and Mulefl/fl;p53+/− (Mule KO;p53+/−) mice were infected with Ad-Cre to obtain control and Mule-deficient MEFs, respectively. We then infected Mule WT;p53+/− or Mule KO;p53+/− MEFs with retrovirus expressing HRasG12V plus either control shRNA or Miz1 shRNA. As expected, Mule was efficiently deleted by Ad-Cre treatment, leading to accumulations of c-Myc and Miz1 and reductions in p21 and p15 in Mule KO;p53+/− MEFs (Fig. 7A). Expression of Miz1 shRNA efficiently decreased Miz1 in both control and Mule-deficient MEFs (Fig. 7A). Strikingly, Miz1 knockdown in Mule KO;p53+/− MEFs abolished c-Myc accumulation and restored p21 and p15 to normal levels (Fig. 7A). Miz1 shRNA did not affect c-Myc mRNA levels (data not shown), raising the possibility that participation in the c-Myc/Miz1 complex stabilizes the c-Myc protein.
Figure 7.
Mule deficiency promotes Ras-induced tumorigenesis in vivo through a mechanism involving Miz1. (A) Immunoblotting to detect the indicated proteins in lysates of primary MEFs from Mulewt/wt;p53+/− (lanes 1–3) or Mulefl/fl(y);p53+/− (lanes 4–6) mice that were exposed to Ad-Cre for 24 h, followed by retroviral infection with pMX-RasG12V-IRES-Puror-control shRNA (lanes 1,4) or pMX-RasG12V-IRES-Puror-Miz1 shRNA (lanes 2,3,5,6). (B) Transformed control or Mule-deficient MEFs from the cultures in A were subcutaneously injected (1 × 106 cells) into the right (Mule WT) (A, lane 1) or left (Mule KO) (A, lane 4) flanks of athymic nude mice. (Top) Representative photos of four mice per group are shown at 3 wk post-injection. (Middle) Gross views of representative tumors isolated from the mice in the top panel. (Bottom) Weights of tumors of injected mice at 3 wk post-injection. Data points are tumor weight per individual mouse. (Horizontal line) Mean weight ± SD for the group (n = 8). P-value is from unpaired Student’s t-test. (C) Transformed control or Mule-deficient MEFs expressing Miz1 shRNA (from the cultures in A) were subcutaneously injected (1 × 106 cells) into the right (Mule WT-Miz1 shRNA) (A, lane 2) or left (Mule KO-Miz1 shRNA) (A, lane 5) flanks of athymic nude mice. (Top) Representative photos of four mice per group at 5 wk post-injection. (Middle) Gross views of representative tumors isolated from the mice in the top panel. (Bottom) Weights of tumors of injected mice at 5 wk post-injection. Data points are tumor weight per individual mouse. (Horizontal line) Mean weight ± SD for the group (n = 6). P-value is from unpaired Student’s t-test. (D) Immunoblotting to detect the indicated proteins in PAM212 cells infected with pMX-RasG12V-IRES-puror-control shRNA (Con) or pMX-RasG12V-IRES-puror-Mule shRNA (Mule). (E) The control or Mule-depleted PAM212 cells in D were subcutaneously injected (1 × 106 cells) into the left (control shRNA) (D, lane 1) or right (Mule shRNA) (D, lane 2) flanks of athymic nude mice. (Top) Representative photos of five control and five Mule-depleted mice at 3 wk post-injection. (Middle) Gross views of representative tumors isolated from the mice in the top panel. (Bottom) Tumors of injected mice at 3 wk post-injection. Data points are tumor weight per individual mouse. (Horizontal line) Mean weight ± SD for the group (n = 9). P-value is from unpaired Student’s t-test. (F) Proposed model for the role of Mule in cancer. (Left) In wild-type cells exposed to DMBA/PMA, oncogenic Ras signaling induces cell cycle arrest, senescence, and/or apoptosis through activation of the p19Arf–p53–p21 and p16/p15–Retinoblastoma (Rb) tumor suppressor pathways. Unlimited cellular proliferation and transformation are blocked. Thus, wild-type mice exposed to DMBA/PMA develop only small benign tumors such as papillomas. (Right) In Mule-deficient cells exposed to DMBA/PMA, c-Myc/Miz1 complexes that have accumulated due to the lack of Mule prevent p21 and p15 induction by oncogenic Ras signaling. This down-regulation of p21 and p15 weakens the cell cycle arrest/senescence/apoptosis barrier against unlimited proliferation and transformation. Thus, Mule-deficient mice exposed to DMBA/PMA develop larger papillomas and KA.
Next, we injected our transformed Mule WT;p53+/− or Mule KO;p53+/− MEFs subcutaneously into athymic nude mice, which were then monitored for tumor growth. Mule-deficient transformed MEFs produced larger growths in nude mice compared with controls (Fig. 7B), demonstrating that Mule deficiency enhances tumor development in vivo in a cell-autonomous manner. To investigate whether Miz1 is absolutely required for Mule’s tumor-suppressive function, transformed Mule WT;p53+/− or Mule KO;p53+/− MEFs expressing Miz1 shRNA were injected into nude mice. Regardless of Mule status, MEFs depleted of Miz1 failed to proliferate efficiently, and at least 4∼5 wk of growth was required to obtain tumors of a size that could be isolated. Nevertheless, we observed that Mule deficiency could not enhance tumor formation in the absence of Miz1 (Fig. 7C; Supplemental Fig. 5A), establishing that Miz1 is critical for Mule’s effects on tumor growth in vivo.
To confirm our results in a keratinocyte context, we infected PAM212 cells with retrovirus expressing HRasG12V plus either Mule shRNA or control shRNA. Mule shRNA successfully decreased Mule protein and, consistent with our previous results, induced increases in Miz1 and c-Myc proteins and a decrease in p21 protein (Fig. 7D). We then injected our transformed PAM212 cells expressing control shRNA or Mule shRNA into nude mice and monitored tumorigenesis. PAM212 cells expressing HRasG12V plus Mule shRNA displayed accelerated tumor growth compared with PAM212 cells expressing HRasG12V plus control shRNA (Fig. 7E; Supplemental Fig. 5B), confirming in vivo our in vitro finding that Mule deficiency in keratinocytes enhances skin tumor growth. Taken together, our results identify Mule as a bona fide in vivo tumor suppressor that controls the stability of c-Myc/Miz1 complexes and thereby influences the effects of Miz1 on the proliferative regulators p21 and p15.
Discussion
It has been difficult to ascertain the precise role of Mule in cancer because both tumor suppressors (p53, ATM, and p19Arf) and oncogenes (c-Myc, N-Myc, and Miz1) either associate with it or are its substrates. Our findings definitively demonstrate that (1) Mule suppresses oncogenic Ras signaling in vivo, (2) the c-Myc/Miz1 complex is a major Mule substrate, and (3) Mule’s tumor-suppressive function depends on its ability to prevent c-Myc/Miz1-mediated down-regulation of p21 and p15.
The role of c-Myc in skin tumorigenesis was previously examined using c-Myc transgenic and c-Myc knockout mice (Oskarsson et al. 2006; Honeycutt et al. 2010). Mice lacking c-Myc showed increased p21 and resistance to DMBA/PMA-induced skin carcinogenesis. Concomitant knockout of p21 fully restored skin tumor formation, indicating that c-Myc is required for HRas-mediated skin tumorigenesis through its regulation of p21 (Oskarsson et al. 2006). However, whether c-Myc itself down-regulates p21 during DMBA/PMA-mediated skin carcinogenesis was not clearly established. Our results suggest that this is not the case.
Miz1 has a dual role in transcriptional regulation of its target genes. On the one hand, Miz1 forms a repressive complex with c-Myc that down-regulates genes encoding CDK inhibitors, including p15, p21, and p57. On the other hand, in the absence of c-Myc, Miz1 supports the transcriptional activation of target genes such as Bcl-2, clusterin, integrins, and cell adhesion-related molecules (Gebhardt et al. 2006). A direct role for Miz1 in cancer was recently revealed using a conditional knockout mouse lacking Miz1 specifically in keratinocytes (K14Cre;Miz1fl/fl) (Honnemann et al. 2012). Upon DMBA/PMA exposure, these Miz1-deficient mutants developed fewer and smaller skin tumors, indicating that Miz1 promotes oncogenesis. PMA-exposed primary keratinocytes lacking Miz1 exhibited up-regulated p21 and p15, reduced proliferation, and enhanced differentiation, all of which were reversed with concomitant knockout of p21 but not p15. Thus, Miz1’s oncogenic function depends on its regulation of p21. p21 is known to have a direct role in preventing skin tumorigenesis, since keratinocytes lacking p21 show increased proliferation in vivo and resistance to differentiation stimuli (Missero et al. 1996). Furthermore, HRas-transformed p21-deficient keratinocytes are more tumorigenic in vivo than controls (Missero et al. 1996).
Our data provide the missing link between c-Myc, Miz1, and p21 and implicate Mule in this pathway. We showed that, in the absence of Mule, c-Myc/Miz1 complexes accumulate and down-regulate p21 and p15. As a result, Mule-deficient cells exhibit increased proliferation and an enhanced ability to initiate tumorigenesis. A model of how we believe Mule exerts its tumor-suppressive function during skin carcinogenesis in vivo is described in Figure 7F.
Several questions remain regarding the role of Mule in cancer. Chief among these is whether Mule’s oncogenic capacity is tissue- and/or oncogene-specific. Indeed, p53, rather than c-Myc/Miz1, is Mule’s predominant substrate in B cells (Hao et al. 2012) and pancreatic β cells (Kon et al. 2012). It is also unclear whether Mule’s tumor-suppressive function is general or Ras-specific and which additional factors are required for full transformation. Although wild-type mice developed only papillomas in our system, Mule deficiency resulted in papillomas or KAs but not the malignant squamous cell carcinomas (SCCs) observed when p53 is absent (Supplemental Table II). While both KAs and SCCs are more rapidly proliferating cutaneous tumors than papillomas and some groups believe that KAs are low-grade SCCs, others point out that KAs differ from SCCs in their morphologic features, tumor kinetics, and potential for regression (Schwartz 1994). It may be relevant that, in addition to driving keratinocyte proliferation, Myc supports the differentiation of epidermal stem cells (Watt et al. 2008). K14Cre-Myc transgenic mice spontaneously develop gross histological abnormalities that were not apparent in our Mule kKO mice (Fig. 5D). These intriguing differences suggest that the function of Mule in epidermal stem cell differentiation should be investigated.
The half-life of c-Myc in keratinocytes is regulated by the E3 ligases Fbxw7 (Ishikawa et al. 2013) and Skp2 (Muller and Eilers 2008). For Fbxw7-mediated c-Myc degradation to occur, c-Myc must be phosphorylated on Thr 58 by ERK and on Ser 62 by GSKβ (Muller and Eilers 2008). However, Mule deficiency does not affect oncogenic Ras-induced phosphorylation of ERK, Akt, or GSK3β (Supplemental Fig. 6A). Similarly, Mule overexpression reduces not only wild-type c-Myc but also a T58A/S62A c-Myc mutant that cannot be phosphorylated (Supplemental Fig. 6B). Moreover, siRNA knockdown of either Fbxw7 or Skp2 further increases c-Myc accumulation in Mule knockdown cells (Supplemental Fig. 7). These data strongly suggest that the Mule/c-Myc axis is independent of Fbxw7 and Skp2.
A recent study implicates c-Myc as a universal amplifier that enhances the transcriptional output of all activated genes, rather than acting on specific “Myc target genes” (Lin et al. 2012). This same group has also questioned the validity of normalizing gene expression data to total cellular RNA content and has proposed normalization to cell number instead (Loven et al. 2012). However, we have preliminary data (data not shown) confirming that neither genetic deficiency nor knockdown of Mule affects cellular RNA content. Thus, our quantitative RT–PCR results (Fig. 3B,C) remain reliable regardless of normalization approach. This finding also suggests that, unlike ectopically overexpressed c-Myc, the c-Myc accumulating in Mule kKO cells does not act as a transcriptional amplifier.
In addition to Miz1’s function in transcriptional regulation in the nucleus, this protein can act in the cytoplasm to suppress inflammatory signaling initiated by tumor necrosis factor α (TNFα). Upon TNFα stimulation, Miz1 is recruited to the TNF receptor 1 (TNFR1) complex at the cell membrane by associating with the adaptor protein TRAF2. TNFα-induced JNK activation and inflammation are suppressed unless Miz1 is removed by Mule-mediated ubiquitination/proteasomal degradation (Yang et al. 2010; Lin 2011; Liu et al. 2012). We undertook preliminary examinations of TNFR1 signaling in PAM212 cells expressing Mule shRNA. TNFα-induced JNK1 activation is slightly reduced by Mule depletion, while cycloheximide/TNFα-induced apoptosis is normal (Supplemental Fig. 8). We are currently investigating TNFR1 signaling in Mule kKO keratinocytes.
A key finding of our work is that Mule acts on Miz1 complexed to c-Myc, with consequent p21 and p15 mRNA down-regulation. Our overexpression experiments have demonstrated that, although Mule acts on both c-Myc and Miz1, its preferred target is c-Myc rather than Miz1 (Fig. 4A,B). On the other hand, our Miz1 siRNA experiments suggest that Miz1 plays a role in stabilizing the c-Myc/Miz1 complex (Fig. 7A). The precise biochemical mechanisms by which Mule targets the c-Myc/Miz1 complex remain to be elucidated.
Although expression profiling and mutational analyses of MULE in human cancers have been performed, the available clinical data do not firmly identify MULE as either a tumor suppressor or a tumor promoter. MULE is up-regulated in cancers of the breast, colon, lung, prostate, larynx, liver, pancreas, and thyroid but down-regulated in stomach and uterine tumors as well as glioblastomas (Adhikary et al. 2005; Confalonieri et al. 2009). The MULE locus is deleted in some glioblastoma patients (Zhao et al. 2009) but commonly amplified in individuals with non-syndromic intellectual disability. These observations suggest that, while normal MULE levels may suppress brain cancer, too much MULE promotes mental retardation (Froyen et al. 2012). According to publicly available databases, low MULE expression correlates with poor prognosis in breast and ovarian cancer patients (Supplemental Fig. 9). In addition, examinations of the Catalogue of Somatic Mutations in Cancer (COSMIC) (Supplemental Fig. 10A) and the cBio Cancer Genomics database (Supplemental Fig. 10B) confirm that somatic mutation, homozygous deletion, or amplification of MULE occurs in many human cancers. Although most of these mutations are located in nonfunctional domains of MULE, mutations in the HECT domain have been identified in a few patients with colon and rectal adenocarcinomas (Supplemental Fig. 10A). Interestingly, homozygous deletion of MULE has been documented in ∼4% of SCC of the head and neck (SCCHN) (Supplemental Fig. 10B). Currently available human tumor sequencing data also provide evidence suggesting that Miz1 and Ras may cooperate in human cancers. MIZ1 amplification occurs most often in soft tissue sarcomas, at a frequency of ∼5% (Supplemental Fig. 11A). Interestingly, there is a strong association between MIZ1 amplification and amplification of RAS family members in that ∼86% of tumors showing RAS amplification also display MIZ1 amplification (P < 0.01) (Supplemental Fig. 11B). Although the biological and clinical consequences of homozygous MULE deletion and concurrent MIZ1/RAS amplification remain to be explored, these observations support the relevance of our model to human tumor development.
In conclusion, our study has demonstrated a crucial in vivo function for Mule in suppressing tumorigenesis induced by oncogenic Ras signaling. Our findings have clarified the perplexing and multifaceted role of Mule and point to Miz1 as a potential new target for novel anti-cancer therapies.
Materials and methods
Mice
The generation of Mulefl/fl(y) mice has been previously described (Hao et al. 2012). Mulefl/fl(y) mice were bred with K14Cre (Jonkers et al. 2001), p53flox/flox (Jonkers et al. 2001), c-Mycflox/flox (de Alboran et al. 2001), p19Arf−/− (Zindy et al. 2003), or p53−/− (Jacks et al. 1994) mice. All breedings were approved by the University Health Network Animal Care Committee (UHN-ACC; ID: AUP985).
Plasmids
Plasmids pRK5-HA-Ub-WT, pRK5-HA-Ub-K48O, and pRK5-HA-Ub-K63O (Addgene, catalog nos. 17608, 17605, and 17606, respectively) were from Addgene. The Mule expression vectors were the kind gift of Dr. Qing Zhang (Zhong et al. 2005). The wild-type and T58A/S62A c-Myc expression vectors (Supplemental Fig. 6B) were the kind gifts of Dr. Linda Penn (University Health Network). To construct the c-Myc expression vector (Fig. 4), full-length c-Myc cDNA was subcloned into pcDNA3.1/Zeo (Invitrogen). To construct the Miz1 expression vector, full-length Miz1 cDNA was subcloned into pCMV6-A-Puro (Origene, catalog no. PS100025). To create the base pMX-HRasG12V plasmid with which to construct shRNA vectors, the HRasG12V insert from pBabe-puro-HRasG12V (Addgene, catalog no. 1768) was subcloned into a pMXs-puro retroviral vector (Cell Biolab, catalog no. RTV-012). To create the pMX-HRasG12V-IRES-Puror-shRNAmir plasmids, IRES-Puror-shRNAmir sequences from pGIPZ-Miz1 shRNA (Open Biosystems, catalog no. RHS4531-NM_003443; clone ID: V3LHS_391462 for Fig. 7C and V3LHS_391464 for Supplemental Fig. 5A), pGIPZ-Mule shRNA (Open Biosystems, catalog no. RHS4531-NM_031407; clone ID: V2LHS_110969 for Fig. 7D,E and V2LHS_262858 for Supplemental Fig. 5B), or pGIPZ-control shRNA (Open Biosystems, catalog no. RHS4346) were amplified and subcloned into pMX-HRasG12V using the In-Fusion HD cloning system (Clontech, catalog no. 639646).
Primary MEFs
Primary MEFs were prepared from E13.5 mouse embryos and cultured in DMEM containing 10% fetal calf serum (FCS) plus 55 μM 2-mercaptoethanol. For Cre-mediated deletion of the floxed Mule allele, primary MEFs (2 × 10 to approximately 2.5 × 104 cells) were infected overnight with 5 × 106 plaque formation unit (pfu) of either control adenovirus (Ad-CMV-Null; Vector Biolaboratories, catalog no. 1300) or adenovirus expressing Cre (Ad-CMV-Cre; Vector Biolaboratories, catalog no. 1045). Retroviral-mediated gene transfer in primary MEFs was performed as previously described (Serrano et al. 1997) with slight modifications. Briefly, early passage primary MEFs (P1 or P2) were infected with pMXs-RasG12V-IRES-Puro retroviral vector expressing either scrambled control shRNA or Miz1 shRNA. Infected cell populations were selected in puromycin (2 μg/mL) for 24 h. Transformed MEFs were cultured for 2–3 wk before use in allograft experiments.
Primary epidermal keratinocytes
Primary epidermal keratinocytes isolated from newborn pups were cultured as described (Lichti et al. 2008). For Cre-mediated deletion of the floxed Mule allele, primary keratinocytes (1 × 104 to approximately 2 × 104 cells per well in six-well collagen I-coated plates; BD, catalog no. 354400) were infected overnight with either Ad-CMV-Null or Ad-CMV-Cre (5 × 106 pfu per well). Cell numbers were counted every 2 d using a Vi-Cell XR cell counter (Beckman Coulter). BrdU incorporation was measured using the APC-BrdU Flow kit (BD Pharmingen). For colony formation assays, primary keratinocytes at P1 were seeded in six-well collagen I-coated plates (2 × 104 cells per well). After 2 wk, cells were stained with 0.5% crystal violet, and primary colonies were counted. For Miz1 knockdown, primary keratinocytes were transfected with 50 nM siRNA directed against Miz1/ZBTB17 (Dharmacon, catalog no. L-046195-01), Fbxw7 (Dharmacon, catalog no. L-041553-01), or Skp2 (Dharmacon, catalog no. L-040845-01) using Lipofectamine 2000 (Invitrogen).
Immunoblotting
Cells were lysed in 1% Triton X-100 lysis buffer (20 mM Tris-HCl at pH 8, 100 mM NaCl, 10% glycerol) containing protease inhibitor (Roche, catalog no. 1183617001) for 30 min on ice. Cell lysates (10∼50 μg) were subjected to standard SDS-PAGE and immunoblotting using the primary Abs listed in Supplemental Table III.
Histology and IHC
Samples of normal dorsal skin and skin tumors were fixed in formalin prior to hematoxylin–eosin (HE) staining. For IHC, normal skin and skin tumor samples were fixed with formalin, and 5-μm paraffin sections were subjected to immunostaining using the primary Abs listed in Supplemental Table IV. For SA-β-gal staining, tissues were mounted in OCT compound (SakuraFinetek) and stained with SA-β-gal as previously described (Sun et al. 2007).
Real-time RT–PCR
Total RNA was extracted using the NucleoSpin RNA II kit (Clontech, catalog no. 740955) and reverse-transcribed using iScript (Bio-Rad, catalog no. 170-8891). The resulting cDNAs were diluted to 1:5∼10 and served as templates for real-time PCR using Power SYBR Green PCR Master Mix (Applied Biosystems) and 500 nM forward primer plus 500 nM reverse primer on an ABI 7700 instrument (Applied Biosystems). All procedures were performed according to the manufacturer’s instructions. The primer sequences used for real-time RT–PCR are listed in Supplemental Table V. Each sample was assayed in triplicate, and data were normalized to the housekeeping gene 18S ribosomal RNA. Results were calculated using the comparative threshold cycle method (2-ΔΔCt).
ChIP assay
ChIP assays were performed using anti-Miz1 (Santa Cruz Biotechnology, H181) and anti-c-Myc (Santa Cruz Biotechnology, N262) Abs as previously described with slight modifications (Staller et al. 2001; Herold et al. 2002; Wu et al. 2003).
Ubiquitination assay
PAM212 cells (2 × 105 cells per well in six-well plates) were cotransfected with pcDNA3.1/Zeo-c-Myc (0.6 μg) and/or pCI.G-Neo-3xFlag-12xHis-mouse Mule FL (1 μg) plus pRK5-HA-Ub-WT, pRK5-HA-Ub-K48O, or pRK5-HA-Ub-K63O (1 μg). At 24 h post-transfection, transfected PAM212 cells were exposed to 5 μM MG132 for 2 h and lysed in 0.5% Triton X-100 buffer. The resulting cell lysates were precleared with protein G beads (GE Healthcare) for 1 h at 4°C, and the supernatants were immunoprecipitated with anti-c-Myc (Santa Cruz Biotechnology, N262) or anti-Miz1 (Santa Cruz Biotechnology, H190) Ab overnight at 4°C. Immunocomplexes were washed five times with lysis buffer and subjected to standard immunoblotting using anti-c-Myc, anti-Miz1, or HRP-conjugated anti-HA Abs.
Skin carcinogenesis and hyperplasia
For skin carcinogenesis, the two-step DMBA/PMA model was employed using mice of the C57B6/FVB (F4) genetic background. Briefly, dorsal skin of 4-wk-old mice was exposed once to DMBA (25 μg in 100 μL of acetone) followed by biweekly exposure to PMA (6.25 μg in 100 μL of acetone) starting at 2 wk post-DMBA exposure. Tumor number and size were measured once per week. For PMA-induced epidermal hyperplasia, dorsal skin of mice was exposed to PMA (6.25 μg in 100 μL of acetone). Skin samples were analyzed on day 5. All skin carcinogenesis experiments were approved by the UHN-ACC (ID: AUP733).
Allografts
MEFs (1 × 106) or PAM212 cells (2 × 106) were suspended in 100 μL of medium and injected subcutaneously into the left or right flank of female NIH athymic nude mice. Mice were monitored for 3–5 wk, and tumors were isolated and weighed. All allograft experiments were approved by the UHN-ACC (ID: AUP732).
Acknowledgments
This study is dedicated to the memory of Dr. Susan Teresa McCracken (1954–2012), whose massive enthusiasm and support for this work are sincerely appreciated. We thank Dr. Stuart Yuspa of the National Cancer Institute for the PAM212 cell line. For their insightful comments and generous assistance, we are grateful to all members of the Mak laboratory, particularly Ms. Irene Ng; Dr. Eleonora Candi from Roma University; and Dr. Gerald M Cohen, Dr. Richard Knight, and Dr. Alessandro Rufini of the MRC Toxicology Unit in the United Kingdom. We also appreciate the extensive efforts of the staff members of the flow cytometer facility, the genotyping facility, and the animal resource center at the Princess Margaret Hospital (Toronto). Finally, we are grateful to Dr. Mary Saunders for scientific editing of this manuscript. This work was supported by a grant to T.W.M. and Z.H. from the Canadian Institutes of Health Research and by grants to DG.M. from the AIRC (AIRC 2011-IG11955 and AIRC 5xmile, #9979).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.214577.113.
References
- Adhikary S, Peukert K, Karsunky H, Beuger V, Lutz W, Elsasser HP, Moroy T, Eilers M 2003. Miz1 is required for early embryonic development during gastrulation. Mol Cell Biol 23: 7648–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. 2005. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123: 409–421 [DOI] [PubMed] [Google Scholar]
- Basu S, Liu Q, Qiu Y, Dong F 2009. Gfi-1 represses CDKN2B encoding p15INK4B through interaction with Miz-1. Proc Natl Acad Sci 106: 1433–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL 1989. ras oncogenes in human cancer: A review. Cancer Res 49: 4682–4689 [PubMed] [Google Scholar]
- Bringold F, Serrano M 2000. Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol 35: 317–329 [DOI] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W 2005. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121: 1071–1083 [DOI] [PubMed] [Google Scholar]
- Confalonieri S, Quarto M, Goisis G, Nuciforo P, Donzelli M, Jodice G, Pelosi G, Viale G, Pece S, Di Fiore PP 2009. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene 28: 2959–2968 [DOI] [PubMed] [Google Scholar]
- D’Arca D, Zhao X, Xu W, Ramirez-Martinez NC, Iavarone A, Lasorella A 2010. Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proc Natl Acad Sci 107: 5875–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14: 45–55 [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Tuveson DA 2009. RAS in cellular transformation and senescence. Eur J Cancer (Suppl 1) 45: 211–216 [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc’s broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Belet S, Martinez F, Santos-Reboucas CB, Declercq M, Verbeeck J, Donckers L, Berland S, Mayo S, Rosello M, et al. 2012. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am J Hum Genet 91: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Frye M, Herold S, Benitah SA, Braun K, Samans B, Watt FM, Elsasser HP, Eilers M 2006. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol 172: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Kosan C, Herkert B, Moroy T, Lutz W, Eilers M, Elsasser HP 2007. Miz1 is required for hair follicle structure and hair morphogenesis. J Cell Sci 120: 2586–2593 [DOI] [PubMed] [Google Scholar]
- Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q, Cook JG 2007. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell 18: 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Duncan GS, Su YW, Li WY, Silvester J, Hong C, You H, Brenner D, Gorrini C, Haight J, et al. 2012. The E3 ubiquitin ligase Mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J Exp Med 209: 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkert B, Eilers M 2010. Transcriptional repression: The dark side of myc. Genes Cancer 1: 580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell 10: 509–521 [DOI] [PubMed] [Google Scholar]
- Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R, Bernards R, Eilers M 2008. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J 27: 2851–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeycutt KA, Waikel RL, Koster MI, Wang XJ, Roop DR 2010. The effect of c-myc on stem cell fate influences skin tumor phenotype. Mol Carcinog 49: 315–319 [DOI] [PubMed] [Google Scholar]
- Honnemann J, Sanz-Moreno A, Wolf E, Eilers M, Elsasser HP 2012. Miz1 is a critical repressor of cdkn1a during skin tumorigenesis. PLoS ONE 7: e34885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Hosogane M, Okuyama R, Aoyama S, Onoyama I, Nakayama KI, Nakayama K 2013. Opposing functions of Fbxw7 in keratinocyte growth, differentiation and skin tumorigenesis mediated through negative regulation of c-Myc and Notch. Oncogene 32: 1921–1932 [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425 [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA 2008. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol 9: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ 2004. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol 2: e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CJ, Donehower LA, Bradley A, Balmain A 1993. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell 74: 813–822 [DOI] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianov GL 2011. The emerging role of Mule and ARF in the regulation of base excision repair. FEBS Lett 585: 2831–2835 [DOI] [PubMed] [Google Scholar]
- Kon N, Zhong J, Qiang L, Accili D, Gu W 2012. Inactivation of arf-bp1 induces p53 activation and diabetic phenotypes in mice. J Biol Chem 287: 5102–5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosan C, Saba I, Godmann M, Herold S, Herkert B, Eilers M, Moroy T 2010. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity 33: 917–928 [DOI] [PubMed] [Google Scholar]
- Kurek D, Garinis GA, van Doorninck JH, van der Wees J, Grosveld FG 2007. Transcriptome and phenotypic analysis reveals Gata3-dependent signalling pathways in murine hair follicles. Development 134: 261–272 [DOI] [PubMed] [Google Scholar]
- Levens D 2002. Disentangling the MYC web. Proc Natl Acad Sci 99: 5757–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U, Anders J, Yuspa SH 2008. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc 3: 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A 2011. Temporal control of TNFα signaling by Miz1. Adv Exp Med Biol 691: 127–128 [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yan J, Jiang S, Wen J, Chen L, Zhao Y, Lin A 2012. Site-specific ubiquitination is required for relieving the transcription factor Miz1-mediated suppression on TNF-α-induced JNK activation and inflammation. Proc Natl Acad Sci 109: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA 2012. Revisiting global gene expression analysis. Cell 151: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markkanen E, van Loon B, Ferrari E, Parsons JL, Dianov GL, Hubscher U 2012. Regulation of oxidative DNA damage repair by DNA polymerase λ and MutYH by cross-talk of phosphorylation and ubiquitination. Proc Natl Acad Sci 109: 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ 2008. Reflecting on 25 years with MYC. Nat Rev Cancer 8: 976–990 [DOI] [PubMed] [Google Scholar]
- Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto GP 1996. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev 10: 3065–3075 [DOI] [PubMed] [Google Scholar]
- Muller J, Eilers M 2008. Ubiquitination of Myc: Proteasomal degradation and beyond. Ernst Schering Found Symp Proc 2008: 99–113 [DOI] [PubMed] [Google Scholar]
- Noy T, Suad O, Taglicht D, Ciechanover A 2012. HUWE1 ubiquitinates MyoD and targets it for proteasomal degradation. Biochem Biophys Res Commun 418: 408–413 [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Essers MA, Dubois N, Offner S, Dubey C, Roger C, Metzger D, Chambon P, Hummler E, Beard P, et al. 2006. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev 20: 2024–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Tait PS, Finch D, Dianova II, Edelmann MJ, Khoronenkova SV, Kessler BM, Sharma RA, McKenna WG, Dianov GL 2009. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J 28: 3207–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba I, Kosan C, Vassen L, Moroy T 2011. IL-7R-dependent survival and differentiation of early T-lineage progenitors is regulated by the BTB/POZ domain transcription factor Miz-1. Blood 117: 3370–3381 [DOI] [PubMed] [Google Scholar]
- Schwartz RA 1994. Keratoacanthomata. J Am Acad Dermatol 30: 1–19 [DOI] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J 2001. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol 3: 400–408 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, et al. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol 3: 392–399 [DOI] [PubMed] [Google Scholar]
- Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, et al. 2007. PRAK is essential for ras-induced senescence and tumor suppression. Cell 128: 295–308 [DOI] [PubMed] [Google Scholar]
- Wanzel M, Herold S, Eilers M 2003. Transcriptional repression by Myc. Trends Cell Biol 13: 146–150 [DOI] [PubMed] [Google Scholar]
- Watt FM, Frye M, Benitah A 2008. MYC in mammalian epidermis: How can an oncogene stimulate differentiation? Nat Rev Cancer 8: 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG 2003. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22: 351–360 [DOI] [PubMed] [Google Scholar]
- Yang Y, Do H, Tian X, Zhang C, Liu X, Dada LA, Sznajder JI, Liu J 2010. E3 ubiquitin ligase Mule ubiquitinates Miz1 and is required for TNFα-induced JNK activation. Proc Natl Acad Sci 107: 13444–13449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuspa SH, Hawley-Nelson P, Koehler B, Stanley JR 1980. A survey of transformation markers in differentiating epidermal cell lines in culture. Cancer Res 40: 4694–4703 [PubMed] [Google Scholar]
- Zhang J, Kan S, Huang B, Hao Z, Mak TW, Zhong Q 2011. Mule determines the apoptotic response to HDAC inhibitors by targeted ubiquitination and destruction of HDAC2. Genes Dev 25: 2610–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A 2008. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol 10: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, D’Arca D, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, et al. 2009. The N-Myc–DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell 17: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X 2005. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121: 1085–1095 [DOI] [PubMed] [Google Scholar]
- Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, Roussel MF, Sherr CJ 2003. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci 100: 15930–15935 [DOI] [PMC free article] [PubMed] [Google Scholar]