Fertilization is a crucial yet poorly characterized event in eukaryotes. Here, Ning et al. use RNA sequencing to identify genes uniquely expressed in Chlamydomonas gametes. They show that the Chlamydomonas gene CrGEX1 and its Plasmodium ortholog are essential for the production of viable meiotic progeny and thus for mosquito transmission of malaria. This comparative transcriptomics approach provides a new resource for studying sexual development and identifies a previously unrecognized family of factors essential for sexual reproduction.
Keywords: RNA sequencing, fertilization, Plasmodium, Chlamydomonas, KAR5/GEX1/Brambleberry, nuclear envelope fusion
Abstract
Fertilization is a crucial yet poorly characterized event in eukaryotes. Our previous discovery that the broadly conserved protein HAP2 (GCS1) functioned in gamete membrane fusion in the unicellular green alga Chlamydomonas and the malaria pathogen Plasmodium led us to exploit the rare biological phenomenon of isogamy in Chlamydomonas in a comparative transcriptomics strategy to uncover additional conserved sexual reproduction genes. All previously identified Chlamydomonas fertilization-essential genes fell into related clusters based on their expression patterns. Out of several conserved genes in a minus gamete cluster, we focused on Cre06.g280600, an ortholog of the fertilization-related Arabidopsis GEX1. Gene disruption, cell biological, and immunolocalization studies show that CrGEX1 functions in nuclear fusion in Chlamydomonas. Moreover, CrGEX1 and its Plasmodium ortholog, PBANKA_113980, are essential for production of viable meiotic progeny in both organisms and thus for mosquito transmission of malaria. Remarkably, we discovered that the genes are members of a large, previously unrecognized family whose first-characterized member, KAR5, is essential for nuclear fusion during yeast sexual reproduction. Our comparative transcriptomics approach provides a new resource for studying sexual development and demonstrates that exploiting the data can lead to the discovery of novel biology that is conserved across distant taxa.
Sexual reproduction and its accompanying ploidy transitions are fundamental processes in eukaryotes, yet the cellular and molecular mechanisms for generation of gametes and the eventual fusion of these specialized cells and their haploid nuclei to reconstitute the diploid genome remain obscure. In spite of the many cellular events common to fertilization and zygote development in almost all eukaryotes, only recently was the first gamete-specific gene identified that is conserved and essential for fertilization in broadly disparate organisms. The gamete membrane protein HAP2(GCS1) is now known to be essential for gamete fusion in the isogamous, unicellular green alga Chlamydomonas; the unicellular malaria organism Plasmodium; and the higher plant Arabidopsis (Mori et al. 2006; von Besser et al. 2006; Hirai et al. 2008; Liu et al. 2008). Based on our results from Chlamydomonas and Plasmodium, the current model is that HAP2 is a gamete membrane fusogen in all three organisms and presumably in the many other protists, plants, and multicellular animals that possess it (Mori et al. 2006; von Besser et al. 2006; Hirai et al. 2008; Liu et al. 2010; Wong and Johnson 2010).
These observations made it likely that a Chlamydomonas/Plasmodium comparative genomics strategy would uncover additional conserved sexual reproduction genes and reveal insights into fundamental events in fertilization. Importantly, discovering new molecules essential for Plasmodium sexual reproduction, which increasingly is recognized as a vulnerable “bottleneck” in its life cycle (Sinden 2009), could offer insights into methods to prevent the spread of this devastating organism.
In spite of the very complex life cycle of Plasmodium (which, in this parasitic pathogen, takes place in two different hosts), many of the events in Chlamydomonas and Plasmodium fertilization are remarkably similar. Both exist as asexually replicating haploid cells for most of their life and produce gametes by mitosis. In Chlamydomonas, gametogenesis is induced by nutrient deprivation of vegetatively growing plus and minus cells. In Plasmodium, gametes arise from gametocytes, dimorphic sexual precursor stages that originate from asexually replicating forms in the blood stream of the vertebrate host, where they remain developmentally arrested until the red blood cells in which they reside are ingested by a mosquito. Upon activation by conditions in the mosquito blood meal, haploid gametocytes egress from their host cells. Microgametocytes differentiate into eight sperm-like microgametes, each capable of fertilizing a macrogamete. In Plasmodium berghei, a malaria parasite infecting rodents, gamete adhesion is dependent on the Plasmodium-specific P48/45 and P47 gamete surface proteins (van Dijk et al. 2001).
In both organisms, the two gametes adhere to each other using species-limited proteins, position themselves appropriately for fusion, and then rapidly undergo HAP2-dependent fusion to form a zygote. Within the zygote, nuclear fusion takes place within minutes to hours. The zygote, however, does not undergo mitosis as in multicellular plants and animals but, after just a brief time as a diploid cell, undergoes meiosis. In Chlamydomonas, the nuclear envelope remains intact during meiosis and production of the four individual haploid cells. In Plasmodium, the two meiotic divisions also take place within an intact nuclear envelope, but the zygote remains a single cell. Concomitantly, the zygote undergoes cellular morphogenesis to become a motile ookinete that penetrates the midgut epithelium of the mosquito and forms an oocyst (Sinden et al. 1996). Over a period of days to weeks, the four haploid genomes replicate many times within the rapidly growing cyst, leading to the thousands of haploid progeny that eventually infect the salivary glands of the mosquito.
Here, we use next-generation sequencing of the Chlamydomonas transcriptome (Wang et al. 2009) to exploit the rare biological phenomenon of isogamy in Chlamydomonas to define clusters of coregulated genes as candidates for sexual reproduction. One gene that we identified in a minus gamete cluster is conserved in Plasmodium (PBANKA_113980) and is similar to an Arabidopsis gene, GEX1, previously shown to be involved in gametophyte development and embryogenesis (Alandete-Saez et al. 2011). We show that the protein localizes to the nuclear envelope of sexual stages of Chlamydomonas and Plasmodium, functions in nuclear fusion in Chlamydomonas, and is essential for production of viable meiotic progeny in both organisms. Our analysis uncovered a new, large GEX1-containing gene family whose first reported member encodes the Saccharomyces cerevisiae nuclear envelope protein KAR5, which is essential for nuclear fusion during sexual reproduction in yeast (Beh et al. 1997). We show that the GEX1/KAR5 family also includes the newly characterized nuclear envelope fusion protein Brambleberry, described recently in Danio rerio (Abrams et al. 2012) as being required for nuclear fusion and early embryo formation in zebrafish. This comparative genomics strategy provides an important resource to the research community. Moreover, it has led to the discovery of novel biology that is conserved across distant taxa and to the characterization of a new Plasmodium gene that functions only in the gut of the mosquito and is essential for mosquito transmission of malaria.
Results
The published reference genome of Chlamydomonas reinhardtii (Merchant et al. 2007) is from a plus laboratory strain (CC-503 cw92 mt+) that is related but not identical to the plus and minus strains used in the present study. We resequenced the genomes of the strains that we used and observed an average frequency of single-nucleotide polymorphisms of 0.42 (plus) and 1.72 (minus) per 1 kb of sequence (Supplemental Table S1), which we considered sufficiently low so as not to interfere significantly with the mapping of RNA sequencing (RNA-seq) reads to the reference genome. More than 20% of sequencing reads failed to map to the reference, highlighting the possibility that one or both laboratory strains contained sequences not present in the reference genome. These reads were provisionally assembled de novo. Putative gene models were predicted and included in the database to map RNA-seq reads. In the final analysis, 131 of these gene models (sequence IDs starting with “cuff1” or “g” in Supplemental Table S3) were supported by transcription data, including the MTA1 gene in the minus mating type locus, which is lacking in the reference (plus) strain (Ferris et al. 2002).
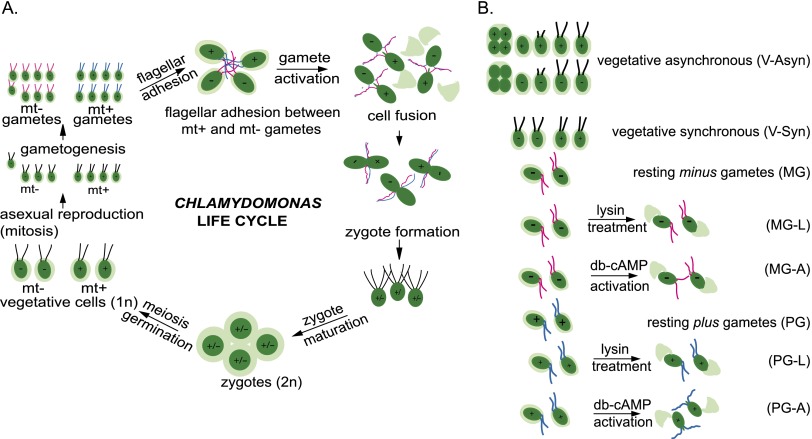
Next, we generated transcriptomes by deep sequencing mRNA extracts from C. reinhardtii plus gametes and minus gametes prepared under three different conditions to stimulate complex expression patterns of sexual development genes (Fig. 1B). Gametes of the plus or minus mating type were taken either at rest or following stimulation with dibutyryl cAMP to induce genes involved in gamete activation, a process that is triggered naturally by flagellar adhesion between plus and minus gametes (Snell and Goodenough 2009). We also treated gametes with the cell wall-degrading enzyme lysin to experimentally remove their cell walls, triggering expression of extracellular matrix (cell wall) genes, a process that occurs during gamete interactions (Buchanan and Snell 1985, 1988; Adair and Apt 1990; Kurvari 1997). To recognize more broadly expressed genes, we analyzed mRNA samples from vegetative cultures that were mixtures of the plus and minus strains. One vegetative sample consisted of flagellated interphase cells from synchronized cultures growing in a 13:11-h light:dark cycle. A second vegetative sample contained cells dividing asynchronously in continuous light, which would contain cells at all stages of the cell cycle. We reasoned that analysis of the transcriptomes of minus and plus strains cultured in such a range of experimental conditions should reveal diverse patterns of gene regulation that would allow us to cluster genes that were coregulated and identify gamete-specific and mating type-specific genes.
Figure 1.
Life cycle of Chlamydomonas and description of treatment groups. (A) Gametes are formed when vegetative plus and minus cells undergo gametogenesis induced by nitrogen deprivation. When gametes of opposite mating type are mixed, they adhere to each other by adhesion molecules on their flagella—SAG1 on plus gametes and SAD1 on minus gametes. Flagellar adhesion induces gamete activation and an increase in cAMP that leads to release of cell walls, erection of mating structures at the apical ends of the gamete cell bodies (shown at top right), and several other cellular and biochemical changes that prepare the gametes for cell fusion. All of the events associated with gamete activation can be induced in gametes of a single mating type by incubating them in dibutyryl cAMP. The mating structure adhesion molecule FUS1 on the plus gamete binds to an unidentified receptor on the minus mating structure, and the membranes of the two structures fuse through the action of the minus gamete-specific fusion protein HAP2. Upon coalescence into a quadriflagellated zygote, the plus gamete-specific homeodomain protein GSP1 interacts with the minus gamete-specific homeodomain protein GSM1 to form a complex essential for activation of the zygote developmental pathway. Within several hours, the plus and minus nuclei congress and undergo fusion to form the diploid zygote nucleus. Upon return to the light after an obligatory period (at least 5 d) in the dark, the zygote undergoes DNA replication and meiosis, and the resulting four haploid progeny are released (germination) from the remnant of the mother cell wall and enter the vegetative part of their life cycle (Snell and Goodenough 2009). (B) The following eight samples were analyzed by RNA-seq: asynchronously growing vegetative cells (V-asyn), synchronously growing vegetative cells (V-syn), resting mating type minus gametes (MG), lysin-treated minus gametes (MG-L), dibutyryl cAMP-activated minus gametes (MG-A), resting mating type plus gametes (PG), lysin-treated plus gametes (PG-L), and dibutyryl cAMP-activated plus gametes (PG-A).
For each experimental condition, we sequenced three biological replicates and two technical replicates, generating >736 million sequence reads, which we mapped onto 17,375 putative transcripts from the reference genome and any newly generated contigs (Supplemental Table S2). One biological replicate of the synchronous and one of the asynchronous vegetative cells were excluded from further analysis, since they yielded suboptimal read numbers. Supplemental Table S3 shows normalized expression values for each transcript as reads per kilobase per million mapped reads (RPKM). Using pairwise comparisons, we first determined Pearson correlation coefficients between all samples (Supplemental Fig. S1). Samples were most highly correlated by treatment group, with the exception of the second biological replicate of activated plus gametes, which was identified as an outlier and excluded from further analysis. In marked contrast, mating type proved not to be a major determinant of the transcriptomes, since even within treatment groups, samples from the same mating type did not generally cluster together. This confirmed our expectation that in Chlamydomonas, only a few transcripts would differentiate between mating types.
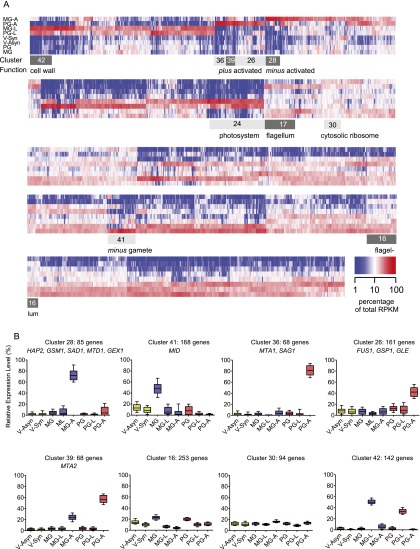
Next, we identified putative fertilization-related transcripts by grouping genes according to their expression patterns. We first normalized expression levels by transforming each RPKM value for a given transcript into a percentage of the sum of the RPKM values for that transcript in all eight treatment groups. Six-thousand-six-hundred-thirty-seven predicted gene models with no or very low expression across all treatment groups (highest RPKM ≤ 4) were eliminated from further analysis, since the data provided insufficient evidence to support specific expression patterns. Ward's hierarchical clustering algorithm was applied to the Euclidean distances between all possible pairs of normalized expression profiles for 10,738 transcripts. Partitioning the resulting tree into 42 clusters revealed compact and mostly well-separated sets of expression profiles (Fig. 2A; Supplemental Table S3). Remarkably, HAP2 and another three of the five known minus gamete-specific genes were in cluster 28 (C28), which comprises 0.5% of all C. reinhardtii genes. All 10 known mating type-specific genes were concentrated in five clusters, which together comprise only 5% of all C. reinhardtii genes.
Figure 2.
Cluster analysis of gene expression patterns in C. reinhardtii. (A) Heat map showing the relative abundance of 10,738 putative transcripts across eight treatment groups as a percentage of total RPKM for each transcript. Transcripts were clustered along the X-axis according to Ward. The light-gray and dark-gray boxes indicate the borders of selected clusters. (B) Relative expression patterns in selected clusters. Boxes show the median and the 25th and 75th percentiles. Whiskers indicate the 10th and 90th percentile. Marker genes with known functions in sexual development are indicated.
The composition of the clusters reflects gene function
Minus gamete activation clusters
C28 contains 85 genes expressed predominantly in minus gametes that are strongly responsive to activation but not to lysin treatment (Fig. 2B). Three of these have known functions specifically in minus gametes: the fusion-essential protein HAP2, the homeodomain protein GSM1, and the minus flagellar adhesion protein SAD1. A fourth, MTD1 (function unknown), is encoded only in the mating type minus locus (Ferris et al. 2002). A subset of genes within C28, but excluding the four minus marker genes, additionally showed milder up-regulation in activated plus gametes (PG-A [plus gamete activation]). The only other known minus-specific gene, the minus dominance gene MID, was present in C41 (Fig. 2B), which is characterized by transcripts with higher expression levels in resting minus gametes (MGs), consistent with the known role for MID in specifying mating type early during gametogenesis (Ferris and Goodenough 1997; Lin and Goodenough 2007).
PG-A clusters
Five genes known to be specific to plus gametes (the flagellar adhesion protein SAG1, the plus mating structure adhesion protein FUS1, the plus homeodomain protein GSP1, and two proteins of unknown function: MTA1 and MTA2) (Kurvari et al. 1998; Ferris et al. 2002; Misamore et al. 2003) were found in three related clusters (C26, C36, and C39) (Fig. 2B; Supplemental Table S3). The 68 genes in C36 (which contained MTA1 and the plus flagellar adhesion gene SAG1) were expressed exclusively in plus gametes and were strictly dependent on activation (Fig. 2B). In C26, FUS1, GSP1, and GLE (which encodes the cell wall-degrading lysin used for the MG-L and PG-L treatment groups) were also induced by activation but with higher backgrounds in one or more of the other samples. C39 was notable because it contained genes whose transcript profiles were relatively high in both the PG-A and MG-A (minus gamete activation) treatment groups. It is not surprising that MTA2 was in this cluster, since previous Northern blot studies showed that it was expressed in both types of gametes (Fig. 2B; Supplemental Table S3; Ferris et al. 2002).
Other cellular processes
To characterize gene expression clusters systematically, we looked for the significant enrichment of gene ontology (GO) terms (Supplemental Table S4). C16 and C17 were both enriched in motor and microtubule-associated proteins and, upon closer examination, were found to contain a striking assemblage of flagella-associated genes (Supplemental Table S5), which were expressed in vegetative cells and gametes, but (for unknown reasons) were negatively regulated upon lysin treatment or gamete activation (Fig. 2B). Further study of the uncharacterized genes in this cluster might uncover new flagella-related genes. C30 was enriched in genes involved in protein translation, which were equally expressed in all eight treatment groups (Fig. 2B). In marked contrast, photosynthesis genes, which were 17.5-fold enriched in C24 (Supplemental Table S4), were expressed nearly exclusively in the vegetative samples (Fig. 2B).
The profiles for the 143 genes in C42 stood out for being strongly induced by lysin treatment of plus and minus gametes, which induces synthesis of cell wall genes (Fig. 2B). The Chlamydomonas cell wall lacks cellulose and is enriched in hydroxyproline-rich glycoproteins. C42 contains known cell wall proteins, such as members of the pherophorin family, best characterized in Volvox (Godl et al. 1997), and several matrix metalloproteinases. Taken together, these data demonstrate that our experimental design succeeded in subtracting the vast majority of expressed genes from a small number of clusters highly enriched in coregulated genes with known functions in mating type-specific gamete activation and interaction.
Some mating type-specific genes have putative orthologs in other eukaryotes
To discover genes with conserved functions in gamete activation or interactions, we first searched the Plasmodium and human genomes for distant homologs of 199 Chlamydomonas genes, which were most closely coexpressed with either HAP2 or FUS1; i.e., genes whose expression was highly mating type-specific and tightly linked with activation. Supplemental Table S6 shows that in addition to the minus gamete fusion-essential gene HAP2, about a third of genes had homologs in humans or malaria parasites at the chosen significance threshold of E < 10−4. The data indicate potentially conserved genes and biological processes that have not previously been linked to sexual development, such as RNA modification by a pseudouridine synthase (Cre02.g086050), a putative ABC transporter (Cre04.g220850), or a putative inositol 1-phosphate synthase (Cre03.g180250). The presence of the latter gene in C39 is intriguing considering the recent implication of an inositol monophosphatase-like gene in sexual development of C. reinhardtii (Nishimura et al. 2012).
Cre06.g280600 is a conserved gene expressed in minus gametes and is similar to the fertilization-related gene GEX1 of Arabidopsis
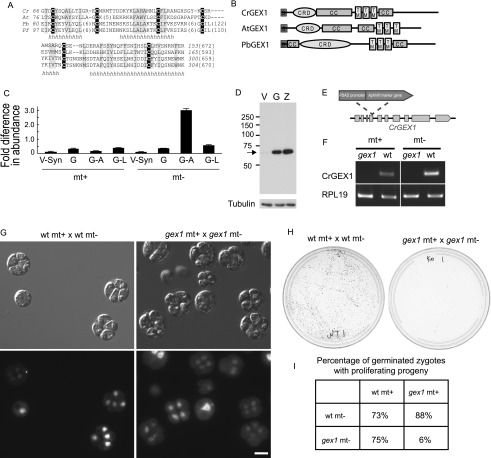
For biological validation of our candidate list, we used a stringent, PCR-based protocol (Gonzalez-Ballester et al. 2011) to screen a library of insertional Chlamydomonas mutants and obtained a mutant for Cre06.g280600 in a minus mating type strain. Cre06.g280600 is a gene from C28, which is enriched in minus gamete-specific genes (Fig. 2B). Not only was an ortholog of Cre06.g280600 present in Plasmodium, but it also was similar to a previously studied Arabidopsis gene with gamete-specific expression, GEX1 (Alandete-Saez et al. 2011). GEX1 proteins from all three species possessed three predicted transmembrane domains in the C-terminal half, each had several coiled-coil regions, and each possessed a conserved N-terminal signal peptide followed by a cysteine-rich domain (CRD) (Fig. 3A,B).
Figure 3.
Characterization of CrGEX1. (A) The 88-residue CRD is present in Chlamydomonas (Cr), P. berghei (Pb), Plasmodium falciparum (Pf), and Arabidopsis (At). (B) GEX1 family members have a predicted N-terminal signal peptide, two coiled-coil domains (CC), three transmembrane domains (TMs), and a CRD. (C) Levels of CrGEX1 transcripts expressed relative to the housekeeping gene RPL19. Error bars show standard deviations of three or more replicates. (D) Immunoblot showing CrGEX1 protein in synchronized mt− vegetative cells (V), gametes (G), and zygotes formed from mt+ and mt− gametes (Z). (E) Structure of the CrGEX1 genomic locus, illustrating insertion of the aminoglycoside 3′-phosphotransferase type VIII-encoding gene (AphVIII) at the second of 10 exons (boxes). (F) Diagnostic RT–PCR of cDNA showing the absence of CrGEX1 transcripts in gex1 gametes. (G) Differential interference contrast and fluorescence microscopy images of germinated zygotes formed from fusion of wild-type plus gametes with a wild-type minus gametes (shown in the left panel) and from gex1 plus gametes fused with gex1 minus gametes showing that most gex1/gex1 zygotes underwent meiosis to form progeny. Bar, 5 μm. (H) Agar plates 5 d after induction of germination showing that most progeny from gex1/gex1 zygotes failed to proliferate. Equal numbers of zygotes were plated. (I) Table showing percentage of zygotes examined 24–30 h after induction of germination whose progeny had begun to proliferate.
Quantitative PCR (qPCR) analysis of GEX1 expression confirmed the validity of the RNA-seq results, documenting low expression in vegetative cells, resting gametes, and lysin-treated gametes of both mating types and up-regulation only in minus gametes activated with db-cAMP (Fig. 3C). In a wild-type strain expressing a transgene encoding a Flag-tagged CrGEX1, the protein was almost undetectable in vegetative cells but was detected in gametes and in slightly increased amounts in 1-h zygotes (Fig. 3D). Because expression of the CrGEX1-Flag transgene was driven by a constitutive RBCS-HSP70 chimeric promoter (Sizova et al. 2001), the transcript or the protein must be unstable in vegetative cells, or sequences within the gene itself regulate expression (Lodha et al. 2008).
gex1/gex1 zygotes are blocked in production of viable meiotic progeny
The gex1 mutant that we identified was from a minus strain and carried an insertion of the AphVIII marker gene in the second predicted exon of the GEX1 gene (Fig. 3E). To assess the phenotype of GEX1 disruptants in both mating types, we crossed the minus gex1 mutant with a wild-type plus strain and isolated a plus gex1 mutant clone from the progeny. RT–PCR analysis showed that GEX1 transcripts were present in wild-type gametes but not in the plus or minus gex1 gametes (Fig. 3F). Vegetative cells of both gex1 strains were indistinguishable from wild-type vegetative cells in cell size, appearance, and growth rate. Similarly, both mutants underwent gametogenesis to form gametes that were indistinguishable from wild-type gametes. Mixtures of wild-type plus/wild-type minus, wild-type plus/gex1 minus, gex1 plus/wild-type minus, and gex1 plus/gex1 minus gametes all underwent flagellar adhesion and formed quadriflagellated zygotes at similar rates (50%–90%).
We observed a strong phenotype, however, when we tested for growth of zygotic progeny after meiosis. Newly formed wild-type/wild-type and gex1/gex1 zygotes plated onto agar were placed in the dark for 5 d and returned to continuous light to induce meiosis. Examination 14–18 h later showed that both the wild-type/wild-type and gex1/gex1 zygotes had initiated meiosis and had germinated and produced progeny. Overall, the gex1 progeny were similar to wild type, although more of the progeny from gex1/gex1 zygotes were uneven in cell and nuclear size, and in a small number of germinated zygotes, only two cells were visible (Fig. 3G). As expected, progeny from wild-type/wild-type crosses proliferated and produced collective colonies containing all four meiotic progeny. In contrast, the progeny from gex1/gex1 zygotes produced very few collective colonies (Fig. 3H).
Confirming the results from the collective colony experiments, when we separated gex1/gex1 tetrads into individual cells under a dissecting microscope, most failed to proliferate. Only four of 30 zygotes produced four progeny that initiated growth, two of 30 had three progeny that initiated growth, and two of 30 produced a single product that initiated growth. Finally, we found that one copy of the wild-type GEX1 gene in the zygote is sufficient and required for meiotic progeny to proliferate (Fig. 3I).
GEX1 is also required for growth of zygotic progeny in Plasmodium
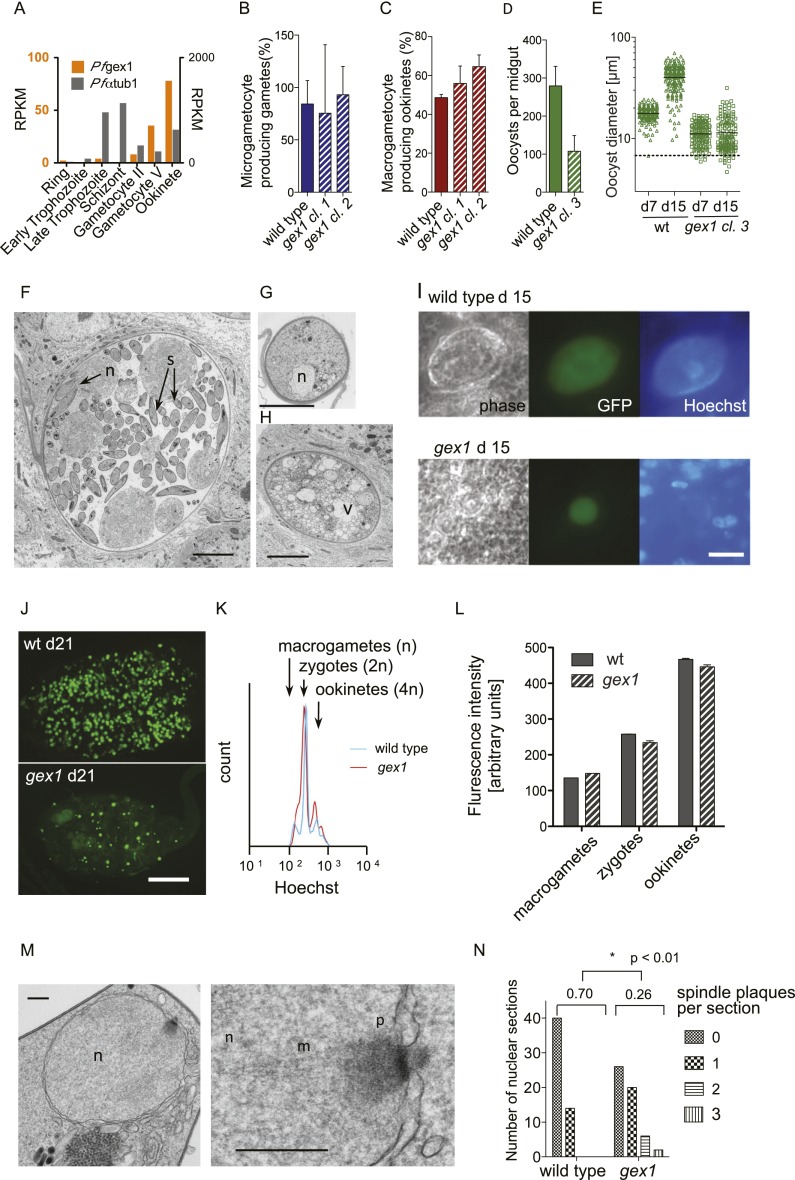
A single GEX1 homolog is present in each completely sequenced Plasmodium genome. In Plasmodium falciparum, the most lethal malaria parasite infecting humans, genome-wide expression analyses found PfGEX1 (PF3D7_1363800) expression very low in asexual blood stages but strongly up-regulated in gametocytes and ookinetes (Le Roch et al. 2003; López-Barragán et al. 2011), which would be consistent with a conserved function in sexual development (Fig. 4A). To investigate the function of GEX1 in Plasmodium, we disrupted the GEX1 gene (PBANKA_113980) in the rodent parasite P. berghei, in which the complete life cycle, including sexual stages, can be readily studied, by replacing the entire protein-coding region with an expression cassette conveying resistance to pyrimethamine (Supplemental Fig. S2A,B). Using two different vector designs, we obtained three independent knockout clones and confirmed their genotypes (Supplemental Fig. S2C,D). GEX1 was not required for asexual parasite growth in erythrocytes of infected mice, and the ability of asexual blood stages to give rise to gametocytes was not affected in the mutants.
Figure 4.
Phenotypic analysis of gex1 knockout clones in P. berghei. (A) Sexual stage-specific expression profile for P. falciparum gex1 from RNA-seq data by López-Barragán et al. (2011). Expression of α-tubulin 1 (PF3D7_0903700) is shown for comparison. (B) Exflagellating microgametocytes as a percentage of all microgametocytes in blood samples from infected mice. B–D show arithmetic means and standard deviations (SD) from three biological replicates. (C) Macrogametocyte-to-ookinete conversion in blood samples cultured for 24 h. (D) Average oocyst numbers on midguts 7 d after the infectious blood meal. Bars show arithmetic means ± SD of three transmission experiments per strain, each derived from analyzing 20–30 mosquitoes. (E) Size distribution of oocysts. Data from clone 3 are representative of observations with the other clones. (F) TEM of wild-type oocyst showing budding sporozoites (s) on day 15 post-feeding. (F–H) Bar, 5 μm. (G) Typical small gex1 oocyst on day 15. (H) Representative larger gex1 oocyst with vacuolation. (I) Immunofluorescence micrographs of oocysts showing replicated nuclei in the center of the cyst and lined up around its periphery in wild type but not in the gex1 cyst. Hoechst staining outside the oocyst is from nuclei of midgut epithelial cells. Bar, 20 μm. (J) Overview of whole, live infected midguts showing strong GFP fluorescence of oocysts and weak background fluorescence of the midgut tissue. Bar, 0.2 mm. (K) Fluorescence intensity profiles of macrogametocyte-derived stages purified from a representative 24-h ookinete culture and stained with Hoechst 33342. (L) Relative DNA content as determined by Hoechst fluorescence intensity of the different populations indicated in K. Error bars show the SD from three independent cultures and infections. (M) TEM of the nucleus of a 16-h-old wild-type ookinete illustrating an electron-dense spindle plaque (p) and emanating spindle microtubules (m) located in a nuclear pore. Bars, 250 nm. (N) Distribution of spindle plaque numbers in 54 ookinete nuclear sections per strain. Means are given and are significantly different by Student's t-test.
We therefore investigated the role of GEX1 in the subsequent stages of sexual development, which naturally occur in the blood meal of the mosquito vector. When gex1 gametocytes were triggered to differentiate into gametes by exposing them to activating conditions in vitro, the release of microgametes was normal (Fig. 4B), as was the frequency at which macrogametes converted to mature ookinetes (Fig. 4C), a process that requires fertilization (Sinden 2009). Consistent with the formation of ookinetes in vitro, all mosquitoes fed on infected mice had developed oocysts on their midgut wall when they were dissected 7 d later. However, the average number of oocysts per midgut appeared much reduced in the mutant (Fig. 4D). Importantly, gex1 oocysts had only half the diameter of wild-type cysts on day 7 and, on average, had not grown by day 15 (Fig. 4E). The apparent reduction in oocyst numbers may simply reflect our reduced ability to detect the smaller cysts of the mutant.
Transmission electron microscopy showed that on day 15, wild-type oocysts were forming sporozoites (Fig. 4F), while gex1 cysts either remained small and retained a compact nucleus (Fig. 4G) or became highly vacuolated but invariably failed to show signs of sporogony (Fig. 4H). On day 15, wild-type oocysts had replicated many times and stained strongly for DNA, while the few gex1 oocysts that increased at all in size had failed to replicate (Fig. 4I). After 21 d, while free sporozoites were readily noted in wild-type infections (data not shown), few gex1 oocysts remained (Fig. 4J), and no sporozoites were observed, indicating that PbGEX1 is essential for mosquito transmission of malaria.
Thus, as in Chlamydomonas, the gex1 phenotype in P. berghei becomes apparent after gamete fusion and after meiosis, at about the time when the oocyst first initiates a new replicative cycle. We therefore asked whether the developmental block in the young oocyst could be a delayed consequence of problems during meiosis. Flow cytometry of sexual stages purified from cultures 18 h after fertilization revealed three intensity peaks when stained with a fluorescent DNA dye, which correspond to unfertilized macrogametes (haploid), unreplicated zygotes (diploid), and replicated ookinetes (4C) (Fig. 4K,L). Neither the average distribution of parasites over these groups nor the average intensity within each developmental category differed between wild type and gex1, suggesting that DNA replication in the zygote does not require GEX1. An ultrastructural analysis of ookinetes 18 h after fertilization, however, suggested that meiosis in the gex1/gex1 zygotes was altered at a step after DNA replication. We detected meiotic spindle poles, marked by electron-dense centriolar plaques on the nuclear envelope (Fig. 4M; Sinden et al. 1976), more than twice as frequently in the gex1/gex1 zygotes as we did in wild-type Plasmodium zygotes (Fig. 4N). Thus, an uncharacterized event in the nuclear envelope was altered in the gex1/gex1 zygotes.
CrGEX1 is required for timely nuclear fusion
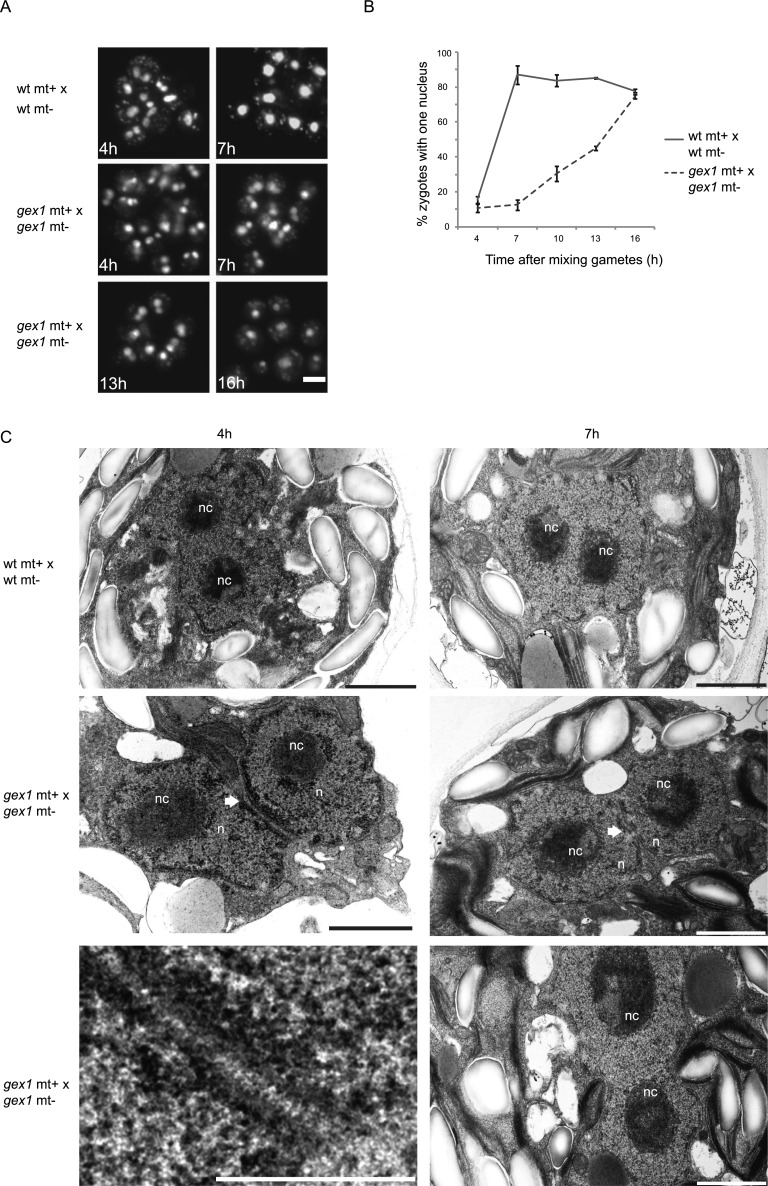
We next returned to Chlamydomonas, where large numbers of zygotes can readily be produced in culture, to ask whether GEX1 had a role in fertilization at a step between gamete fusion and meiosis. The gex1/gex1 zygotes withdrew their flagella and formed large collections of adhering zygotes similarly to wild-type cells (data not shown). On the other hand, nuclear fusion was strongly impaired in the gex1/gex1 zygotes. Four hours after mixing gametes, many of the nuclei appeared to be in close contact in both wild-type and gex1 zygotes when observed by light microscopy, but the two nuclei remained distinct. At 7 h, nuclear fusion was nearly complete in wild-type zygotes, and >85% of the cells had a single large nucleus. In 7-h gex1/gex1 zygotes (Fig. 5A,B), however, the two nuclei in most of the cells were closely apposed to each other but had failed to coalesce into a single nucleus. A small proportion of gametes that remained unfertilized accounted for a baseline value of 10% cells with single nuclei 4 h after mixing gametes. Thereafter, nuclear fusion began to occur in the gex1/gex1 zygotes but at a much lower rate than in wild-type zygotes. Transmission electron microscopy analysis was consistent with the light microscopy results (Fig. 5C). At 4 h, we found fused nuclei in wild-type zygotes, although each of the fused nuclei partially retained their shape, and at 7 h, the fused nuclei had completely coalesced into a single round nucleus with two nucleoli. In the 4-h gex1/gex1 zygotes, however, some nuclei were closely apposed to each other, but we did not detect fusion. Rather, the nuclei appeared to be adhering to each other and were connected by fine striations (Fig. 5C). In addition, in the 7-h gex1 zygotes, although we detected early stages of nuclear fusion, the two nuclei retained their individual shapes, which was consistent with the light microscopic results. Thus, CrGEX1 was essential for the rapid nuclear fusion that typifies wild-type zygotes.
Figure 5.
GEX1 is required for timely nuclear fusion in Chlamydomonas. (A) Fluorescence images of groups of zygotes. DNA of zygotes was labeled with SYTOX Green before gametes were mixed. Bar, 10 μm. (B) Nuclear fusion expressed as a percentage of zygotes with a single nucleus as determined by observing >100 SYTOX Green-stained cells per time point. Error bars show the standard error of the mean (SEM) from three independent experiments. (C) Transmission electron micrographs of wild-type and gex1/gex1 zygotes at the indicated times after plus and minus gametes were mixed together to undergo fusion. (n) Nucleus; (nc) nucleolus. Arrows point to sites where nuclear envelopes are in close apposition but not fused. The bottom left panel is a higher-magnification view of the interacting nuclei from the panel above it. Bars: bottom left panel, 0.5 μm; other panels, 1 μm.
CrGEX1 and PbGDEX1 are nuclear envelope proteins
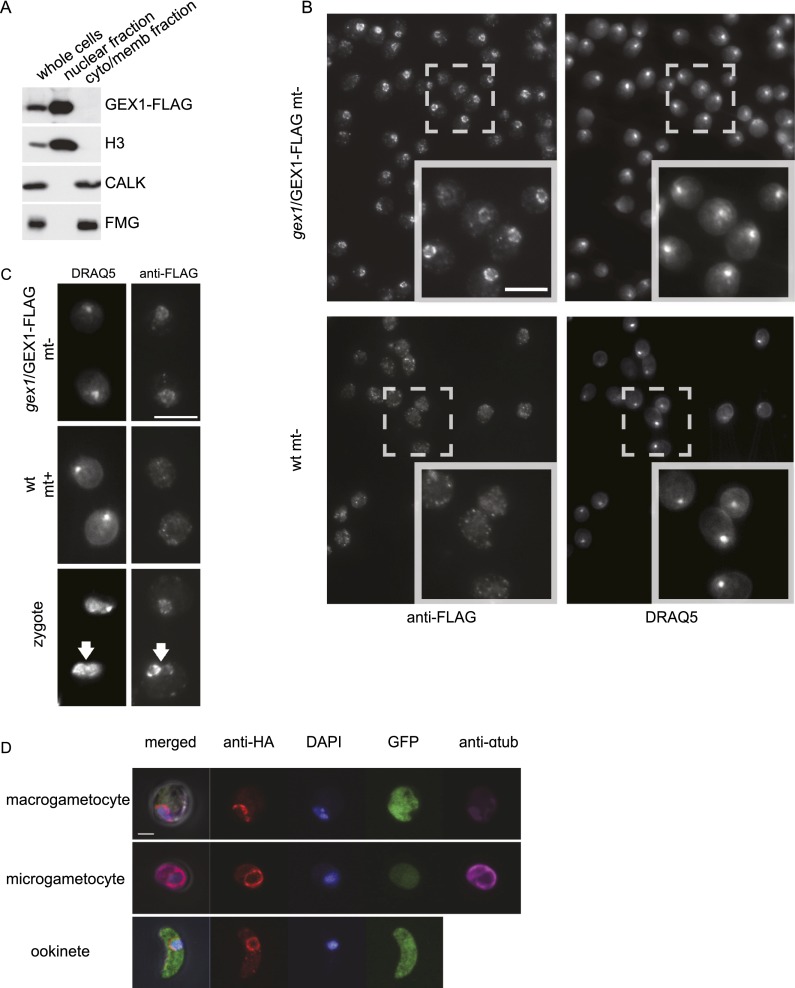
Because the above results suggested that CrGEX1 functions in the nucleus, we used cell fractionation and immunoblotting to test whether CrGEX1 was enriched in nuclei. Confirming the effectiveness of the cell fractionation method, a cytoplasmic protein kinase, CALK, and a cell body/flagellar membrane protein, FMG, were enriched in the cytoplasm/membrane fraction, whereas a nuclear protein, histone 3, was enriched in the nuclear fraction (Fig. 6A). Importantly, GEX1-Flag was also enriched in the nuclear fraction. Indirect immunofluorescence provided striking confirmation of the biochemical fractionation results (Fig. 6B). Almost every cell displayed a nucleus brightly stained with anti-Flag M2 antibody. In addition, the CrGEX1-Flag was present as a ring on the periphery of the nuclei, rather than within the organelles. This peripheral nuclear localization of GEX1, its enrichment in isolated nuclei, and its predicted transmembrane domains are consistent with the properties of a nuclear envelope protein.
Figure 6.
GEX1 is in the nuclear envelope in Chlamydomonas and Plasmodium. (A) Immunoblots of total cellular protein (whole cells) and the nuclear and cytoplasmic/membrane fractions prepared from minus gametes expressing a Flag-tagged GEX1 protein. CALK and FMG are cytoplasmic and flagellar/membrane proteins, respectively, and histone 3 (H3) is a nuclear protein. Equal amounts of protein were loaded in each lane. (B) Confocal immunofluorescence microscopy of wild-type and gex1/GEX1-Flag-expressing gametes stained with anti-Flag antibody and the DNA dye DRAQ5. Bar, 10 μm. (C) Anti-Flag immunostaining of zygotes fixed ∼20 min after fusion of wild-type plus gametes with gex1/GEX1-Flag minus gametes. GEX1-Flag can be detected in both nuclei. Bar, 10 μm. (D) Confocal immunofluorescence analysis of P. berghei sexual stages expressing a C-terminally HA epitope-tagged protein from the endogenous gex1 locus. Gametocytes were from peripheral blood. To distinguish sexes, fixed gametocytes were immunolabeled for α-tubulin (expressed highly in males) and GFP (expressed more highly from the eef1αa promoter in females). Bar, 2 μm.
Because both gamete nuclei come to occupy the common cytoplasm of the zygote, we tested whether a wild-type nucleus in a newly formed zygote whose nuclei had not yet fused could incorporate GEX1 contributed by a gex1 gamete expressing GEX1-Flag. As shown in Figure 6C, within 20 min after fusion, we detected zygotes in which both nuclei were stained with the Flag antibody. These results showing that the GEX1 protein from one gamete could be incorporated into the other nucleus in the zygote cytoplasm provided an explanation for the results in Figure 3H showing that zygotes formed from one wild-type and one mutant gamete produced wild-type levels of progeny.
To localize the GEX1 protein in P. berghei, we fused a triple HA epitope tag to the 3′ end of the endogenous gene. Consistent with the transcription data from P. falciparum, no protein was detected in asexually replicating blood stages. PbGEX1-HA was detected in a circular pattern surrounding the nucleus of the microgametocyte and macrogametocyte. Circumnuclear staining persisted to the ookinete stage (Fig. 6D). The gex1/PbGEX1-HA progeny behaved as wild type, demonstrating that the tagged protein was functional. We conclude that GEX1 is a nuclear envelope protein in both organisms.
GEX1s are members of an ancient family of nuclear fusion proteins
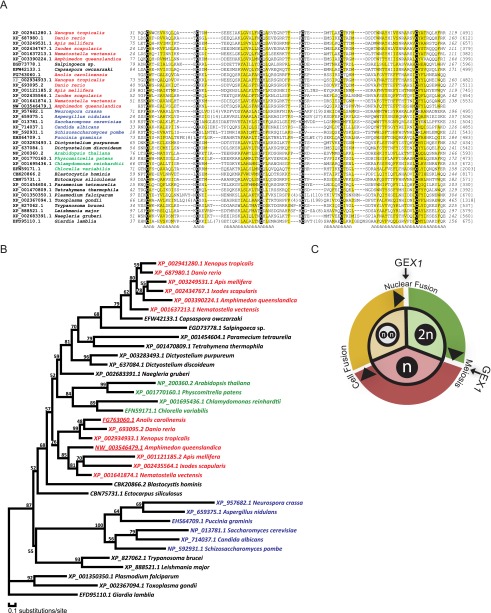
Because of the conserved locations and functions of GEX1, we carried out a more extensive analysis of GEX1 amino acid sequences from Chlamydomonas, Plasmodium, and Arabidopsis using PSI-BLAST. We discovered that the GEX1s are related to KAR5, a yeast nuclear fusion protein. Using additional bioinformatics methods, we determined that the KAR5/GEX1 family is large and is broadly conserved in eukaryotes (Fig. 7A). Family members were found in protists, including several vertebrate pathogens in addition to Plasmodium; in fungi; and in higher plants, including rice, maize, barley, soybean, and grape. Multicellular animals also possess KAR5/GEX1 orthologs, including the sponge Amphimedon; cnidarians, including Hydra and Nematostella; arthropods, including mosquito and deer tick; and chordates, including Ciona, Brachiostoma, and Danio (Fig. 7A,B). Of particular relevance to our findings in Chlamydomonas and Plasmodium and to that in Arabidopsis was that one of the two Danio KAR5-CRD-containing family members, Brambleberry, was noted to be related to KAR5 and is required for fusion of the nuclear envelope during fertilization (Abrams et al. 2012). Thus, our findings evolutionarily and functionally link the fungal, protist, plant, and vertebrate proteins. Although we found two family members in most of the multicellular animals, only a single copy was evident in protists, fungi, and plants. For each of the multicellular animal species examined, one of the two members was more related to plant proteins than to the other animal family member. The KAR5/GEX1 family proteins differ widely in size and degree of sequence similarity, but all have a well-defined CRD (KAR5-CRD) in their N-terminal portions and two or three transmembrane domains.
Figure 7.
Phylogenetic analysis of KAR5-CRD-containing proteins. (A) Alignment of KAR5-CRD-containing proteins. Sequences are denoted by NCBI gene accession numbers followed by species information. Starting and ending residue numbers (italics) are shown before and after the sequences, respectively. The lengths of proteins are shown in brackets. Insertions are replaced by the numbers of residues in parentheses. Conserved cysteines are highlighted on a black background. Positions with mainly hydrophobic residues are shaded in yellow. Consensus secondary structure predictions are shown below the sequences. (h) Predicted α-helix. XP_687980.1 is the Brambleberry protein (Abrams et al. 2012). (B) Phylogenetic tree generated using MOLPHY illustrating the relationships of KAR5-CRD-containing proteins. (C) Diagram illustrating that GEX1 has been used during evolution for both nuclear fusion and meiosis.
Discussion
The studies in this paper demonstrate that clustering Chlamydomonas genes based on their expression patterns not only identifies species-limited genes essential for sexual reproduction, but also can reveal new core elements of fertilization that are broadly conserved across species. Hierarchical clustering of expression profiles for nearly 11,000 Chlamydomonas genes placed four out of five previously identified minus gamete genes, including the conserved, fusion-essential HAP2 gene, in the 85-member C28 cluster whose transcripts were up-regulated during activation of minus gametes. Sets of genes involved in cell wall biosynthesis, protein translation, photosynthesis, and flagellar function, in contrast, sorted out into separate clusters whose study promises to yield new genes required for each of these cellular processes. Importantly, investigation of the previously uncharacterized Cre06.g280600 gene in C28 validated the power of this comparative genomics strategy. The approach revealed a previously unrecognized, broadly conserved family of nuclear envelope proteins, uncovered a conserved function for the GEX1 gene product in Chlamydomonas and Plasmodium sexual reproduction, and showed that Plasmodium GEX1 is essential for mosquito transmission of malaria.
Our cell fractionation results in Chlamydomonas, along with the GEX1 predicted transmembrane domains and the immunolocalization studies in both Chlamydomonas and Plasmodium, indicated that GEX1 is present in the nuclear envelope of both organisms. These results, along with the requirement for CrGEX1 in timely nuclear fusion, were consistent with the sequence analysis that showed that the proteins are part of a large, ancient protein family. Family members function in the nuclear envelope and are defined by the presence of the KAR5-CRD, whose best-characterized member is the yeast nuclear fusion protein KAR5. Although the similarity among the KAR5-CRD-containing protein family members outside of the KAR5-CRD is low, the overall architecture of the proteins is similar. All have at least two predicted transmembrane domains near the C terminus and possess the KAR5-CRD in the N-terminal portion. In their recent studies of Brambleberry, which we show here is one of the two D. rerio KAR5-CRD-containing proteins, Abrams et al. (2012) reported that it is essential for fusion of pronuclei to form the diploid nucleus in the zygote. Furthermore, they found that zebrafish exploited this KAR5-CRD-containing family member for fusion of the nuclear envelopes of chromatin masses in the early embryonic cell divisions.
In their analysis of the Brambleberry protein sequence, Abrams et al. (2012) further reported low but significant overall sequence similarity between yeast KAR5 and the Brambleberry protein. Moreover, they identified a domain in Brambleberry—the Brambleberry homology domain (BHD)—that they reported was absent from KAR5 and was found only in several metazoans. Possibly our use of PSI-BLAST with low stringency settings followed by use of nonmetazoan proteins as queries were important to uncover the much larger, ancient gene family that we report here. The KAR5-CRD includes the BHD region but is slightly larger because of the two most N-terminal cysteines.
The finding that Amphimedon, an early metazoan, contains two copies of the gene indicates that gene duplication occurred soon after appearance of the metazoa. The finding that the Arabidopsis GEX1 forms a complex, probably a dimer (Alandete-Saez et al. 2011), raises that possibility that KAR5-CRD-containing proteins in zebrafish and other metazoa also form complexes, possibly even heterodimers composed of both gene products. Disruption of the second KAR5-CRD-containing zebrafish family member and biochemical studies of Brambleberry should help to distinguish between the two possibilities.
The most striking phenotype that we observed in the Chlamydomonas and Plasmodium disruptants was a nearly complete growth arrest at a stage after meiosis, when mitosis of the haploid cells begins. A similar phenotype was observed for a Plasmodium mutant lacking a male-derived nuclear formin-like protein of the zygote, MISFIT (Bushell et al. 2009). The oocyst growth phenotype of the misfit and gex1 mutants can be mimicked by blocking premeiotic genome replication in the zygote with aphidicolin (Bushell et al. 2009), a treatment that does not interfere with the formation of a motile ookinete, highlighting that the cell cycle can be uncoupled from the nuclear cycle. It is tempting to speculate, therefore, that the developmental block at the oocyst stage in gex1 oocysts is the late manifestation of an irregularity in the nuclear envelope that occurs before or during meiosis in Plasmodium. Interestingly, Arabidopsis GEX1 is also required upon fertilization, and without the protein, zygote progeny (in this case, diploid) fail to proliferate (Alandete-Saez et al. 2011). Taken together, a simple explanation for these results from widely disparate species is that KAR5-CRD-containing proteins in all five organisms are required for nuclear fusion and that interfering with nuclear fusion can lead to defects in zygote development that are manifest in the inability of zygote progeny (diploid or haploid) to undergo cell proliferation.
As a counterpoint, however, studies of GEX1 mutants in Arabidopsis demonstrated that the protein is expressed during both fertilization and meiosis and that it is required for both events (Alandete-Saez et al. 2011). Because Arabidopsis meiosis is widely separated in time and space from nuclear fusion, these results argue strongly that evolution has exploited KAR5-CRD-containing family members for a second independent function in ploidy transitions that is distinct from its function in fusion of the two nuclear envelopes in the zygote (Fig. 7C; Supplemental Fig. S3). Thus, it remains possible that the failure of gex1 meiotic progeny to proliferate in Chlamydomonas and Plasmodium is related to a second function for GEX1 in meiosis in these two organisms, which could explain the increased number of spindle plaques in the nuclear envelopes of Plasmodium gex1 ookinetes at 18 h post-fertilization. Both of these protists undergo closed mitosis and meiosis, and, for example, remodeling of the nuclear envelope after meiosis might require GEX1.
Our results, taken together with studies in Danio, yeast, and Arabidopsis, indicate that KAR5-CRD-containing proteins function in cells undergoing ploidy transitions in widely diverse organisms (Supplemental Fig. S3) and suggest that this gene family appeared concomitantly with the origins of syngamy and meiosis. Finally, our findings emphasize the power of comparative biological analysis of conserved processes to identify new gene functions. The initial success of this strategy bodes well for its continued use to dissect molecular mechanisms of fertilization and sexual reproduction.
Materials and methods
Genomic DNA sequencing and analysis
C. reinhardtii strains 21gr (mt+) (CC-1690) and 6145C (mt−) (CC-1691) are available from the Chlamydomonas Resource Center, University of Minnesota. For whole-genome sequencing, genomic DNA was prepared from both strains. Genomic DNA was sheared into 300- to 400-base-pair (bp) fragments using Covaris Adaptive Focused Acoustics technology (AFA). Standard Illumina libraries were then prepared following the manufacturer's protocol (Bentley et al. 2008). The libraries were sequenced on the Illumina GAIIx for 76 paired-end cycles following the manufacturer's standard cluster generation and sequencing protocols (Bentley et al. 2008). We obtained 52 million paired reads of 76 bp (average fragment size 380 bp), which were processed with iCORN (Otto et al. 2010) using the reference genome Chlamydonomas sequence version 4 (http://genome.jgi-psf.org/Chlre4/Chlre4.home.htm). Data accession numbers and sequencing statistics are shown in Supplemental Table S1. Nonmapping reads from both strains were assembled separately with Velvet using the following parameters: k-mer 55; -exp_cov auto; -ins_length 400; -cov_cutoff 4. The resulting contigs >500 bp were merged with a custom Perl script, and Augustus (Stanke et al. 2008) was used to predict 1044 putative gene models using the default training set for C. reinhardtii and including the hints file from Specht et al. (2011).
Sample preparation and cDNA library construction
Unsynchronized vegetative cells were grown under continuous light in liquid M medium, and synchronized vegetative cells were cultured with aeration at 23°C in a 13:11-h light:dark cycle in M medium and harvested at 4 h into the light part of the cycle for RNA extraction. Synchronized vegetative cells 9 h into the light part of the light:dark cycle were induced to become gametes by transferring them to medium without nitrogen (N-free medium) followed by culturing in continuous light with aeration for 18 h at room temperature before harvest.
Lysin was prepared by centrifugation of crude lysin (Buchanan and Snell 1988) at 50,000g for 30 min at 4°C to remove cell debris. Lysin-treated gametes were incubated with lysin for 1 h, activated gametes were incubated with 15 mM db-cAMP and 0.15 mM papaverine for 1 h in N-free medium (Pasquale and Goodenough 1987), and activation was confirmed by use of a cell wall loss assay as described previously (Snell 1982). For isolation of RNA, samples were harvested by centrifugation and resuspended in lysis buffer (50 mM Tris-Cl at pH 7.5, 150 mM NaCl, 15 mM EDTA, 2% SDS, 40 μg/mL Proteinase K). Following organic extraction (25:24:1 [v/v] phenol/chloroform/isoamyalcohol), RNA was recovered by precipitation with an equal volume of isopropanol. Total RNA resuspended in water was further purified with TRIzol reagent (Invitrogen) following the manufacturer's instructions. mRNA was extracted from total RNA using Dynabeads Oligo-dT (Invitrogen Dynal) following the manufacturer's instructions. After elution from the beads, first and second strand cDNA was generated using the Just cDNA Double-Stranded cDNA Synthesis kit (Agilent) following the manufacturer's directions.
RNA extraction and sequencing
Oligo-dT primers were used to enrich mRNA species from complex total RNA pools, and cDNA was subsequently synthesized using random primers. Complex cDNA samples representing the transcriptomes of each of the eight treatment groups were sheared into 200- to 300-bp fragments, and libraries were prepared as for the genomic DNA with the addition of index tags to allow multiplexing of the 24 samples representing three biological replicates of each treatment group. These were split into two pools (technical replicates), each of which was sequenced for 100 paired-end cycles on the Illumina HiSeq 2000. Sequencing data (submitted to ArrayExpress with accession no. E-ERAD-124) were mapped with TopHat (Trapnell et al. 2009) to the combined genome. See Supplemental Table S2 for mapping statistics. Using the TopHat mapping, we ran Cufflinks to further improve gene annotation in the newly assembled contigs, predicting additional gene models where they did not overlap with the Augustus predictions. A Perl script calculated the RPKM value for each gene model based on annotation version 10 and the newly predicted models.
Profiles and clustering of RNA-seq data
For the initial sample clustering, we calculated Pearson correlations for pairwise comparisons of raw sequence tag counts for each gene in all samples followed by complete linkage clustering, resulting in Supplemental Figure S1. Following the elimination of outliers, as explained in the Results, the median of all remaining replicates was used to obtain a single RPKM value per gene and condition, which is shown in Supplemental Table S3. To generate expression patterns for cluster analysis, the expression of each gene was expressed as a percentage of the sum of all RPKM values across all treatment groups for that gene. Expression profiles were clustered using the Ward method (Ward 1963) on Euclidian distances between row-wise normalized RPKM profiles, setting the cutoff for the number of final clusters to 42.
GO term enrichment and detection of distant homologs
Since GO annotation was not available for version 10 of the C. reinhardtii genome assembly, annotations for proteins in version 4 (http://genome.jgi-psf.org/Chlre4/Chlre4.download.ftp.html) were mapped to version 10 proteins using BLAST and custom scripts. GO term enrichment analysis was performed in R using the topGO package (http://www.bioconductor.org/packages/2.12/bioc/html/topGO.html) on each of the 42 gene profile clusters. Fisher's exact test and the “parent–child” algorithm (Grossmann et al. 2007) were used to assign statistical significance to GO term enrichments.
To search for conserved families among sexual development genes, predicted amino acid sequences of candidate genes from C. reinhardtii were used for a similarity search (blastp -W 2) against the UniProt eukaryotic database (Apweiler et al. 2004). An alignment was generated with T_coffee (Notredame et al. 2000) for each query sequence and its hits (E-value cutoff 1 × 10−20). From the alignments, hidden Markov models were constructed with HMMer3 (Finn et al. 2011) and used to search for distant homologs in the UniProt eukaryotic database. Following manual inspection of selected alignments, we chose a 1 × 10−4 cutoff for the HMMer search. A custom Perl script extracted all of the hits with an E-value >1 × 10−4 of the human, P. berghei, and Chlamydomonas sequences.
Real-time qRT–PCR
To analyze the relative abundance of transcripts using qRT–PCR, 1 μg of total RNA was reverse-transcribed for 45 min at 42°C in a 20-μL reaction volume using the High Fidelity SuperScript III RT–PCR kit (Invitrogen) according to the manufacturer's instructions. Gene-specific PCR primers were designed using a stringent set of criteria, including a predicted melting temperature of 60°C ± 5°C, primer lengths of 20–24 nucleotides (nt), and PCR amplicon lengths of 80–150 bp. qRT–PCR was performed in optical 96-well plates using Bio-Rad CFX96 Real-Time PCR systems. Reactions were performed in a final volume of 20 μL containing 10 μL of 2× SYBR Green Master Mix, 0.5 μM each primer, and 10 ng of cDNA. PCR conditions were as follows: 30 sec at 95°C, followed by 45 cycles of 5 sec at 95°C and 30 sec at 60°C. Fluorescence threshold data (Ct) were analyzed using Bio-Rad CFX Manager software (version 2.0). For each reaction, the threshold cycle was determined by setting the threshold within the logarithmic amplification phase. Quantitative reactions were done in triplicate and averaged. Relative expression levels in each cDNA sample were normalized to a RPL19 reference gene under the same conditions. Real-time qPCR was performed using the following primers: Gex1RTF2 (5′-GCTCGTCGTCGCCAACCAA-3′), Gex1RTR2 (5′-GCAGTCCGTGCCGAACAAT-3′), RPL19F3 (5′-CGCAAGGTTTGGCTGGAC-3′), and RPL19R3 (5′-TGACGGGCTGCTTACGGA-3′).
Insertional mutagenesis and determination of percentage of germinated zygotes with proliferating progeny
We screened 10,000 insertional mutants generated in the 6145c mt− strain as described (Gonzalez-Ballester et al. 2011). Insertional mutants were PCR-screened with CrGEX1 gene-specific primer gex1scrR2 (5′-CAGCAAAGGAGTGAAAGCACAGAT-3′) and marker gene primer RB1 (5′-ATGGGGCGGTATCGGAGGAAAAG-3′) (Gonzalez-Ballester et al. 2011). Nested PCR with CrGEX1 gene-specific primer gex1scrR1 (5′- GCGCTCGAAGTTGCGGTTT-3′) and marker gene primer RB2 (5′-TACCGGCTGTTGGACGAGTTCTTCTG-3′) was performed to confirm the mutant. Zygote generation and maturation were performed as described (Wegener et al. 1989; Wegener and Beck 1991). After maturation, zygotes harvested from agar plates were sonicated for 40 sec at 40 W. After centrifugation at 3000g for 1 min at room temperature, the zygotes were transferred to TAP medium agar plates and exposed to chloroform vapor for 30 sec to kill vegetative cells. The locations of 70–100 zygotes on the plates were marked before the plates were exposed to continuous light. The marked locations were reinspected 24–30 h later to identify those with more than four meiotic progeny, which indicated that the meiotic progeny had begun to divide. The percentage of germinated zygotes with proliferating progeny was defined as (number of germinated zygotes with proliferating progeny)/(number of zygotes) × 100.
Determination of nuclear fusion efficiency
Plus gametes were mixed with minus gametes for 4, 7, 10, 13, and 16 h with the live-cell impermeant, nucleic acid fluorochrome SYTOX Green (1 μM) in N-free medium, and fixed in 2% paraformaldehyde. The samples were washed with N-free medium once, placed on a microscope slide, and viewed by fluorescence and differential interference contrast microscopy. Only the cells in zygote aggregates were counted. The percentage of nuclear fusion was defined as (number of cells with single nucleus)/(total number of fixed cells counted) × 100. At least 100 cells from randomly chosen aggregates were counted. The data shown are averages from three independent experiments, and the error bars are standard error of the mean (SEM). Transmission electron microscopy with 4-h and 7-h zygotes was performed as previously described (Liu et al. 2008).
Expressing the CrGEX1-Flag transgene in the Chlamydomonas gex1 mutant
The plasmid pHSP70A-RBCS2-CrGEX1-Flag contains an AphVIII gene under the control of the PSAD promoter (Gonzalez-Ballester et al. 2011) and a CrGEX1 genomic fragment that begins with the ATG coding for the initiation methionine of CrGEX1, with a 3× Flag at the C terminus. CrGEX1 is driven by a RBCS-HSP70 chimeric promoter and contains the RBCS terminator from Chlamydomonas (Schroda et al. 2000). One microgram of linearized plasmid was introduced into 21gr (mt+) cells by electroporation (Shimogawara et al. 1998). Colonies were screened by PCR and immunoblotting. Plus gex1/GEX1-Flag cells were crossed with a gex1 minus strain to obtain minus gex1 cells that contained the GEX1-Flag transgene.
Isolation of Chlamydomonas nuclei
Nuclei were prepared with a CelLytic PN Isolation/Extraction kit (Sigma-Aldrich) using the protocol described by Winck et al. (2011) with some modifications. Briefly, ∼8 × 108 gametes were harvested by centrifugation at 3000g for 2 min at 4°C, resuspended in 50 mL of medium containing the cell wall-degrading enzyme lysin (Buchanan and Snell 1988), and incubated for 15 min at room temperature, when a detergent sensitivity assay indicated that they had lost their walls. The wall-less cells were harvested by centrifugation as above and resuspended in 5 mL of 1× NIBA solution (Sigma-Aldrich) for 10 min on ice. Cells were harvested by centrifugation at 1260g for 10 min and resuspended in 10 mL of NIBA solution with 1% NP-40 for cell lysis. The samples were kept for 10 min on ice and then centrifuged at 1000g for 30 min at 4°C. The harvested nuclear pellets were washed twice with 1 mL of 1× NIBA solution and resuspended in 600 μL of 1× NIBA solution.
Indirect immunofluorescence microscopy
For Chlamydomonas, cells were affixed to coverslips coated with poly-L-lysine (Sigma). After being fixed with methanol for 15 min at −20°C, the coverslips were blocked with blocking solution (5% BSA, 1% cold fish gelatin in PBS) for 30 min at 37°C. Then, the coverslips were exposed to anti-Flag M2 mouse antibody (1:100 dilution; Sigma) for 2 h at 37°C. After further washing three times with PBS for 5 min, the samples were incubated with Alexa Fluor 488 goat anti-mouse IgG (1:200 dilution; Invitrogen) and DRAQ5 (Cell Signaling) for 2 h at 37°C. After washing with PBS three times for 5 min, the samples were mounted in Fluoromount G (Southern Biotech). Images were acquired with an Axioplan2 microscope (Zeiss).
P. berghei gametocytes and ookinetes were fixed in suspension with 3% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 2% BSA in PBS. Primary antibodies were diluted in blocking solution (mouse monoclonal Tat1 against tubulin [1:500; Sigma], rat anti-HA [1:200; Roche], and rabbit anti-GFP [1:500; Abcam]). Anti-mouse Alexa633, anti-rat Alexa555, and anti-rabbit Alexa488 were used as secondary antibodies together with DAPI (all from Invitrogen), all diluted 1:200 in blocking solution. Stained cells were mounted on glass slides using Fluoromount (Sigma). Confocal images were acquired with an LSM510 laser-scanning confocal microscope (Zeiss). Wild-type gametocytes and ookinetes showed no background signal when stained with anti-HA antibody as control. Oocysts were imaged live by staining dissected midguts with PBS containing Hoechst 33342. Transmission electron microscopy was performed as previously described (Moon et al. 2009).
P. berghei maintenance and transmission
P. berghei ANKA parasites were maintained in female Theiler's Original outbred mice by intraperitoneal injection of infected blood. Parasitemia was monitored on Giemsa-stained thin blood smears. Parasites were transmitted to Anopheles stephensi (strain SD500) mosquitoes by allowing 40–50 female mosquitoes to feed for 20 min on an anesthetized mouse that had been infected 3 d earlier. Unfed mosquitoes were removed 1 d later, and fed mosquitoes were maintained at 19°C on a solution of fructose. On days 7, 15, and 21 post-infection, mosquitoes were dissected in a drop of PBS to obtain midguts for imaging. Blood with high gametocyte counts for in vitro cultures of sexual stages was obtained by infecting mice 3 d after treating with phenyl hydrazine at 50 mg/kg intraperitoneally, which maximizes gametocyte numbers by inducing mild anemia and reticulocytosis. These mice were used on days 4–6 post-infection.
Phenotyping P. berghei sexual stages
To measure microgametocyte activation, 4 μL of blood from a superficial vein of a high-gametocytemia mouse was mixed with 150 μL of ookinete medium (RPMI1640 containing 25 mM HEPES, 20% FCS, 100 μM xanthurenic acid at pH 7.5) and a drop placed in an improved Neubauer hemocytometer. After 12–15 min at 19°C, microgamete release was quantified as number of exflagellation centers using a 40× objective. The number of morphologically mature microgametocytes in the same volume of blood was calculated from the red blood cell count and the microgametocytemia in a Giemsa-stained blood smear prepared from the same infected mouse. Microgametocyte activation was determined by establishing the percentage of morphologically mature microgametocytes that underwent exflagellation.
Fertilization and ookinete formation were assessed by culturing blood from gametocytemic mice on days 4–6 post-infection in nine volumes of ookinete medium (as above). After 24 h at 19°C, live cells were labeled in suspension with Cy3-conjugated mouse monoclonal antibody 13.1 against the P28 protein, which marks the surface of activated macrogametocytes, zygotes, and ookinetes. The conversion efficiency of activated macrogametocytes to mature ookinetes was determined as the ratio between elongate ookinetes to all P28-positive cells.
To assess oocyst development, infected midguts were dissected from 15–30 mosquitoes on day 7 post-infection, flattened under a coverslip in a drop of PBS, and imaged using a Leica M205A stereomicroscope. Recorded images of entire midguts were analyzed using the “3D objects counter” plug-in of the ImageJ software to count the total number of fluorescent oocysts per gut. Uninfected midguts were used initially to calibrate the software. Oocyst diameters were determined using ImageJ software on higher-resolution images recorded with a 20× or 63× objective.
For flow cytometric determination of DNA content, macrogametes, zygotes, and ookinetes were purified from overnight cultures using paramagnetic anti-mouse IgG beads (Dynabeads, Invitrogen) coated with anti-13.1 antibodies against the P28 surface protein and stained for 15 min with Hoechst 33342 DNA dye (Invitrogen). Samples were filtered on a 35-μm mesh to remove aggregates and examined with a 488-nm blue laser and a 355-nm UV laser on a BD LSRFortessa flow cytometer (BD Biosciences). GFP fluorescence was detected by a 530/30 filter, and Hoechst was detected by a 450/50 filter. BD FACSDiva software (BD Biosciences) was used to collect 10,000 events of interest for each sample. The data collected were further analyzed with FlowJo (Tree Star). Hoechst stain and GFP fluorescence were used to gate on parasite populations, and Hoechst fluorescence intensity was then determined in the gated population.
P. berghei genetic modification
All transgenic P. berghei parasites were generated in a selectable marker-free reporter strain expressing GFP (RMgm-7) from the eef1αa promoter (Janse et al. 2006a). gex1 knockout clones 1 and 2 were generated with a conventional gene targeting vector. For this, two fragments of ∼500 bp flanking the coding region were amplified from genomic DNA using primer pairs olSA00021 (GCGCgggcccAAACAATTACAAAAGATAGAA)/olSA00328 (GGGgctagcATTTAACTTATGTTGGCGTG) and olSA000320 (GCGCgaattcGTTCTACATATTTGTTCTTATCG)/olSA000321 (CGCggatccGGTGGTGGTAATATGGCTTAAACG). These fragments were inserted on either side of the Tgdhfr/ts expression cassette of plasmid pOB90 (Billker et al. 2004), which confers resistance to pyrimethamine, using ApaI/NheI and EcoRI/BamHI restriction sites, respectively (shown in lowercase in the primer sequences). The integration cassette was released as a linear fragment by an ApaI/BamHI digest. For independent confirmation of the phenotype, a third mutant clone was created using a gex1 deletion vector from the PlasmoGEM resource (Pfander et al. 2011) with the design number PbGEM-45681 (see http://plasmogem.sanger.ac.uk for details of vector design). To add a triple HA epitope tag C-terminally to the genomic copy of GEX1, we used PlasmoGEM vector 45689. Purified P. berghei schizonts were transfected as described previously (Janse et al. 2006b), except that electroporated merozoites were allowed to reinvade reticulocytes for 20 min at 37°C before being injected intraperitoneally into a naïve mouse. Transgenic parasites were selected in vivo and cloned by dilution cloning. Correct integration into the gex1 locus and absence of wild-type parasites were confirmed by integration-specific PCR and pulse-field gel electrophoresis.
Acknowledgments
We are grateful to Michel Theron for help with flow cytometry. We thank Mary Collins for her generosity in preparing illustrations. The C. reinhardtii reference genome was sequenced by the Department of Energy Joint Genome Institute (JGI) and the Chlamydomonas Genome Consortium. Portions of this work were performed in laboratories constructed with support from NIH grant C06 RR 30414. This work was funded by grants from the National Institutes of Health (GM56778 and GM25661) to W.J.S., the Wellcome Trust (098051) to the Sanger Institute, the Medical Research Council (G0501670) to O.B., and a Marie Curie Fellowship (PIEF-GA-2009-253899) and an EMBO Long Term Fellowship (ALTF 45-2009) to M.B. T.D.O. was supported by the EVIMalaR European Union 7th framework grant number 242095.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.212746.112.
References
- Abrams EW, Zhang H, Marlow FL, Kapp L, Lu S, Mullins MC 2012. Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development. Cell 150: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair WS, Apt KE 1990. Cell wall regeneration in Chlamydomonas: Accumulation of mRNAs encoding cell wall hydroxyproline-rich glycoproteins. Proc Natl Acad Sci 87: 7355–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alandete-Saez M, Ron M, Leiboff S, McCormick S 2011. Arabidopsis thaliana GEX1 has dual functions in gametophyte development and early embryogenesis. Plant J 68: 620–632 [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. 2004. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res 32: D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Brizzio V, Rose MD 1997. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J Cell Biol 139: 1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, Brinkmann V 2004. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117: 503–514 [DOI] [PubMed] [Google Scholar]
- Buchanan MJ, Snell WJ 1985. Characterization of the wall-degrading enzyme, lysin, released by Chlamydomonas gametes. J Cell Biol 101: 379a [Google Scholar]
- Buchanan MJ, Snell WJ 1988. Biochemical studies on lysin, a cell wall degrading enzyme released during fertilization in Chlamydomonas. Exp Cell Res 179: 181–193 [DOI] [PubMed] [Google Scholar]
- Bushell ES, Ecker A, Schlegelmilch T, Goulding D, Dougan G, Sinden RE, Christophides GK, Kafatos FC, Vlachou D 2009. Paternal effect of the nuclear formin-like protein MISFIT on Plasmodium development in the mosquito vector. PLoS Pathog 5: e1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW 1997. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Armbrust EV, Goodenough UW 2002. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160: 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR 2011. HMMER Web server: Interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godl K, Hallmann A, Wenzl S, Sumper M 1997. Differential targeting of closely related ECM glycoproteins: The pherophorin family from Volvox. EMBO J 16: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, Pootakham W, Mus F, Yang W, Catalanotti C, Magneschi L, de Montaigu A, Higuera JJ, Prior M, Galvan A, et al. 2011. Reverse genetics in Chlamydomonas: A platform for isolating insertional mutants. Plant Methods 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann S, Bauer S, Robinson PN, Vingron M 2007. Improved detection of overrepresentation of gene-ontology annotations with parent child analysis. Bioinformatics 23: 3024–3031 [DOI] [PubMed] [Google Scholar]
- Hirai M, Arai M, Mori T, Miyagishima SY, Kawai S, Kita K, Kuroiwa T, Terenius O, Matsuoka H 2008. Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr Biol 18: 607–613 [DOI] [PubMed] [Google Scholar]
- Janse CJ, Franke-Fayard B, Waters AP 2006a. Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat Protoc 1: 614–623 [DOI] [PubMed] [Google Scholar]
- Janse CJ, Ramesar J, Waters AP 2006b. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat Protoc 1: 346–356 [DOI] [PubMed] [Google Scholar]
- Kurvari V 1997. Cell wall biogenesis in Chlamydomonas—molecular characterization of a novel protein whose expression is up-regulated during matrix formation. Mol Gen Genet 256: 572–580 [DOI] [PubMed] [Google Scholar]
- Kurvari V, Grishin NV, Snell WJ 1998. A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J Cell Biol 143: 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, et al. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301: 1503–1508 [DOI] [PubMed] [Google Scholar]
- Lin H, Goodenough UW 2007. Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1. Genetics 176: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, Grishin NV, Steele RE, Sinden RE, Snell WJ, et al. 2008. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev 22: 1051–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Misamore MJ, Snell WJ 2010. Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas. Development 137: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha M, Schulz-Raffelt M, Schroda M 2008. A new assay for promoter analysis in Chlamydomonas reveals roles for heat shock elements and the TATA box in HSP70A promoter-mediated activation of transgene expression. Eukaryot Cell 7: 172–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Barragán MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ 2011. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misamore MJ, Gupta S, Snell WJ 2003. The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for a critical membrane adhesion event during fusion with minus gametes. Mol Biol Cell 14: 2530–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RW, Taylor CJ, Bex C, Schepers R, Goulding D, Janse CJ, Waters AP, Baker DA, Billker O 2009. A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS Pathog 5: e1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T 2006. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol 8: 64–71 [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Shikanai T, Nakamura S, Kawai-Yamada M, Uchimiya H 2012. Gsp1 triggers the sexual developmental program including inheritance of chloroplast DNA and mitochondrial DNA in Chlamydomonas reinhardtii. Plant Cell 24: 2401–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Otto TD, Sanders M, Berriman M, Newbold C 2010. Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology. Bioinformatics 26: 1704–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale SM, Goodenough UW 1987. Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J Cell Biol 105: 2279–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander C, Anar B, Schwach F, Otto TD, Brochet M, Volkmann K, Quail MA, Pain A, Rosen B, Skarnes W, et al. 2011. A scalable pipeline for highly effective genetic modification of a malaria parasite. Nat Methods 8: 1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Blocker D, Beck CF 2000. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J 21: 121–131 [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RE 2009. Malaria, sexual development and transmission: Retrospect and prospect. Parasitology 136: 1427–1434 [DOI] [PubMed] [Google Scholar]
- Sinden RE, Canning EU, Spain B 1976. Gametogenesis and fertilization in Plasmodium yoelii nigeriensis: A transmission electron microscope study. Proc R Soc Lond B Biol Sci 193: 55–76 [DOI] [PubMed] [Google Scholar]
- Sinden RE, Butcher GA, Billker O, Fleck SL 1996. Regulation of infectivity of Plasmodium to the mosquito vector. Adv Parasitol 38: 53–117 [DOI] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P 2001. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Snell WJ 1982. Study of the release of cell wall degrading enzymes during adhesion of Chlamydomonas gametes. Exp Cell Res 138: 109–119 [DOI] [PubMed] [Google Scholar]
- Snell WJ, Goodenough UW 2009. Flagellar adhesion: Flagellar generated signaling and gamete fusion during mating. In The Chlamydomonas sourcebook (ed. Witman GB), pp. 369–394. Elsevier, San Diego [Google Scholar]
- Specht M, Stanke M, Terashima M, Naumann-Busch B, Janssen I, Hohner R, Hom EF, Liang C, Hippler M 2011. Concerted action of the new Genomic Peptide Finder and AUGUSTUS allows for automated proteogenomic annotation of the Chlamydomonas reinhardtii genome. Proteomics 11: 1814–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JA, Dodemont HJ, Stunnenberg HG, van Gemert GJ, Sauerwein RW, Eling W 2001. A central role for P48/45 in malaria parasite male gamete fertility. Cell 104: 153–164 [DOI] [PubMed] [Google Scholar]
- von Besser K, Frank AC, Johnson MA, Preuss D 2006. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ 1963. Hierarchical grouping to optimize an objective function. Amer StatistAssoc 58: 236–244 [Google Scholar]
- Wegener D, Beck CF 1991. Identification of novel genes specifically expressed in Chlamydomonas reinhardtii zygotes. Plant Mol Biol 16: 937–946 [DOI] [PubMed] [Google Scholar]
- Wegener D, Treier U, Beck CF 1989. Procedures for the generation of mature Chlamydomonas reinhardtii zygotes for molecular and biochemical analyses. Plant Physiol 90: 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winck FV, Kwasniewski M, Wienkoop S, Mueller-Roeber B 2011. An optimized method for the isolation of nuclei from Chlamydomonas reinhardtii (chlorophyceae). J Phycol 47: 333–340 [DOI] [PubMed] [Google Scholar]
- Wong JL, Johnson MA 2010. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol 20: 134–141 [DOI] [PubMed] [Google Scholar]