Abstract
In this study, we determined the function of a novel non-ribosomal peptide synthetase (NRPS) system carried by a streptococcal integrative conjugative element (ICE), ICESe2. The NRPS shares similarity with the yersiniabactin system found in the high-pathogenicity island of Yersinia sp. and is the first of its kind to be identified in streptococci. We named the NRPS product ‘equibactin’ and genes of this locus eqbA–N. ICESe2, although absolutely conserved in Streptococcus equi, the causative agent of equine strangles, was absent from all strains of the closely related opportunistic pathogen Streptococcus zooepidemicus. Binding of EqbA, a DtxR-like regulator, to the eqbB promoter was increased in the presence of cations. Deletion of eqbA resulted in a small-colony phenotype. Further deletion of the irp2 homologue eqbE, or the genes eqbH, eqbI and eqbJ encoding a putative ABC transporter, or addition of the iron chelator nitrilotriacetate, reversed this phenotype, implicating iron toxicity. Quantification of 55Fe accumulation and sensitivity to streptonigrin suggested that equibactin is secreted by S. equi and that the eqbH, eqbI and eqbJ genes are required for its associated iron import. In agreement with a structure-based model of equibactin synthesis, supplementation of chemically defined media with salicylate was required for equibactin production.
Introduction
Horizontal gene transfer has facilitated greatly microbial evolution and rapid adaptation to new niches. The term ICE has been adopted to describe all self-transmissible mobile genetic elements with predicted or demonstrated integrative and conjugative properties (Burrus et al., 2002a, b). ICEs excise from the donor chromosome to form a non-replicative circular molecule, then promote self-transfer to a new host by conjugation and subsequently insert into a new target site. This transfer can occur between genetically unrelated bacteria.
Tn916, identified in Enterococcus faecalis, was the first ICE to be completely sequenced (Flannagan et al., 1994). Recently, the number of sequenced ICEs has risen considerably, particularly with the advent of genome sequencing (Burrus et al., 2002a, b; Sebaihia et al., 2006). Comparisons between ICEs of low G+C Gram-positive bacteria have revealed that these elements are widespread and possess a modular organization (Garnier et al., 2000; Roberts et al., 2001; Burrus et al., 2002b). Each of these modules includes all of the coding sequences required for a specific biological function, namely site-specific recombination, conjugation and regulation. This is not dissimilar to the combination of functional modules seen in other bacterial mobile elements such as phages and plasmids (Toussaint and Merlin, 2002) and allows these structures, even when unrelated or distantly related, to evolve by exchange of modules (Burrus et al., 2002b). Consequently, a diverse set of mosaic ICEs are emerging; most encode a tyrosine recombinase (like the majority of integrated prophage) but a small number have been identified that utilize a serine recombinase to mediate site-specific recombination (Wang et al., 2000; Burrus et al., 2002a).
ICEs have played an important role in the distribution of antibiotic resistance loci. Other functions encoded by ICEs include toxic aromatic compound degradation pathways, sucrose utilization, bacteriocin biosynthesis, heavy metal resistance, type III and type IV secretion systems, capsule antigen and a metalloprotease virulence factor (Burrus and Waldor, 2004; Franco, 2004). A novel ICE in the ECOR31 strain of Escherichia coli carries the high-pathogenicity island (HPI) encoding the yersiniabactin siderophore system, which is critical to the virulence of Yersinia sp. and is widely distributed among E. coli strains and other Enterobacteriaceae that cause extraintestinal infections (Schubert et al., 2004).
The acquisition of iron is an essential process for all pathogenic bacteria. In mammalian host tissue iron is sequestered by transferrin, lactoferrin or in red blood cells (Wooldridge and Williams, 1993). Bacterial pathogens access this limited iron supply through the production of cell surface receptors for transferrin and lactoferrin, utilizing haem-containing compounds, iron transporters, or by the synthesis and secretion of iron-sequestering siderophores (Wandersman and Delepelaire, 2004). Streptococcus pneumoniae encodes multiple iron transporters that are important for virulence (Brown et al., 2002) and a cell-surface lactoferrin receptor (Hakansson et al., 2001). Streptococcus pyogenes utilizes haemoprotein binding and transport proteins (Lei et al., 2002) and a multimetal transport system (Janulczyk et al., 2003). Streptococcus mutans transports ferrous iron and produces a Mn/Fe uptake system that is important in vivo (Evans et al., 1986). Recently, a siderophore-iron(III) transport gene was identified in S. pyogenes, S. agalactiae and S. pneumoniae (Hanks et al., 2005; Clancy et al., 2006; Pramanik and Braun, 2006), but siderophore biosynthesis has not been previously identified in any streptococci (Eichenbaum et al., 1996).
Streptococcus equi is the causative agent of strangles, one of the most commonly diagnosed and important infectious diseases of horses worldwide. The disease is characterized by pyrexia followed by profuse nasal discharge and abscessation of the lymph nodes of the head and neck. These can restrict the airway and it is this clinical feature that gave the disease ‘strangles’ its name. Approximately 10% of horses that recover from strangles become persistently infected and may intermittently shed S. equi for several years. This ‘carrier state’ is believed to be fundamental to the epidemiology of strangles and originates from incomplete drainage of the guttural pouches and/or sinuses following rupture of abscesses formed in the retropharyngeal lymph nodes (Newton et al., 1997; 2000; Waller and Jolley, 2007).
Genome sequencing of S. equi and its close ancestral relative Streptococcus zooepidemicus has enabled us to identify possible virulence determinants in the former that could contribute to its increased pathogenicity (data to be reported elsewhere). In this study, our aim was to determine the function of a yersiniabactin-like non-ribosomal peptide synthetase (NRPS) system encoded by a novel ICE, ICESe2 identified in S. equi. Although we have not yet identified the peptide made by this NRPS, data consistent with its role in iron acquisition have been established. This is the first example of a NRPS system involved in the acquisition of iron to be identified in streptococci.
Results
Identification and prevalence of a novel ICE encoding a siderophore-like NRPS in S. equi
The genomes of S. equi 4047 and S. zooepidemicus H70 share > 97% DNA sequence identity although their in vivo virulence differs markedly. Comparison of the S. equi 4047 genome and S. zooepidemicus H70 unfinished genome using the Artemis Comparison Tool (Carver et al., 2005) led to the identification of a large 63 kb locus in S. equi 4047 that was absent from the S. zooepidemicus H70 genome, had a significant decrease in G+C content (31% compared with 42% across the whole S. equi genome) and was bordered by 108 bp perfect inverted repeat elements.
blastp and fasta analysis against UniProt determined that the locus contained coding regions with similarity and conserved gene order with coding sequences (CDSs) contained within the conjugative transposons CDTn2 and CDTn5 in the genome of Clostridium difficile strain 630 (Sebaihia et al., 2006) and the plasmid-borne Tn1549 in Enterococcus spp. (Garnier et al., 2000) (Fig. S1). Consequently we have named this S. equi integrative conjugative element, ICESe2, according to the suggested nomenclature (Burrus et al., 2002a). Like Tn916, these elements appear modular, with CDSs likely to be associated with conjugation and site-specific recombination located in the left and right extremities respectively. The recombination module of ICESe2 encodes a single serine recombinase [284 amino acids (aa)] and has homology (43–47% aa sequence identity) to the N-terminal regions (first 277–303 aa) of similar proteins in CDTn2 and CDTn5 and also TndX (533 aa, 43% aa sequence identity), which is responsible for the integration and excision of the conjugative transposon Tn5397 (Wang et al., 2000). A closer homologue is also present in the genome of S. pyogenes strain MGAS10750 (557 aa, 64% aa sequence identity). The sequence and annotation of ICESe2 has been submitted to GenBank/EMBL with the Accession No. AM909652.
The ‘cargo’ region between the conjugation and recombination modules that encodes vancomycin resistance in Tn1549 carries transport genes of unknown function in CDTn2/5 and CDSs in ICESe2 with most overall similarity to the NRPS cluster 1 of Clostridium kluyveri, which is proposed to biosynthesize a putative siderophore (Seedorf et al., 2008). Several of the encoded proteins were also similar to the NRPS complex of Yersinia sp. that produces the ferric iron-binding siderophore yersiniabactin. Therefore, we hypothesized that the ICESe2 cargo region which contains the ‘eqb’ gene cluster is responsible for the biosynthesis of a novel thiazoline-containing non-ribosomal peptide that we have tentatively named ‘equibactin’ although it is possible that the product from this NRPS could be identical to previously identified molecules. The eqb cluster contains 14 coding sequences (eqbA–N), the putative functions of which include a DtxR-like repressor (eqbA), non-ribosomal peptide biosynthetic proteins (eqbB–G, eqbMN), a ferric-siderophore-like importer (eqbH–J) and ABC transporters (Fig. 1A).
Fig. 1.
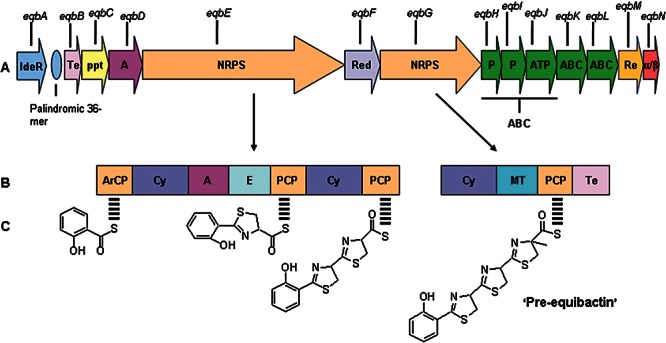
The equibactin locus and predicted functions of the eqb gene products. A. Predicted functions of CDSs in the ICESe2 eqb cluster [IdeR, iron-dependent regulator; Te, type II thioesterase; ppt, 4′-phosphopantetheinyl transferase; A, salicylate-AMP ligase; NRPS, non-ribosomal peptide synthetase; Red, thiazoline reductase; P, permease (component of ABC transporter); ATP, ATPase (component of ABC transporter); ABC, ABC transporter; Re, putative oxidoreductase; α/β, putative α/β hydrolase]. See Table S2 for homology to other NRPS systems and transporters. B. Organization of modules and domains in the Eqb NRPS (ArCP, aryl acid carrier protein; Cy, heterocyclization; A, adenylation; E, epimerization; PCP, peptidyl carrier protein; MT, methyl transferase; Te, type I thioesterase). C. Proposed intermediates in equibactin biosynthesis.
The prevalence of the equibactin locus in diverse S. equi and S. zooepidemicus populations was determined by Southern blot with a probe specific to the eqbN gene and PCR of the eqbE gene using primers listed in Table S1. The eqbN and eqbE genes were detected in 18 of 18 S. equi isolates but 0 of 73 S. zooepidemicus isolates. In addition, the location of the ICESe2 as determined by Southern blot and PCR across flanking regions was identical in all 18 S. equi strains.
Functions of the equibactin gene cluster as predicted by genetic analysis
Dissection of the modular structure revealed a remarkable similarity between the C. kluyveri, yersiniabactin and equibactin NRPS systems (Table S2). The first of the two peptide synthetases, EqbE, shares 45% and 30% aa sequence identity with CKL_1505 and HMWP2 respectively, while EqbG shares 25% aa sequence identity with CKL_1511 and HMWP1. Analagous to the characterized HMWP1 and HMWP2, EqbG and EqbE are comprised of four carrier domains, three cyclization domains, an epimerization domain, a single adenylation domain, a methyltransferase domain and a type I thioesterase domain (Fig. 1B). The EqbG protein has 1272 aa, but lacks the N-terminal 1895 aa of HMWP1 involved in polyketide synthesis (Miller et al., 2002). EqbC shares 23% aa identity with CKL_1523 (Table S2). Although lacking sequence homology, eqbC like ybtD, which is located outside of the Yersinia pestis HPI (Bobrov et al., 2002), encodes a putative 4′-phosphopantetheinyl transferase. EqbM shares 31% aa identity with CKL_1509 of C. kluyveri (Seedorf et al., 2008) and aligned to the Pfam Saccharopine dehydrogenase family, a member of the FAD/NAD(P)-binding Rossmann fold superfamily of redox enzymes. EqbN, a putative membrane protein, has a predicted N-terminal transmembrane helix and shares 31% aa identity with a hypothetical protein Csac_2163 of Caldicellulosiruptor saccharolyticus. EqbN and Csac_2163 aligned to the Pfam α/β hydrolase family of enzymes that contain a catalytic triad associated with diverse catalytic activity.
The three predicted ABC transporters encoded by eqbHIJ, eqbK and eqbL are homologous (34–54% aa sequence identity) to unknown ABC transporters in Clostridium beijerinckii (Cbei_3098, Cbei_3097, Cbei_3096, Cbei_3094 and Cbei_3093 respectively) (Table S2). EqbI, EqbJ, EqbK and EqbL also share 24–45% aa identity with the putative C. kluyveri proteins CKL_1516, CKL_1517, CKL_1512 and CKL_1513 respectively (Seedorf et al., 2008). EqbK and EqbL share 55–57% aa sequence identity with SAS2320 and SAS2319 in Staphylococcus aureus (Holden et al., 2004) and have a fused N-terminal permease and C-terminal ATPase. Normally this would suggest involvement in efflux of extracellular toxins (Saurin et al., 1999). However, EqbK and EqbL share 28–30% aa sequence identity with YbtP and YbtQ of Y. pestis which, like IrtA and IrtB of Mycobacterium tuberculosis, are involved in siderophore-mediated iron import and are structurally unique among the subfamily of ABC transporters associated with iron transport (Fetherston et al., 1999; Rodriguez and Smith, 2006).
Substrate predictions
An important characteristic of the equibactin and yersiniabactin biosynthetic clusters is the presence of a single A-domain in HMWP2/EqbE that is probably responsible for the aminoacylation of all three different PCP domains (two in HMWP2/EqbE and one in HMWP1/EqbG), two of which are not linked physically to the A-domain. By comparison to the GrsA APhe prototype, critical residues lining the substrate-binding pocket of the EqbE A-domain active site were identified (Stachelhaus et al., 1999; Challis et al., 2000) and found to match those mediating cysteine recognition with the exception of a change from asparagine to aspartate at residue 278 (Table 1).
Table 1.
Prediction of EqbD and EqbE A-domain substrate specificity
| Residue (according to APhe numbering) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A-domaina | Substrate | 235 | 236 | 239b | 278 | 299 | 301 | 322 | 330b | Accession number |
| HMWP2 | Cys | D | L | Y | N | M | S | M | I | Q9Z399 |
| BacA | Cys | D | L | Y | N | L | S | L | I | O68006 |
| AngR | Cys | D | L | Y | N | M | S | M | I | P19828 |
| PchE | Cys | D | L | F | N | L | S | L | I | Q9RFM8 |
| PchF | Cys | D | L | Y | N | L | S | L | I | Q9RFM7 |
| CtaC | Cys | D | L | Y | N | M | S | L | V | Q5MD35 |
| MtaC | Cys | D | L | Y | N | M | S | L | I | Q9RFK9 |
| BlmIV | Cys | D | L | Y | N | L | S | L | I | Q9FB18 |
| EqbE | Cys? | D | L | Y | D | M | S | M | I | |
| DhbE | DHB | N | Y | S | A | Q | G | V | V | P40871 |
| EntE | DHB | N | Y | S | A | Q | G | V | V | P10378 |
| MxcE | DHB | N | F | S | A | Q | G | V | V | Q9F638 |
| VibE | DHB | N | F | S | A | Q | G | V | V | O07899 |
| AngE | DHB | N | F | S | A | Q | G | V | V | Q5DK17 |
| YbtE | Sal | N | F | C | A | Q | G | V | L | Q56950 |
| PchD | Sal | N | F | C | A | Q | G | V | I | Q9RFM9 |
| EqbD | Sal? | N | F | C | G | Q | G | I | I | |
| SnbA | 3-Hydroxy-picolinic acid | N | F | C | S | Q | G | V | L | P95819 |
Protein name: HMWP2, yersiniabactin-NRPS, Y. pestis (Gehring et al., 1998b); BacA, bacitracin NRPS, Bacillus licheniformis (Konz et al., 1997); AngR, anguibactin NRPS, Vibrio anguillarum (Tolmasky et al., 1993); PchE and PchF, pyochelin NRPSs, Pseudomonas aeruginosa (Quadri et al., 1999); CtaC, cystothiazole A NRPS, Cystobacter fuscus (Feng et al., 2005); MtaC, myxothiazol NRPS, Stigmatella aurantiaca (Silakowski et al., 1999); BlmIV, bleomycin NRPS, Streptomyces verticillus (Du et al., 2000); DhbE, bacillibactin DHB-AMP ligase, Bacillus subtilis (May et al., 2002); EntE, enterochelin synthase, Escherichia coli (Gehring et al., 1997); MxcE, myxochelin DHB-AMP ligase, S. aurantiaca (Silakowski et al., 2000); VibE, vibriobactin DHB-AMP ligase, Vibrio cholerae (Wyckoff et al., 1997); AngE, anguibactin DHB-AMP ligase, V. anguillarum (Alice et al., 2005); YbtE, yersiniabactin salicyl-AMP ligase, Y. pestis (Gehring et al., 1998b); PchD, pyochelin salicyl-AMP ligase, P. aeruginosa (Quadri et al., 1999); SnbA, pristinamycin I 3-hydroxy-picolinic acid-AMP ligase, Streptomyces pristinaespiralis (de Crecy-Lagard et al., 1997).
Amino acids at positions 239, 330 discriminate Sal from DHB (May et al., 2002). S. equi residues defined in bold differ from the consensus code of characterized substrate-activating proteins.
The predicted amino acid sequences between core A4 and A5 sequence motifs of the EqbE A-domain and EqbD were aligned, using clustalw, to A-domains or aryl-AMP ligase homologues with > 30% sequence identity to EqbE or EqbD respectively (Stachelhaus et al., 1999). Based on the structural data of DhbE and GrsA, residues conferring substrate specificity were identified (Stachelhaus et al., 1999; May et al., 2002).
EqbD shares 42% aa identity to the YbtE salicylate-AMP ligase (Gehring et al., 1998a), 41% sequence identity with 2,3-dihydroxybenzoate (DHB) AMP ligase, DhbE (May et al., 2002), and 55% aa identity with the uncharacterized CKL_1504 of C. kluyveri (Seedorf et al., 2008). The adapted specificity conferring code for aryl acid-activating domains discriminates between DHB and salicylate-activating enzymes (May et al., 2002). EqbD is predicted to activate salicylate since it has a cysteine at position 239, which would impede access of the 3′-OH group of DHB and a more sterically demanding isoleucine at position 330 that replaces the conserved valine in DHB-activating enzymes (Table 1). However, a small change from alanine to glycine was identified relative to residue 278 according to GrsA Aphe numbering, which differs from other NRPS systems that utilize salicylate.
Proposed mechanism of equibactin biosynthesis
Three condensation (C) domains identified in the two peptide synthetases, EqbE and EqbG, are strongly modified, diverging from classical C-domains involved in peptide bond formation. Instead, C-domains shared the highest homology (29–32% aa sequence identity) with cyclization (Cy) domains that catalyse thiazoline ring formation in yersiniabactin and bacitracin biosynthesis (De Crecy-Lagard et al., 1995; Konz et al., 1997; Bobrov et al., 2002). Seven signature regions identified in Cy domains of Yersinia enterocolitica (yersiniabactin), Bacillus licheniformis (bacitracin) and Vibrio anguillarum (anguibactin) by Konz et al. (1997) were also mostly conserved in the three putative Cy domains of EqbE and EqbG (Fig. 2). Some differences in these sequence alignments were apparent although the contribution of residues in these regions to catalytic activity is not yet known. However, the highly conserved Cy motif D-x-x-x-x-D-x-x-S that corresponds in location to the highly conserved condensation domain catalytic core, H-H-x-x-x-D-G-x-S (Konz et al., 1997; Keating et al., 2000), is present in all three putative Cy domains of S. equi. This suggests an involvement of these putative Cy domains in heterocylization activity since the two asparate residues of this Cy core are essential to both amide bond formation and heterocyclization by Cy1 of HMWP2 in Y. pestis (Keating et al., 2000).
Fig. 2.
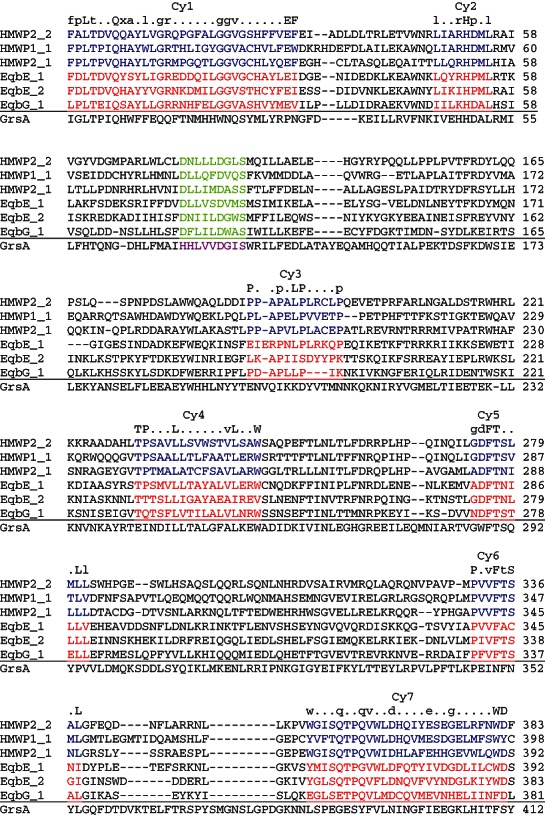
Bioinformatic prediction of eqb NRPS substrates. clustalw alignment of amino acid sequences of the putative Cy domains of EqbE, EqbG S. equi, HMWP1, HMWP2 Y. pestis and the classical condensation domain of GrsA Bacillus brevis (below line). Highlighted areas (red: S. equi, blue: Y. pestis) represent the seven conserved signature sequences (Cy1–Cy7) with the consensus shown above (Konz et al., 1997). The DX4DX2S catalytic core containing the two aspartate residues critical to amide bond formation and heterocyclization (Keating et al., 2000) is shown in green. The HHX3DGXS catalytic core of classic condensation domains is shown in purple for GrsA (Konz et al., 1997).
The presence of a putative thiazoline reductase (EqbF) with 32% aa identity with YbtU (Gehring et al., 1998a) and 32% and 31% aa identity with CKL_1506 and CKL_1510 of C. kluyveri, respectively (Seedorf et al., 2008), supports a prediction of thiazoline rings in the chemical structure of equibactin. Thiazoline rings of yersiniabactin and pyochelin are reduced to thiazolidine by the activity of these thiazoline reductase enzymes (Patel and Walsh, 2001; Miller et al., 2002). These enzymes also have a methyl transferase module, which, although apparently non-functional in yersiniabactin biosynthesis, introduce a methyl group to the N-atom of the thiazolidine residue in pyochelin (Patel and Walsh, 2001; Miller et al., 2002). A prediction of the NRPS intermediates is presented in Fig. 1C.
Non-ribosomal peptides are usually released from the last carrier domain in the assembly line through the activity of a terminal thioesterase (type I) (Challis and Naismith, 2004). The C-terminal domain of EqbG has similarity with type I thioesterases. However, sequence alignment of this domain to thioesterases in Pfam showed a S1092 to A mutation in the critical serine residue (GXSXG motif) of the predicted serine, histidine, aspartate catalytic triad essential for function (Bobrov et al., 2002; Reimmann et al., 2004). Alignment of the type II thioesterase encoded by eqbB showed an intact catalytic triad.
Regulation of the eqb NRPS
Upstream of the eqb operon is a CDS predicted to encode a putative metal ion-dependent repressor, EqbA. Close homologues of eqbA and eqbD gene products are present in the genome of S. agalactiae serotype III NEM316, although the latter gene appears to be a pseudogene in this group B streptococcus. EqbA is predicted to have the characteristic N-terminal DNA binding, central metal ion binding and dimerization domains of the MntR and IdeR family of manganese and iron-dependent repressors to which it has 33% and 28% sequence identity respectively (Schmitt and Holmes, 1991; Que and Helmann, 2000). DtxR, a member of the growing family of IdeR global iron-dependent regulators in Gram-positive bacteria, is activated on binding of divalent iron. Subsequent binding of the homodimeric form to a 21 bp DNA duplex blocks the transcription of downstream genes (Schmitt and Holmes, 1991; Pohl et al., 1999a). A unique 36 bp imperfect palindrome (underlined) was identified in the promoter region of eqbB and represents a potential operator sequence for EqbA:
5′-AACTATTATTGTTAGATGTATCTAACAATAATAGTTCTAGTAGTATATTAATAATCAGATGGAAGGTGTTTTGATG-3′ (−35, −10 promoter sites and the ATG translational start are highlighted in bold).
To determine the role of EqbA on the regulation of the eqb NRPS, we generated a series of allelic replacement mutants in S. equi strain 4047. Deletion of eqbE had no effect on the growth rate of S. equi on Todd–Hewitt agar (THA). However, deletion of eqbA produced very small colonies (Fig. 3A). We hypothesized that this phenotype resulted from iron toxicity resulting from over-production of the product(s) of the eqb cluster. Over-expression of eqbE was confirmed since deletion of eqbA resulted in a 13-fold increase in eqbE transcript levels (Fig. 3B). The generation of an ΔeqbA, ΔeqbE double deletion strain (ΔeqbAE), which had a large colony phenotype on THA, established that the slow growth of the ΔeqbA strain was as a direct consequence of the function of the eqbE gene product (Fig. 3A). Transformation of the ΔeqbA or ΔeqbAE strains with the pGhost9 plasmid containing a second copy of eqbA under the control of the eqbA promoter or a second copy of eqbE under the control of the eqbB promoter complemented the ΔeqbA and ΔeqbE and induced a large or small colony phenotype respectively (Fig. S2). The lack of an effect on the colony phenotype of S. equi grown in vitro following deletion of eqbE may be due to continued import of cations through the activity of several alternative cation transport systems, which include an HtsABC haem-binding system (SEQ0445 to SEQ0448) (Nygaard et al., 2006), a putative MtsABC Mn2+ and Fe3+ metal transport system (SEQ1658 to SEQ1660) with 80–91% aa sequence identity to that of S. pyogenes (Janulczyk et al., 1999) and a putative FtsABCD Fe3+ ferrichrome transport system (SEQ1836 to SEQ1839) with 59–77% aa sequence identity to that of S. pyogenes (Hanks et al., 2005) (http://www.sanger.ac.uk).
Fig. 3.
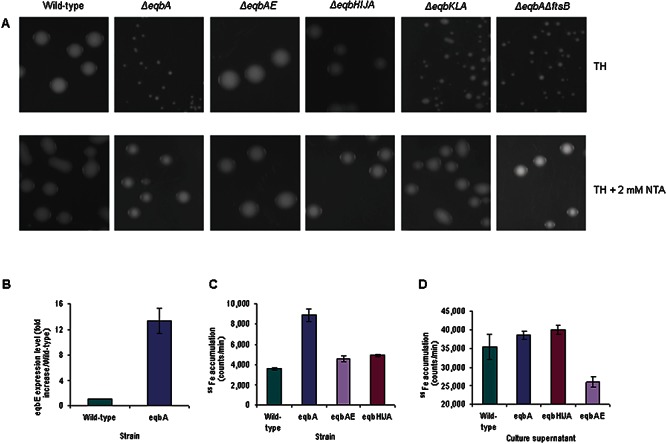
Phenotypic effects of deletions in the eqb gene cluster. A. Photographs show colonies of wild-type, ΔeqbA, ΔeqbAE, ΔeqbHIJA, ΔeqbKLA and ΔeqbAΔftsB S. equi strains grown overnight on THA and the increase in colony size of the ΔeqbA, ΔeqbKLA and ΔeqbAΔftsB strains grown on THA supplemented with 2 mM NTA. B. Fold increase in eqbE transcript level in the ΔeqbA strain relative to the wild-type 4047 strain, which has been normalized to one (mean ± standard error mean, n = 3). C. Quantification of 55Fe accumulation by different S. equi strains (mean ± standard error mean, n = 3). The difference in 55Fe accumulation between the ΔeqbA strain and the ΔeqbAE, ΔeqbHIJA and wild-type strains was found to be statistically significant using two-sample Wilcoxon rank-sum (Mann–Whitney) tests (P = 0.05, n = 3). D. Quantification of 55Fe accumulation by S. equi strain ΔeqbAE cross-fed with filter-sterilized culture supernatant from different S. equi strains grown to stationary phase.
Further studies supported the hypothesis that the eqbA deletion phenotype resulted from increased iron uptake. Supplementation of THA with 2 mM nitrilotriacetic acid (NTA), a known chelator of iron, restored near normal colony size in the ΔeqbA strain (Fig. 3A). Susceptibility to streptonigrin was used as an indirect measure of intracellular iron concentration (Brown et al., 2002). Growth of wild-type S. equi was prevented by a minimal inhibitory concentration of 0.06 μM streptonigrin in THB. The ΔeqbA mutant was 16 times more sensitive to streptonigrin, while strains predicted to be unable to produce the NRPS product(s) through deletion of the biosynthetic gene eqbE (ΔeqbE and ΔeqbAE) were twice more resistant to the antibiotic (Table 2A). There was no difference in susceptibility of these strains to the antibiotic erythromycin. Therefore, increased sensitivity of the ΔeqbA mutant to streptonigrin compared with wild-type, ΔeqbE or ΔeqbAE strains suggests a greater intracellular iron pool in the former most likely as a result of de-repression of eqb non-ribosomal peptide synthesis. The same differences in streptonigrin sensitivity were conferred when the ΔeqbAE mutant was cross-fed with filter-sterilized stationary-phase culture supernatant from the wild-type and various mutant strains (Table 2B), consistent with an accumulation of iron being mediated by an excreted, soluble factor.
Table 2.
Production of the eqb NRPS product by allelic replacement mutants of S. equi and E. coli strains expressing different complements of the Eqb proteins
| A | ||
|---|---|---|
| Strain | Streptonigrin MIC(μM) | Erythromycin MIC (μg ml−1) |
| Wild type | 0.06 | 0.016 |
| ΔeqbA | 0.004 | 0.016 |
| ΔeqbE | 0.125 | 0.016 |
| ΔeqbHIJ | 0.06 | 0.016 |
| ΔeqbKL | 0.06 | 0.016 |
| ΔeqbAE | 0.125 | 0.016 |
| ΔeqbHIJA | 0.03 | 0.016 |
| B | ||
|---|---|---|
| Conditioned THB from strain | Streptonigrin MIC of strain ΔeqbAE (μM) | Erythromycin MIC of strain ΔeqbAE (μg ml−1) |
| Wild type | 0.03 | 0.016 |
| ΔeqbA | 0.004 | 0.016 |
| ΔeqbE | 0.125 | 0.016 |
| ΔeqbAE | 0.125 | 0.016 |
| ΔeqbHIJ | 0.016 | 0.016 |
| ΔeqbKL | 0.06 | 0.016 |
| ΔeqbHIJA | 0.002 | 0.016 |
| C | |||
|---|---|---|---|
| Conditioned CDM from strain | Supplementation | Streptonigrin MIC of strain ΔeqbAE (μM) | Erythromycin MIC of strain ΔeqbAE (μg ml−1) |
| ΔeqbAE | 0.125 | 0.03 | |
| ΔeqbHIJA | 0.125 | 0.03 | |
| ΔeqbAE | 10 μM salicylate | 0.125 | 0.03 |
| ΔeqbHIJA | 10 μM salicylate | 0.0005 | 0.03 |
| D | |||
|---|---|---|---|
| Conditioned media from E. coliexpressing Eqb proteins | Media | Streptonigrin MIC of strain ΔeqbAE (μM) | Erythromycin MIC of strain ΔeqbAE (μg ml−1) |
| EqbBCDEFGMN | LB | 0.06 | 0.004 |
| EqbBCDEFGMN | LB + 1 mM salicylate | 0.002 | 0.004 |
| EqbBCDFGMN + pET21a (no EqbE) | LB + 1 mM salicylate | 1 | 0.004 |
| EqbBCDEFMN (no EqbG) | LB + 1 mM salicylate | 1 | 0.004 |
| EqbBCDEFGN (no EqbM) | LB + 1 mM salicylate | 1 | 0.004 |
| EqbBCDEFGM (no EqbN) | LB + 1 mM salicylate | 0.5 | 0.004 |
| EqbBCDEFGMN | MM + 1 mM salicylate | 0.002 | 0.016 |
| EqbBCDFGMN + pET21a (no EqbE) | MM + 1 mM salicylate | 0.06 | 0.016 |
A. Sensitivity of wild-type and allelic replacement strains of S. equi to streptonigrin and erythromycin. MIC refers to the minimum inhibitory concentration of antibiotic required to prevent growth. The twofold difference in the streptonigrin MIC of wild type versus ΔeqbE was found to be statistically significant using a two-sample Wilcoxon rank-sum (Mann–Whitney) test (P = 0.008, n = 4).
B. Streptonigrin and erythromycin sensitivity in the ΔeqbAE strain cross-fed with filter-sterilized culture supernatant from wild-type and allelic replacement strains grown to stationary phase in THB. The twofold difference in ΔeqbAE streptonigrin MIC conferred by cross-feeding with ΔeqbA-conditioned CDM relative to ΔeqbHIJA-conditioned CDM was found to be statistically significant using a two-sample Wilcoxon rank-sum (Mann–Whitney) test (P = 0.008, n = 4).
C. Streptonigrin and erythromycin sensitivity in the ΔeqbAE strain cross-fed with filter-sterilized culture supernatant from ΔeqbAE and ΔeqbHIJA allelic replacement strains grown to stationary phase in CDM ± 10 μM salicylate.
D. Streptonigrin and erythromycin sensitivity in the ΔeqbAE strain cross-fed with filter-sterilized culture supernatant from E. coli strains expressing different complements of Eqb proteins grown to stationary phase in LB ± 1 mM salicylate or MM + 1 mM salicylate.
The increased production of equibactin in the ΔeqbA mutant was further supported by 55FeCl3 incorporation assays, which showed an almost twofold increase in intracellular iron in the repressor deletion strain, compared with wild type or the ΔeqbAE mutant (Fig. 3C). ΔeqbAE cross-fed with filter-sterilized stationary-phase culture supernatant from the ΔeqbA strain but not the ΔeqbAE strain had a similar increase in 55FeCl3 accumulation (Fig. 3D).
The mechanism for import of the NRPS product(s)
We exploited the small colony phenotype of the ΔeqbA mutant to identify other CDSs likely to be involved in eqb product function. Deletion of the putative ABC transporters encoded by eqbK and eqbL or the ftsB gene, which lies outside the eqb locus and encodes a putative ferric-siderophore receptor (Hanks et al., 2005; Clancy et al., 2006), did not prevent the generation of the small colony phenotype on subsequent deletion of eqbA (Fig. 3A). However, deletion of the eqbH, eqbI and eqbJ genes, followed by deletion of eqbA (ΔeqbHIJA), did prevent the generation of the small colony phenotype (Fig. 3A). Reduced levels of 55Fe in the ΔeqbHIJA strain also suggested that the eqbH, eqbI and/or eqbJ gene products are essential for the majority of eqb product-dependent iron accumulation (Fig. 3C).
To determine if eqbH, eqbI and eqbJ were important to the export or import of the NRPS product(s), filter-sterilized culture supernatant from mutant strains was added to the ΔeqbAE strain and its susceptibility to streptonigrin was quantified. Media from the ΔeqbHIJA strain increased the sensitivity of the ΔeqbAE strain by 64-fold compared with a 32-fold increased sensitivity conferred by media from the ΔeqbA strain (P = 0.008, Table 2B). These data suggest that the ΔeqbHIJA strain secretes, but is unable to import the NRPS product(s) resulting in a build-up of this product in the culture media. In addition, ΔeqbAE cross-fed with filter-sterilized culture supernatant from the ΔeqbHIJA strain had an increase in 55FeCl3 accumulation slightly greater, although not significantly so, than that observed when ΔeqbAE was cross-fed with supernatant from the ΔeqbA strain (Fig. 3D).
The biochemical requirements for the eqb NRPS
A homologue of the salicylate synthases encoded by ybtS in Yersinia sp. (Miller et al., 2002) or pchA and pchB of Pseudomonas sp. (Serino et al., 1995) was not identified in S. equi strain 4047. S. equi cultured in Todd–Hewitt medium prepared from bovine heart infusion and a tryptic digest of animal tissue required no supplementation for equibactin production. However, production of the eqb product by the ΔeqbHIJA strain in chemically defined media (CDM) required salicylate supplementation consistent with salicylate being a substrate for the equibactin NRPS (Table 2C). The CDM contains 0.2 mM l-cysteine.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were used to analyse recombinant EqbA (rEqbA) binding to a 227 bp DNA fragment (A) containing the upstream region of the eqb operon (Peqb) (−237 to −11 bp) (Fig. 4). Unbound DNA ran as two distinct bands on non-denaturing polyacrylamide gels and was unaffected by the presence of different cations (lanes 1, 3, 5, 7, 9, 11 and 13). Binding of rEqbA caused a shift in the mobility of the lower band (lane 2). Pre-treatment of rEqbA with EDTA resulted in a partial shift in the mobility of the lower band (lane 4), which could be enhanced in the presence of additional Fe2+ or Zn2+ (lanes 6 and 12) and to a lesser degree by Mn2+ (lane 8), but not by Fe3+ or Cu2+ (lanes 10 and 14). The presence of the −73 bp to −38 bp DNA palindrome was essential for rEqbA binding as no shift was observed using DNA fragments containing the −237 to −73 bp (B) (lanes 15 and 16) or −237 to −135 bp (C) (lanes 17 and 18) upstream regions of the eqb operon. Incubation of a control DNA fragment [LightShift® Chemiluminescent EMSA Kit (Pierce)] with rEqbA did not result in a shift in mobility and a control crude extract [LightShift® Chemiluminescent EMSA Kit (Pierce)] did not bind to Peqb (data not shown), suggesting that rEqbA binds specifically to Peqb.
Fig. 4.
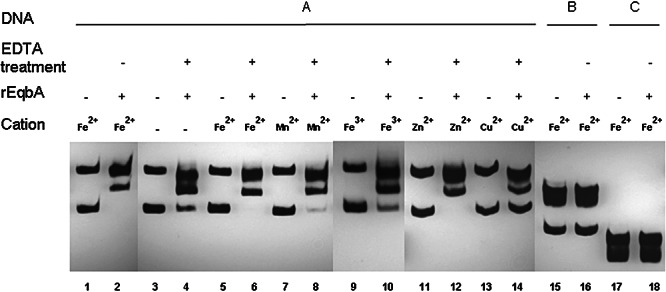
Electrophoretic mobility shift assay. Biotin end-labelled DNA fragments containing upstream regions of eqbB, A (227 bp, −237 to −11 bp), B (165 bp, −237 to −73 bp) or C (103 bp, −237 to −135 bp), were incubated in the presence of Fe2+, Mn2+, Fe3+ Zn2+, Cu2+ or control (−) as indicated with (+) or without (−) rEqbA and with (+) or without (−) prior treatment of rEqbA with EDTA. DNA fragments B and C lack the palindromic promoter region (−73 to −38 bp) immediately upstream of eqbB. Experimental conditions are described in Experimental procedures.
Reconstitution of the equibactin NRPS in E. coli
To determine if all of the genes necessary for production of equibactin were present in the eqb locus, and to produce equibactin in a less complex media, we cloned eqbB, eqbC, eqbD, eqbE, eqbF, eqbG, eqbM and eqbN into three expression plasmids with compatible replicons. Production of all eight Eqb proteins was induced by IPTG treatment of transformed E. coli strain BL21(DE3) cells grown to log phase in LB broth or a minimal medium (MM) in the presence of 1 mM salicylate as in Experimental procedures.
Filter-sterilized supernatant from E. coli containing all eight eqb CDSs and grown in LB increased the sensitivity of strain ΔeqbAE to streptonigrin from 1 μM to 0.002 μM, but had no effect on the sensitivity to erythromycin in cross-feeding assays (Table 2D). Exclusion of eqbE, eqbG, eqbM or eqbN prevented the associated increased streptonigrin sensitivity suggesting that all four of these proteins are required for reconstitution of the equibactin NRPS in E. coli (Table 2D).
An increased sensitivity to streptonigrin on cross-feeding strain ΔeqbAE with filter-sterilized MM following growth of E. coli containing all eight eqb CDSs from 0.06 to 0.002 was also observed (Table 2D).
However, despite these effects on the streptonigrin sensitivity of strain ΔeqbAE in cross-feeding assays, repeated LC-MS analysis of culture supernatants, including conditioned MM, failed to identify equibactin.
Discussion
Available evidence suggests that S. equi evolved from an ancestral S. zooepidemicus strain (Jorm et al., 1994; Webb, 2008), an opportunistic zoonotic pathogen largely associated with subclinical inflammatory airway disease in young horses (Wood et al., 2005). Given the close genetic similarity between these two streptococci, we used comparative genomics to identify differences between the species with a view to distinguishing key determinants of their differing pathogenic properties. The full results of our genome comparison will be reported elsewhere. However, in addition to previously described genetic differences (Galan and Timoney, 1987; Timoney et al., 1997; Lindmark et al., 1999; Artiushin et al., 2002; Proft et al., 2003; Alber et al., 2005) we report here the identification of a streptococcal ICE, ICESe2, and the function of the siderophore-like NRPS system encoded by this element. This element was previously noted in a broad screen for ICEs in sequenced bacterial genomes, but no putative function was assigned (Burrus et al., 2002b).
ICESe2 was present in all strains of S. equi, but absent from all strains of S. zooepidemicus examined. These strains were selected to be a diverse population based on their SeM allele (Kelly et al., 2006; Waller and Jolley, 2007) or 16S−23S RNA gene intergenic spacer/SzP PCR subtype (Newton et al., 2008) and were highly diverse based on their MLST sequence type (Webb, 2008). Our results also indicated that ICESe2 was located at the same genome position in all 18 S. equi strains, suggesting that its integration was an early and stable event in the evolution of S. equi.
The similarity of ICESe2 CDSs predicted to be involved in conjugation and site-specific recombination to those from C. difficile CDTn2 and CDTn5 is extremely interesting and raises the possibility of acquisition of the equibactin NRPS locus via a conjugative-like mechanism from a genetically unrelated bacterium. Although we predict integrative and conjugative genes are present in ICESe2, to date we failed to detect a circular intermediate by PCR through repeated experiments suggesting that this element may no longer retain the ability to recircularize and transfer to other bacteria.
ICESe2, CDTn2 and CDTn5 have putative conjugation modules that are closely related to the conjugation module of Tn1549 in Enterococcus spp., suggesting that they have inherited their similar structure from a common module ancestor. However, the serine recombinase encoded at the right flank of these elements differs from the Int (tyrosine recombinase) and Xis of Tn1549, which taken together with the differences in accessory functions highlights the importance of module exchange in the evolution of novel mosaic ICE structures. Serine recombinases are less common and genetically unrelated to the tyrosine recombinases and use a different mechanism for the site-specific recombination of mobile elements (Grindley et al., 2006). However, this family is rather heterogeneous and ranges in size from 180 to nearly 800 aa residues. ICESe2 encodes a small recombinase (SEQ1229) with homology to the N-terminal half of the large TndX resolvase, which mediates excision and insertion of Tn5397 by introducing 2 bp staggered cuts at the 3′ ends of GA dinucleotides at the ends of the element (excision) or in the target site (integration) (Wang et al., 2000). Consequently, direct GA dinucleotide repeats delineate the ends of Tn5397 and one copy is also present at the joint of the circular form. CDTn5 also has 5 bp direct repeats at these corresponding sites, whereas no direct repeats were identified in the flanks of ICESe2. However, particularly large 108 bp perfect inverted repeats were present at the flanks of the element in S. equi, which like the much smaller imperfect inverted repeats (19–20 bp) in Tn5397 and other ICEs may represent DNA binding sequences for the recombinase during the formation of the synaptic complex.
Reconstitution of the equibactin NRPS in E. coli was successfully achieved using three compatible expression plasmids containing eqbB, eqbC, eqbD, eqbE, eqbF, eqbG, eqbM and eqbN. Production in E. coli was enhanced by supplementation of growth media with 1 mM salicylate, was abolished in the absence of eqbE, eqbG and eqbM and was reduced in the absence of eqbN. Therefore, each of these genes plays a role in the biosynthesis of the eqb NRPS product(s) and the equibactin locus contains all of the genes unique to S. equi that are required for the biosynthesis of NRPS product(s).
The sequence of a NRPS has been used previously to predict the structure of the peptide produced (Challis and Ravel, 2000; Lautru et al., 2005). The similarity in type and organization of functional modules in the equibactin NRPS and the yersiniabactin biosynthetic system (Miller et al., 2002) leads us to suggest a tentative model for equibactin biosynthesis (Fig. 1). Initially, we propose activation of the aryl acid carrier protein (ArCP) domain of EqbE and the peptide carrier domains (PCP) of EqbE and EqbG by the putative 4′-phosphopantetheinyl transferase encoded by eqbC. EqbD is proposed to activate salicylate, which is then transferred to the phosphopantotheine thiol of the ArCP domain of EqbE. Our observation that addition of salicylate is required for production of equibactin in CDM provides evidence in support of the proposed involvement of salicylate. Salicylate was also required for optimum production of equibactin in E. coli containing the reconstituted NRPS, but was not required for production of equibactin by S. equi in Todd–Hewitt medium. This medium is prepared from bovine heart infusion and a tryptic digest of animal tissue and we propose that an appropriate NRPS substrate is released from host tissue during infection with S. equi. The single adenylation (A)-domain of EqbE is predicted to activate cysteine which is then transferred to the phosphopantotheine thiol of each of the three PCP domains of the NRPS (two in EqbE and one in EqbG). We predict that the cysteinyl thioesters attached to each PCP domain will be condensed and cyclized as the growing chain translocates to each of the three PCP domains. The presence of a putative thiazoline reductase (EqbF) suggests that as in yersiniabactin biosynthesis, the second thiazoline ring of equibactin is likely to be reduced to a thiazolidine to prevent the auto-oxidation of the first ring to a thiazole. However, the available data do not rule out the possibility that the first or third thiazoline ring could be reduced to a thiazolidine instead of or as well.
Yersiniabactin is released from the last PCP domain by a C-terminal thioesterase domain. However, the C-terminal thioesterase domain of EqbG has the critical serine residue (S1092) of the predicted serine, histidine, aspartate catalytic triad essential for function (GXSXG motif) mutated to alanine. The type II thioesterase encoded by eqbB has an intact catalytic triad but to date these type II TEs have only a presumed editing role in maintaining the efficiency of non-ribosomal peptide synthesis via removal of inappropriate substrates (Marahiel et al., 1997; Schneider and Marahiel, 1998; Butler et al., 1999; Bobrov et al., 2002; Reimmann et al., 2004). Interestingly, the tetrapeptide coelichelin is synthesized by a trimodular NRPS lacking a TE domain and is proposed to be released through the action of a separately encoded hydrolase (Lautru et al., 2005). EqbN encodes a putative α/β hydrolase which could provide another potential mechanism for hydrolytic chain release similar to coelichelin. The eqbN gene was required for optimal production of the eqb NRPS product(s) by E. coli and induction of increased streptonigrin sensitivity in cross-feeding assays. However, absence of eqbN did not abolish production of the NRPS product(s) in this system suggesting that E. coli may produce an endogenous hydrolase that can complement the activity of EqbN or that direct hydrolysis with water without the formation of an acyl enzyme intermediate is also possible, either at a low background level or through a base-catalysed attack of water rather than nucleophilic attack of an active site serine (Li and Bugg, 2007).
Regulation of eqb non-ribosomal peptide synthesis is achieved through the action of the IdeR-like transcriptional repressor EqbA. Deletion of eqbA led to increased transcription of the NRPS operon, over-production of the NRPS product(s) and increased import of iron causing toxicity and a small-colony phenotype. Deletions of ideR and other ideR/fur homologues have been lethal in M. tuberculosis and several species of Pseudomonas, Vibrio and Neisseria (Berish et al., 1993; Tolmasky et al., 1994; Venturi et al., 1995). In M. tuberculosis, a rare deletion of ideR was obtained when the lethal effects of ideR inactivation were alleviated by a second-site suppressor mutation which restricted iron assimilation capacity (Rodriguez et al., 2002). We propose that like other IdeR family members, EqbA is activated on binding iron, leading to homodimerization and binding to a 36 bp DNA palindrome located immediately upstream of the NRPS operon to block its transcription. EqbA has a number of novel features compared with other IdeR family members. The suggested operator site is unlike other palindromic DNA iron boxes and this is reflected in the substitution of N-terminal helix–turn–helix residues involved in DNA contact (Fig. S3). Some of the conserved metal binding and dimerization residues differ in EqbA. In addition, the SH3-like fold of DtxR and IdeR, which functions to stabilize the DNA binding conformation of these repressors (Pohl et al., 1999b; Oram et al., 2005), is lacking in EqbA. EqbA is therefore likely to represent a new subclass of the IdeR family. Electrophoretic mobility shift assays confirmed that EqbA does indeed bind to the eqbB promoter in a Fe2+-, Zn2+- and Mn2+-responsive manner. EqbA binding was not affected by Cu2+ or Fe3+ and was dependent on the presence of the eqbB promoter's DNA palindrome. The shift due to addition of Mn2+ was not complete and may reflect a reduced affinity of EqbA for Mn2+. These data are in broad agreement with the activation of IdeR by different cations rather than MntR, which is Mn2+ specific (Schmitt et al., 1995; Que and Helmann, 2000; Guedon and Helmann, 2003; Chou et al., 2004). Further comparison of the putative residues involved in the co-ordination of the regulatory metal (Guedon and Helmann, 2003) revealed that EqbA shares a cysteine at aa position 104 with DtxR, which confers Fe2+ responsiveness rather than the Mn2+-responsive glutamate possessed by MntR and TroR at the homologous position (Fig. S3). The Fe2+-responsive methionine of DtxR has been replaced by asparagine at aa position 13 of EqbA, which is similar to the homologous residue in the Mn2+-responsive TroR.
No iron-chelating activity could be detected in the ΔeqbHIJA mutant of S. equi using the universal chrome azurol S (CAS) assay for iron chelators (Schwyn and Neilands, 1987). We have also been unable to identify equibactin by comparative metabolic profiling of the ΔeqbA, ΔeqbAE and ΔeqbHIJA strains using LC-MS. Furthermore, examination of biologically active conditioned LB and MM from E. coli containing the reconstituted equibactin NRPS by LC-MS also failed to identify equibactin. These data suggest that the concentration of equibactin in culture supernatant is low or the product of the eqb cluster may have a low affinity for iron and a structure different from that proposed. Pyochelin, a siderophore that lacks the third incorporated thiazoline and malonyl linker of yersiniabactin, but otherwise shares the salicyl-bis-thiazonyl core has a relatively low affinity for iron. A role for pyochelin in the uptake of other essential metals has been proposed (Visca et al., 1992) and this may also represent an alternative function for the eqb NRPS product(s). The host catecholamine norepinephrine, although unable to remove iron from CAS, can increase the availability of free iron to bacteria through an interaction that interferes with host glycoprotein iron sequestration (Freestone et al., 2000).
These data are also consistent with a signalling role for the NRPS product(s) in the upregulation of iron transport system(s). A number of siderophores are able to function as signalling molecules, influencing the expression of their own biosynthetic genes and cell surface receptors as well as other secreted virulence factors (Crosa, 1997; Pelludat et al., 1998; Beare et al., 2003; Michel et al., 2005; 2007; Miethke and Marahiel, 2007). Ferric-pyochelin acts as an intracellular effector via a direct interaction with an AraC-type regulator (Michel et al., 2005). The AraC-type YbtA regulator also induces yersiniabactin operons involved in yersiniabactin biosynthesis, Fe-yersiniabactin uptake and salicylate synthesis, and the Fe-yersiniabactin receptor. An interaction of the YbtA with its DNA promoter target and Fe-yersiniabactin is proposed to be a prerequisite for the recruitment of the RNA polymerase complex (Fetherston et al., 1996; Perry et al., 1999; Anisimov et al., 2005a, b; Miethke and Marahiel, 2007).
Siderophore-mediated iron uptake operons, similar to the ferric hydroxamate uptake (fhu) systems of S. aureus and Bacillus subtilis, have been identified in other streptococci (Hanks et al., 2005; Clancy et al., 2006; Pramanik and Braun, 2006). FhuD, the associated lipoprotein receptor in S. agalactiae, is able to bind a range of siderophores of both hydroxamate and catecholate classes. The homologous operon (42–55% aa sequence identity) is also present in the S. equi genome and we hypothesized that this could be involved in acquisition of the eqb NRPS product(s). However, deletion of the ftsB gene did not prevent iron toxicity due to over-production of the NRPS product(s) on the further deletion of eqbA (Fig. 3A). This indicates that FtsB is not absolutely required for import of the NRPS product(s) and further studies are necessary in order to determine if surface receptors play a role in import of the NRPS product(s). EqbK and EqbL share 29% aa sequence identity with YbtP and YbtQ of Y. pestis which, like IrtA and IrtB of M. tuberculosis, are involved in siderophore-mediated iron import and are structurally unique among the subfamily of ABC transporters associated with iron transport (Fetherston et al., 1999; Rodriguez and Smith, 2006). Surprisingly, deletion of eqbK and eqbL did not prevent the generation of a small colony phenotype on deletion of eqbA (Fig. 3A). Similar fused permease-ATPase type proteins have been associated with the efflux of substrates and the small-colony phenotype observed for eqbKLA triple mutant could result from a failure to secrete the siderophore (Saurin et al., 1999). Supernatant cross-feeding studies have consistently demonstrated that strain ΔeqbKL confers twofold decreased sensitivity to streptonigrin relative to wild type (Table 2B) suggesting that eqbK and/or eqbL could be involved in efflux of the NRPS product(s). The ABC transporter encoded by the eqbH, eqbI and eqbJ genes plays a major role in the eqb-dependent incorporation of iron. Purification and elucidation of the eqb NRPS product structure is warranted to better understand the role played by the NRPS product(s) in vivo.
Siderophore biosynthesis has not been identified in any streptococci examined to date (Eichenbaum et al., 1996). A non-ribosomal peptide synthesis system has been noted in the published genome sequence of the oral pathogen S. mutans. Unlike the eqb locus, this NRPS appears to incorporate five diverse residues into a molecule that is predicted to have more of an antibiotic-like structure (gramicidin/bacitracin family). However, homologues of eqbA and eqbD (pseudogene) are present in the genome of S. agalactiae serotype III, suggesting that a locus with similarity to the eqb NRPS may have been important to this organism at some point in its history.
ICESe2 is a key feature in the evolved genome of S. equi. While we have not yet identified the peptide made by the equibactin NRPS or shown direct iron binding, data consistent with its role in iron acquisition have been established. Given the importance of iron acquisition to other streptococcal pathogens (Brown and Holden, 2002), the acquisition of ICESe2 may have contributed to the increased pathogenesis of this important streptococcus. We hypothesize that more efficient acquisition of iron enhances retropharyngeal lymph node abscessation, which is critical to the establishment of long-term carriage and vital to the success of S. equi. Further studies to quantify the in vivo effects of the equibactin NRPS are now required to better understand its pathological significance.
Experimental procedures
Bacterial strains, media and growth conditions
Streptococcus equi strain 4047 was isolated in 1990 from the submandibular abscess of a New Forest pony. Strains were cultured in Todd–Hewitt broth (THB) (Sigma) or on THA (Sigma) at 37°C with 5% CO2 unless otherwise stated. CDM for Group A Streptococci (SAFC Biosciences) was used to produce conditioned supernatant for siderophore assays and to identify the additional biochemical requirements of eqb NRPS product production on supplementation with 10 μM salicylate (Sigma) (CDMs). To measure the influence of free cation concentration on the growth of wild-type and mutant S. equi strains, THA was supplemented with 2 mM NTA (Sigma).
Identification of S. equi-restricted genes
The Artemis Comparison Tool (Carver et al., 2005) was used to view blastn alignments of the publicly available sequence data from the S. equi and S. zooepidemicus genome sequencing projects (http://www.sanger.ac.uk). Comparisons of predicted CDSs with Uniprot were performed using blastp and fasta. Southern blot and PCR analyses were used to determine the genome location, circularization and prevalence of ICESe2 across 18 strains of S. equi and 73 strains of S. zooepidemicus (Table S1). Diverse strains were selected based on their SeM allele (Kelly et al., 2006) and RNA intergenic spacer/M protein subtype (Newton et al., 2008) respectively.
Allelic replacement
Internal gene deletions were introduced into S. equi 4047 through an allelic replacement strategy previously described for the production of a ΔprtM mutant (Hamilton et al., 2006). Table S1 shows primers used and details internal frame deletions in the construction of plasmids pGeqbAΔ, pGeqbEΔ, pGeqbHIJΔ, pGeqbKLΔ and pGftsBΔ. The ΔeqbAE double deletion mutant was generated by transformation of S. equi strain ΔeqbE with the pGeqbAΔ plasmid followed by recombination and selection as described previously. Similarly, for other multiple deletion strains, eqbA was always the last gene to be deleted. PCR and sequencing were used to confirm the presence of gene deletions.
RNA isolation
Total RNA was extracted from bacteria using RNAprotect, RNeasy and DNase Kits according to the manufacturer's instructions (Qiagen). Bacteria grown to mid-log phase were pelleted by centrifugation for 10 min at 5000 g. The pellet was re-suspended in 200 μl of TE buffer (10 mM Tris-Cl, 1 mM EDTA pH 8.0) containing 3 mg of lysozyme (Sigma) and 500 U mutanolysin (Sigma). After incubation at room temperature for 45 min with repeated vortexing, 700 μl of RLT buffer containing β-mercaptoethanol was added, and the tube was vortexed. The mixture was transferred to a 2 ml reaction tube containing 0.05 g of 100-μm-diameter acid-washed glass beads (Sigma) and vortexed for 5 min. The mixture was then centrifuged and total RNA was extracted from the supernatant using an RNeasy Midi Kit. Five micrograms of recovered RNA was treated with DNase to remove any contaminating DNA followed by clean-up on an RNeasy mini column. The quantity and purity of RNA was determined by optical density measurements at 260 and 280 nm using a NanoDrop® ND1000 spectrophotometer (NanoDrop Technologies).
Reverse transcription and quantitative real-time PCR
Transcriptional regulation of the NRPS gene (eqbE) by the putative repressor (EqbA) was assessed through the comparison of eqbE transcript levels in the ΔeqbA and parent 4047 S. equi strains grown to mid-log phase in THB. A quantitative two-step reverse transcription (RT) PCR procedure was used to analyse levels of eqbE gene transcription relative to the housekeeping gene gyrA. RT was performed using the Verso cDNA kit (Abgene). The RT reaction mixture (20 μl) contained 96 ng of total RNA, 2 μM gene-specific antisense primer (Table S1), 500 μM dNTP mix, 16 U RNase inhibitor (RNASIN, Promega), 1× cDNA synthesis buffer, 1 μl of RT enhancer and 1 μl of Verso enzyme mix. RT was performed at 50°C for 30 min and terminated by heating to 95°C for 2 min. Quantitative real-time PCR (QPCR) was performed with a Techne Quantica instrument and data were analysed using Quansoft software (Techne). For the QPCR, 6 μl of RT reaction mixture diluted 1/1000 was mixed with 0.3 μM forward and reverse primers (Table S1), 1× ABsolute QPCR SYBR green mix (Abgene) and 40 nM ROX in a total volume of 20 μl and subjected to thermocycling at 95°C for 15 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Dissociation curves were analysed, following a final ramp step from 60°C to 90°C with reads at 0.5°C increments, to rule out non-specific amplification. No-template-negative controls and reverse transcriptase-negative controls were included to confirm the absence of contaminating DNA from RNA samples. Standard curves (Cp versus log gene copy number) with an efficiency of 1.9 (R2 = 0.999) were generated from gDNA for each target gene and used to calculate gene copy number in cDNA samples generated from three independent RNA extractions. eqbE gene copy number was normalized to gyrA reference gene copy number to correct for differences in the amount of starting material. Data were expressed as fold increase in normalized eqbE transcript level in the ΔeqbA mutant strain relative to the wild-type 4047 strain.
Streptonigrin sensitivity
The minimum inhibitory concentration (MIC) of streptonigrin and erythromycin was determined following overnight incubation of bacterial strains in a standard 96-well microtitre plate assay. One hundred microlitres of re-suspended colony material corresponding to 104 colony-forming units (cfu) ml−1 in THB, CDMs, MM or conditioned THB/CDMs/MM was added to 100 μl of THB supplemented with streptonigrin (concentration ranging from 2 μM to 0.5 nM through twofold serial dilutions) in duplicate wells of a microtitre plate. Sensitivity to erythromycin was determined using the same procedure (0.5 μg ml−1 to 0.5 ng ml−1 erythromycin).
55FeCl3 accumulation
Wild-type and mutant S. equi strains were depleted of iron by growth overnight on THA supplemented with 4 mM NTA. Colony material from these plates was re-suspended in PBS with the volume adjusted to give an optical density at 600 nm of 1.0. Half millilitre of this bacterial suspension was added to 0.5 ml of THB supplemented with 2.0 μCi ml−1 55FeCl3 (Promega) and incubated at 37°C with 5% CO2. After 3 h, cell growth was monitored through measurement of optical density and a volume of cells equivalent to 6 × 107 cfu was collected in triplicate wells of a 96-well UniFilter® GF/CR (Perkin Elmer) with a Packard FilterMate™ Harvester. Cells were washed on the filter with 10 mM NTA and then methanol. The filter was dried for 1 h at room temperature prior to the addition of 20 μl of Microscint 20 (Packard). Bacterial iron accumulation was measured by liquid scintillation counting on a TopCount NXT™ microplate counter (Packard).
A modification of this method was used to analyse the accumulation of 55FeCl3 in the S. equi mutant strain ΔeqbAE when cross-fed with filter-sterilized culture supernatant from wild-type, ΔeqbAE, ΔeqbHIJA or ΔeqbA strains grown to stationary phase overnight. Bacteria were re-suspended in 1 ml of 2× THB supplemented with 2.0 μCi ml−1 55FeCl3 to an optical density at 600 nm of 0.2 and mixed with an equal volume of filter-sterilized culture supernatant. Cells were incubated at 37°C with 5% CO2 until they reached an optical density at 600 nm of 0.5 and then a volume of cells equivalent to 1 × 108 cfu was collected in triplicate wells of a 96-well UniFilter® GF/CR and processed as described above.
Complementation of ΔeqbA and ΔeqbE
Full-length copies of the eqbA or eqbE genes were cloned into the plasmid pGhost9 under the control of the eqbA and eqbB promoters using primers listed in Table S1 to generate constructs pGpAeqbA and pGpBeqbE respectively. The ΔeqbA and ΔeqbAE mutant strains were transformed with pGpAeqbA, pGpBeqbE or pGhost9 control plasmid. Transformants were selected with erythromycin and grown at 28°C, which permits extrachromosomal replication of pGhost9, for 48 h to enable comparison of transformant phenotypes.
LC-MS analyses of culture supernatants
Comparative metabolic profiling of the ΔeqbA, ΔeqbAE and ΔeqbHIJA strains was performed using LC-MS. Culture supernatants (50 ml) were extracted with ethyl acetate (3 × 50 ml) and both the combined organic extract and the residual culture supernatant were concentrated to dryness and re-dissolved in 20% acetonitrile/0.1% formic acid in water (1 ml). Each sample was split into two equal portions and ferric chloride was added to one portion of each sample to a final concentration of 5 mM. Fifty microlitres of each sample was analysed on an Agilent Zorbax C-18 reverse phase column (150 × 4.6 mm) connected to an Agilent 1100 HPLC instrument. The outflow was transferred via a splitter (90% to waste, 10% to mass spectrometer) to a Bruker HCT+ mass spectrometer, equipped with an electrospray source, operating in positive ion mode. The column was eluted as follows (mobile phase A: water with 0.1% formic acid, mobile phase B: acetonitrile with 0.1% formic acid): 100% mobile phase A for 5 min; a gradient from 100% mobile phase A to 100% mobile phase B over 25 min; 100% mobile phase B for 5 min; a gradient from 100% mobile phase B to 100% mobile phase A over 5 min; 100% mobile phase A for 5 min.
Chrome azurol S assay
Iron-chelating activity in supernatant samples was monitored by the chrome azurol S assay (Schwyn and Neilands, 1987). Bacteria were grown overnight in 10 ml of CDMs to stationary phase and then filter-sterilized through a 0.22 μm filter (Millipore) to recover supernatant. Supernatant (0.5 ml) was mixed with an equal volume of CAS assay solution and change in absorbance at 630 nm was measured over time. Desferoxamine (Sigma) was included as a standard.
Overexpression and purification of EqbA
The S. equi eqbA gene was amplified by PCR from 4047 chromosomal DNA with Phusion polymerase (NEB), primer ZM329 and primer ZM330 (Table S1). The PCR product was cloned into the BamHI/EcoRI sites of the pGEX-3X vector (GE Healthcare). The resulting construct, pGEX-eqbA, contains an N-terminal fusion of eqbA to a glutathione S-transferase (GST) tag driven by a tac promoter. DH10B E. coli cells harbouring the plasmid pGEX-eqbA were grown at 37°C with shaking (220 r.p.m.) in 2× YT containing 50 μg ml−1 ampicillin. Once the cells reached an optical density at 600 nm of 0.6, 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to the medium and the culture was incubated for 4 h at 28°C with shaking (220 r.p.m.). Cells were then harvested and lysed by sonication, and the GST–EqbA fusion was purified over glutathione sepharose 4B beads according to the supplier's protocol (GE Healthcare). Recombinant EqbA was cleaved from the beads by Factor Xa protease cleavage in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl and 1 mM CaCl2 according to the supplier's protocol (GE Healthcare). The purified EqbA protein appeared homogenous by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining.
Electrophoretic mobility shift assay
5′ Biotin end-labelled DNA fragments A, B and C containing the upstream region of the eqb operon were amplified from the S. equi 4047 chromosome using the primers indicated in Table S1. Fragment A contains the −237 to −11 bp region upstream of the eqb operon (Peqb), while fragments B and C contain regions −237 to −73 bp and −237 to −135 bp, respectively, and lack the DNA palindrome of Peqb (−73 to −38 bp). The PCR products were purified from a 1.5% agarose gel using a gel extraction kit (Qiagen). Binding reactions (20 μl) were carried out at room temperature for 20 min in 22.5 mM Tris-HCl (pH 7.5), 67.5 mM KCl, 5 mM MgCl2, 1.45 mM dithiothreiotol (DTT), 5% glycerol and 1 μg of dIdC. Biotin-labelled target-DNA (20 fmol) was mixed with 300 pmol of EqbA with or without the addition of 125 μM freshly prepared FeSO4, ZnSO4, MnCl2, CuSO4 or FeC6H5O7. To remove divalent metal ion from the purified EqbA, the protein was incubated with 2 mM EDTA for 2.5 h with mixing at room temperature and then buffer exchanged against 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM DTT using a vivaspin500 column (Sartorius). Samples were analysed by electrophoresis at 200 V on a 5.5% non-denaturing polyacrylamide gel containing 2.5% glycerol in 0.5× Tris-borate-EDTA buffer. Transfer to a positively charged nylon membrane (Roche) and the subsequent detection of biotin-labelled DNA by chemiluminescence were performed using the LightShift® Chemiluminescent EMSA Kit (Pierce) according to the manufacturer's instructions. Kit controls were used to analyse the binding of the Peqb fragment and EqbA to an alternative protein extract and target DNA respectively.
Reconstitution of the equibactin NRPS in E. coli
Novagen expression plasmids with compatible replicons were used for the coexpression of eqbBCD, eqbMN, eqbF, eqbG (multicystronic) and eqbE from T7 promoters in E. coli strain BL21(DE3). DNA fragments containing eqbBCD, eqbMN, eqbF, eqbG and eqbE genes were amplified by PCR from 4047 chromosomal DNA with Phusion polymerase (NEB), using primer pairs indicated in Table S1. eqbBCD was cloned into multiple cloning site (MCS)-1 of pACYCDuet-1 using NcoI/BamHI to generate pACYC-BCD. eqbMN was cloned into MCS-2 of pACYC-BCD using BglII/AatII to generate pACYC-BCD-MN (chloramphenicol resistant). eqbF was cloned into MCS-1 of pCDFDuet-1 using NcoI/BamHI to generate pCDF-F. eqbG was cloned into MCS-2 of pCDF-F using FseI/AatII to generate pCDF-F-G (spectinomycin resistant). eqbE was cloned into pET21a using NheI/BamHI to generate pET21a-E (ampicillin resistant).
pACYC-BCD-MN, pCDF-F-G and pET21a-E were introduced to BL21(DE3) via electroporation. The resultant E. coli strain was grown in Luria–Bertani (LB) broth media or a MM containing: 1.5 mg ml−1 KH2PO4, 4.34 mg ml−1 K2HPO4, 0.4 mg ml−1 (NH4)2SO4, 0.22 mg ml−1 MgSO4·7H2O, 5 mg ml−1 glucose, 24.5 mg ml−1 FeC6H5O7, 2.76 mg ml−1 ZnSO4·7H2O, 1 mg ml−1 CaCl2, 2 mg ml−1 Na2MoO4·2H2O, 1.21 mg ml−1 CuSO4 and 0.5 mg ml−1 H3BO3. Medium was supplemented with 50 μg ml−1 ampicillin, 50 μg ml−1 spectinomycin and 34 μg ml−1 chloramphenicol. Cultures were inoculated 4% (v/v) with a starter culture and growth was carried out at 37°C on a rotary shaker (220 r.p.m.) to an optical density at 600 nm between 0.6 and 0.8. IPTG (0.4 mM) was added together with 1 mM salicylate and the culture was incubated for a further 20 h at 28°C with shaking (220 r.p.m.). Supernatant was filter-sterilized and collected for LC-MS analysis or to determine its effect on streptonigrin sensitivity following cross-feeding to the ΔeqbAE strain as described above.
Additional combinations of plasmids were constructed in order to assess the contribution of EqbE, EqbG, EqbM and EqbN to the biosynthesis of the eqb NRPS product(s) in E. coli and also to provide appropriate negative controls for this system of production. eqbM and eqbN were cloned separately into MCS-2 of pACYC-BCD using BglII/AatII to generate pACYC-BCD-M and pACYC-BCD-N respectively (Table S1). Two E. coli strains lacking either EqbE or EqbG contained pACYC-BCD-MN, pCDF-F-G, pET21a or pACYC-BCD-MN, pCDF-F, pET21a-E respectively. Another two strains lacking EqbM or EqbN contained pACYC-BCD-N, pCDF-F-G, pET21a-E or pACYC-BCD-M, pCDF-F-G, pET21a-E respectively.
Acknowledgments
We thank Richard Newton for help with statistical analyses and Zehava Eichenbaum for kindly releasing the CDM. The Horse Trust funded the genome sequencing of S. equi strain 4047 and The Horserace Betting Levy Board funded the genome sequencing of S. zooepidemicus strain H70. This work was supported by the Animal Health Trust.
Supplementary material
References
- Alber J, El-Sayed A, Estoepangestie S, Lammler C, Zschock M. Dissemination of the superantigen encoding genes seeLseeMszeL and szeM in Streptococcus equi subsp. equi and Streptococcus equi subsp. zooepidemicus. Vet Microbiol. 2005;109:135–141. doi: 10.1016/j.vetmic.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Alice AF, Lopez CS, Crosa JH. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J Bacteriol. 2005;187:2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov R, Brem D, Heesemann J, Rakin A. Molecular mechanism of YbtA-mediated transcriptional regulation of divergent overlapping promoters ybtA and irp6 of Yersinia enterocolitica. FEMS Microbiol Lett. 2005a;250:27–32. doi: 10.1016/j.femsle.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Anisimov R, Brem D, Heesemann J, Rakin A. Transcriptional regulation of high pathogenicity island iron uptake genes by YbtA. Int J Med Microbiol. 2005b;295:19–28. doi: 10.1016/j.ijmm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Artiushin SC, Timoney JF, Sheoran AS, Muthupalani SK. Characterization and immunogenicity of pyrogenic mitogens SePE-H and SePE-I of Streptococcus equi. Microb Pathog. 2002;32:71–85. doi: 10.1006/mpat.2001.0482. [DOI] [PubMed] [Google Scholar]
- Beare PA, For RJ, Martin LW, Lamont IL. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol Microbiol. 2003;47:195–207. doi: 10.1046/j.1365-2958.2003.03288.x. [DOI] [PubMed] [Google Scholar]
- Berish SA, Subbarao S, Chen CY, Trees DL, Morse SA. Identification and cloning of a fur homolog from Neisseria gonorrhoeae. Infect Immun. 1993;61:4599–4606. doi: 10.1128/iai.61.11.4599-4606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov AG, Geoffroy VA, Perry RD. Yersiniabactin production requires the thioesterase domain of HMWP2 and YbtD, a putative phosphopantetheinylate transferase. Infect Immun. 2002;70:4204–4214. doi: 10.1128/IAI.70.8.4204-4214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JS, Holden DW. Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 2002;4:1149–1156. doi: 10.1016/s1286-4579(02)01640-4. [DOI] [PubMed] [Google Scholar]
- Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW. Characterization of pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect Immun. 2002;70:4389–4398. doi: 10.1128/IAI.70.8.4389-4398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guedon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002a;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, Guedon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002b;48:77–97. doi: 10.1016/s0147-619x(02)00102-6. [DOI] [PubMed] [Google Scholar]
- Butler AR, Bate N, Cundliffe E. Impact of thioesterase activity on tylosin biosynthesis in Streptomyces fradiae. Chem Biol. 1999;6:287–292. doi: 10.1016/S1074-5521(99)80074-X. [DOI] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Challis GL, Naismith JH. Structural aspects of non-ribosomal peptide biosynthesis. Curr Opin Struct Biol. 2004;14:748–756. doi: 10.1016/j.sbi.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL, Ravel J. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol Lett. 2000;187:111–114. doi: 10.1111/j.1574-6968.2000.tb09145.x. [DOI] [PubMed] [Google Scholar]
- Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Chou CJ, Wisedchaisri G, Monfeli RR, Oram DM, Holmes RK, Hol WG, Beeson C. Functional studies of the Mycobacterium tuberculosis iron-dependent regulator. J Biol Chem. 2004;279:53554–53561. doi: 10.1074/jbc.M407385200. [DOI] [PubMed] [Google Scholar]
- Clancy A, Loar JW, Speziali CD, Oberg M, Heinrichs DE, Rubens CE. Evidence for siderophore-dependent iron acquisition in group B streptococcus. Mol Microbiol. 2006;59:707–721. doi: 10.1111/j.1365-2958.2005.04974.x. [DOI] [PubMed] [Google Scholar]
- De Crecy-Lagard V, Blanc V, Gil P, Naudin L, Lorenzon S, Famechon A, et al. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis: molecular characterization of the first two structural peptide synthetase genes. J Bacteriol. 1997;179:705–713. doi: 10.1128/jb.179.3.705-713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Crecy-Lagard V, Marliere P, Saurin W. Multienzymatic non ribosomal peptide biosynthesis: identification of the functional domains catalysing peptide elongation and epimerisation. C R Acad Sci III. 1995;318:927–936. [PubMed] [Google Scholar]
- Du L, Sanchez C, Chen M, Edwards DJ, Shen B. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- Eichenbaum Z, Muller E, Morse SA, Scott JR. Acquisition of iron from host proteins by the group A streptococcus. Infect Immun. 1996;64:5428–5429. doi: 10.1128/iai.64.12.5428-5429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SL, Arceneaux JE, Byers BR, Martin ME, Aranha H. Ferrous iron transport in Streptococcus mutans. J Bacteriol. 1986;168:1096–1099. doi: 10.1128/jb.168.3.1096-1099.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Qi J, Tsuge T, Oba Y, Kobayashi T, Suzuki Y, et al. Construction of a bacterial artificial chromosome library for a myxobacterium of the genus Cystobacter and characterization of an antibiotic biosynthetic gene cluster. Biosci Biotechnol Biochem. 2005;69:1372–1380. doi: 10.1271/bbb.69.1372. [DOI] [PubMed] [Google Scholar]
- Fetherston JD, Bearden SW, Perry RD. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- Fetherston JD, Bertolino VJ, Perry RD. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol Microbiol. 1999;32:289–299. doi: 10.1046/j.1365-2958.1999.01348.x. [DOI] [PubMed] [Google Scholar]
- Flannagan SE, Zitzow LA, Su YA, Clewell DB. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- Franco AA. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J Bacteriol. 2004;186:6077–6092. doi: 10.1128/JB.186.18.6077-6092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone PP, Lyte M, Neal CP, Maggs AF, Haigh RD, Williams PH. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J Bacteriol. 2000;182:6091–6098. doi: 10.1128/jb.182.21.6091-6098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Timoney JF. Molecular analysis of the M protein of Streptococcus equi and cloning and expression of the M protein gene in Escherichia coli. Infect Immun. 1987;55:3181–3187. doi: 10.1128/iai.55.12.3181-3187.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier F, Taourit S, Glaser P, Courvalin P, Galimand M. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology. 2000;146(Part 6):1481–1489. doi: 10.1099/00221287-146-6-1481. [DOI] [PubMed] [Google Scholar]
- Gehring AM, Bradley KA, Walsh CT. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, et al. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998a;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- Gehring AM, Mori I, Perry RD, Walsh CT. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998b;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Guedon E, Helmann JD. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol Microbiol. 2003;48:495–506. doi: 10.1046/j.1365-2958.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- Hakansson A, Roche H, Mirza S, McDaniel LS, Brooks-Walter A, Briles DE. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect Immun. 2001;69:3372–3381. doi: 10.1128/IAI.69.5.3372-3381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A, Robinson C, Sutcliffe IC, Slater J, Maskell DJ, Davis-Poynter N, et al. Mutation of the maturase lipoprotein attenuates the virulence of Streptococcus equi to a greater extent than does loss of general lipoprotein lipidation. Infect Immun. 2006;74:6907–6919. doi: 10.1128/IAI.01116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks TS, Liu M, McClure MJ, Lei B. ABC transporter FtsABCD of Streptococcus pyogenes mediates uptake of ferric ferrichrome. BMC Microbiol. 2005;5:62. doi: 10.1186/1471-2180-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulczyk R, Pallon J, Bjorck L. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol Microbiol. 1999;34:596–606. doi: 10.1046/j.1365-2958.1999.01626.x. [DOI] [PubMed] [Google Scholar]
- Janulczyk R, Ricci S, Bjorck L. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun. 2003;71:2656–2664. doi: 10.1128/IAI.71.5.2656-2664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorm LR, Love DN, Bailey GD, McKay GM, Briscoe DA. Genetic structure of populations of beta-haemolytic Lancefield group C streptococci from horses and their association with disease. Res Vet Sci. 1994;57:292–299. doi: 10.1016/0034-5288(94)90120-1. [DOI] [PubMed] [Google Scholar]
- Keating TA, Miller DA, Walsh CT. Expression, purification, and characterization of HMWP2, a 229 kDa, six domain protein subunit of Yersiniabactin synthetase. Biochemistry. 2000;39:4729–4739. doi: 10.1021/bi992923g. [DOI] [PubMed] [Google Scholar]
- Kelly C, Bugg M, Robinson C, Mitchell Z, Davis-Poynter N, Newton JR, et al. Sequence variation of the SeM gene of Streptococcus equi allows discrimination of the source of strangles outbreaks. J Clin Microbiol. 2006;44:480–486. doi: 10.1128/JCM.44.2.480-486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konz D, Klens A, Schorgendorfer K, Marahiel MA. The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: molecular characterization of three multi-modular peptide synthetases. Chem Biol. 1997;4:927–937. doi: 10.1016/s1074-5521(97)90301-x. [DOI] [PubMed] [Google Scholar]
- Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- Lei B, Smoot LM, Menning HM, Voyich JM, Kala SV, Deleo FR, et al. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect Immun. 2002;70:4494–4500. doi: 10.1128/IAI.70.8.4494-4500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Bugg TD. Investigation of a general base mechanism for ester hydrolysis in C–C hydrolase enzymes of the alpha/beta-hydrolase superfamily: a novel mechanism for the serine catalytic triad. Org Biomol Chem. 2007;5:507–513. doi: 10.1039/b615605c. [DOI] [PubMed] [Google Scholar]
- Lindmark H, Jonsson P, Engvall E, Guss B. Pulsed-field gel electrophoresis and distribution of the genes zag and fnz in isolates of Streptococcus equi. Res Vet Sci. 1999;66:93–99. doi: 10.1053/rvsc.1998.0250. [DOI] [PubMed] [Google Scholar]
- Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci USA. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L, Gonzalez N, Jagdeep S, Nguyen-Ngoc T, Reimmann C. PchR-box recognition by the AraC-type regulator PchR of Pseudomonas aeruginosa requires the siderophore pyochelin as an effector. Mol Microbiol. 2005;58:495–509. doi: 10.1111/j.1365-2958.2005.04837.x. [DOI] [PubMed] [Google Scholar]
- Michel L, Bachelard A, Reimmann C. Ferripyochelin uptake genes are involved in pyochelin-mediated signalling in Pseudomonas aeruginosa. Microbiology. 2007;153:1508–1518. doi: 10.1099/mic.0.2006/002915-0. [DOI] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- Newton JR, Wood JL, Dunn KA, DeBrauwere MN, Chanter N. Naturally occurring persistent and asymptomatic infection of the guttural pouches of horses with Streptococcus equi. Vet Rec. 1997;140:84–90. doi: 10.1136/vr.140.4.84. [DOI] [PubMed] [Google Scholar]
- Newton JR, Verheyen K, Talbot NC, Timoney JF, Wood JL, Lakhani KH, Chanter N. Control of strangles outbreaks by isolation of guttural pouch carriers identified using PCR and culture of Streptococcus equi. Equine Vet J. 2000;32:515–526. doi: 10.2746/042516400777584721. [DOI] [PubMed] [Google Scholar]
- Newton JR, Laxton R, Wood JL, Chanter N. Molecular epidemiology of Streptococcus zooepidemicus infection in naturally occurring equine respiratory disease. Vet J. 2008;175:338–345. doi: 10.1016/j.tvjl.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Nygaard TK, Liu M, McClure MJ, Lei B. Identification and characterization of the heme-binding proteins SeShp and SeHtsA of Streptococcus equi subspecies equi. BMC Microbiol. 2006;6:82. doi: 10.1186/1471-2180-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram DM, Must LM, Spinler JK, Twiddy EM, Holmes RK. Analysis of truncated variants of the iron dependent transcriptional regulators from Corynebacterium diphtheriae and Mycobacterium tuberculosis. FEMS Microbiol Lett. 2005;243:1–8. doi: 10.1016/j.femsle.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Patel HM, Walsh CT. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry. 2001;40:9023–9031. doi: 10.1021/bi010519n. [DOI] [PubMed] [Google Scholar]
- Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Balbo PB, Jones HA, Fetherston JD, DeMoll E. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology. 1999;145(Part 5):1181–1190. doi: 10.1099/13500872-145-5-1181. [DOI] [PubMed] [Google Scholar]
- Pohl E, Holmes RK, Hol WG. Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied. J Mol Biol. 1999a;285:1145–1156. doi: 10.1006/jmbi.1998.2339. [DOI] [PubMed] [Google Scholar]
- Pohl E, Holmes RK, Hol WG. Crystal structure of a cobalt-activated diphtheria toxin repressor–DNA complex reveals a metal-binding SH3-like domain. J Mol Biol. 1999b;292:653–667. doi: 10.1006/jmbi.1999.3073. [DOI] [PubMed] [Google Scholar]
- Pramanik A, Braun V. Albomycin uptake via a ferric hydroxamate transport system of Streptococcus pneumoniae R6. J Bacteriol. 2006;188:3878–3886. doi: 10.1128/JB.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T, Webb PD, Handley V, Fraser JD. Two novel superantigens found in both group A and group C Streptococcus. Infect Immun. 2003;71:1361–1369. doi: 10.1128/IAI.71.3.1361-1369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri LE, Keating TA, Patel HM, Walsh CT. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4, 2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry. 1999;38:14941–14954. doi: 10.1021/bi991787c. [DOI] [PubMed] [Google Scholar]
- Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- Reimmann C, Patel HM, Walsh CT, Haas D. PchC thioesterase optimizes nonribosomal biosynthesis of the peptide siderophore pyochelin in Pseudomonas aeruginosa. J Bacteriol. 2004;186:6367–6373. doi: 10.1128/JB.186.19.6367-6373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Johanesen PA, Lyras D, Mullany P, Rood JI. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology. 2001;147:1243–1251. doi: 10.1099/00221287-147-5-1243. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Smith I. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J Bacteriol. 2006;188:424–430. doi: 10.1128/JB.188.2.424-430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin W, Hofnung M, Dassa E. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol. 1999;48:22–41. doi: 10.1007/pl00006442. [DOI] [PubMed] [Google Scholar]
- Schmitt MP, Holmes RK. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect Immun. 1991;59:3903–3908. doi: 10.1128/iai.59.11.3903-3908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MP, Predich M, Doukhan L, Smith I, Holmes RK. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect Immun. 1995;63:4284–4289. doi: 10.1128/iai.63.11.4284-4289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Marahiel MA. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch Microbiol. 1998;169:404–410. doi: 10.1007/s002030050590. [DOI] [PubMed] [Google Scholar]
- Schubert S, Dufke S, Sorsa J, Heesemann J. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol Microbiol. 2004;51:837–848. doi: 10.1046/j.1365-2958.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- Seedorf H, Fricke WF, Veith B, Bruggemann H, Liesegang H, Strittmatter A, et al. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci USA. 2008;105:2128–2133. doi: 10.1073/pnas.0711093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino L, Reimmann C, Baur H, Beyeler M, Visca P, Haas D. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol Gen Genet. 1995;249:217–228. doi: 10.1007/BF00290369. [DOI] [PubMed] [Google Scholar]
- Silakowski B, Schairer HU, Ehret H, Kunze B, Weinig S, Nordsiek G, et al. New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J Biol Chem. 1999;274:37391–37399. doi: 10.1074/jbc.274.52.37391. [DOI] [PubMed] [Google Scholar]
- Silakowski B, Kunze B, Nordsiek G, Blocker H, Hofle G, Muller R. The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur J Biochem. 2000;267:6476–6485. doi: 10.1046/j.1432-1327.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- Timoney JF, Artiushin SC, Boschwitz JS. Comparison of the sequences and functions of Streptococcus equi M-like proteins SeM and SzPSe. Infect Immun. 1997;65:3600–3605. doi: 10.1128/iai.65.9.3600-3605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky ME, Actis LA, Crosa JH. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect Immun. 1993;61:3228–3233. doi: 10.1128/iai.61.8.3228-3233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky ME, Wertheimer AM, Actis LA, Crosa JH. Characterization of the Vibrio anguillarum fur gene: role in regulation of expression of the FatA outer membrane protein and catechols. J Bacteriol. 1994;176:213–220. doi: 10.1128/jb.176.1.213-220.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A, Merlin C. Mobile elements as a combination of functional modules. Plasmid. 2002;47:26–35. doi: 10.1006/plas.2001.1552. [DOI] [PubMed] [Google Scholar]
- Venturi V, Ottevanger C, Bracke M, Weisbeek P. Iron regulation of siderophore biosynthesis and transport in Pseudomonas putida WCS358: involvement of a transcriptional activator and of the Fur protein. Mol Microbiol. 1995;15:1081–1093. doi: 10.1111/j.1365-2958.1995.tb02283.x. [DOI] [PubMed] [Google Scholar]
- Visca P, Colotti G, Serino L, Verzili D, Orsi N, Chiancone E. Metal regulation of siderophore synthesis in Pseudomonas aeruginosa and functional effects of siderophore-metal complexes. Appl Environ Microbiol. 1992;58:2886–2893. doi: 10.1128/aem.58.9.2886-2893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller AS, Jolley KA. Getting a grip on strangles: recent progress towards improved diagnostics and vaccines. Vet J. 2007;173:492–501. doi: 10.1016/j.tvjl.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Wang H, Roberts AP, Lyras D, Rood JI, Wilks M, Mullany P. Characterization of the ends and target sites of the novel conjugative transposon Tn5397 from Clostridium difficile: excision and circularization is mediated by the large resolvase, TndX. J Bacteriol. 2000;182:3775–3783. doi: 10.1128/jb.182.13.3775-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb K, Jolley KA, Mitchell Z, Robinson C, Newton JR, Maiden MCJ, Waller A. Development of an unambiguous and discriminatory multilocus sequence typing scheme for the Streptococcus zooepidemicus group. Microbiology. 2008;154:3016–3024. doi: 10.1099/mic.0.2008/018911-0. [DOI] [PubMed] [Google Scholar]
- Wood JL, Newton JR, Chanter N, Mumford JA. Association between respiratory disease and bacterial and viral infections in British racehorses. J Clin Microbiol. 2005;43:120–126. doi: 10.1128/JCM.43.1.120-126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge KG, Williams PH. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Wyckoff EE, Stoebner JA, Reed KE, Payne SM. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


