Abstract
Overcoming antigenic variation is one of the major challenges in the development of an effective vaccine against Plasmodium falciparum, a causative agent of human malaria. Inclusion of multiple antigen variants in subunit vaccine candidates is one strategy that has aimed to overcome this problem for the leading blood-stage malaria vaccine targets, merozoite surface protein 1 (MSP1) and apical membrane antigen 1 (AMA1). However previous studies, utilizing malaria antigens, have concluded that inclusion of multiple allelic variants, encoding altered peptide ligands (APL), in such a vaccine may be detrimental to both the priming and in vivo re-stimulation of antigen-experienced T cells. Here we analyze the T cell responses to two alleles of MSP1 and AMA1 induced by vaccination of malaria-naïve adult volunteers with bi-valent viral vectored vaccine candidates. We show a significant bias to the 3D7/MAD20 allele compared to the Wellcome allele for the 33kDa region of MSP1, but not for the 19kDa fragment or the AMA1 antigen. Whilst this bias could be caused by ‘immune interference’ at priming, the data don’t support a significant role for ‘immune antagonism’ during memory T cell re-stimulation, despite observation of the latter at a minimal epitope level in vitro. A lack of class I HLA epitopes in the Wellcome allele that are recognized by vaccinated volunteers may in fact contribute to the observed bias. We also show that controlled infection with 3D7 strain P. falciparum parasites neither boosts existing 3D7-specific T cell responses nor appears to ‘immune divert’ cellular responses towards the Wellcome allele.
INTRODUCTION
The development of highly effective cross-strain immunity against infectious pathogens remains the universal goal for all vaccine developers. This is no less true in the case of the apicomplexan parasite Plasmodium falciparum – the causative agent of the most severe and deadly form of human malaria. Like most difficult infectious pathogens, the high level of antigenic variation and polymorphism displayed by this parasite in endemic areas frequently poses a huge challenge in the context of effective subunit vaccine development (1, 2).
Antigens expressed during the blood-stage parasite infection, such as merozoite surface protein 1 (MSP1) (3) and apical membrane antigen 1 (AMA1) (4), remain leading targets for inclusion in subunit vaccine candidates. These two antigens have been associated with protective immunity in naturally-exposed individuals (5-7), as well as proving efficacious in pre-clinical vaccine studies of mice (8-10) and non-human primates (11-14). Although protective blood-stage immunity has been widely associated with antibody responses, a growing body of evidence in both animal and human studies supports a contributing role for cellular immunity (15, 16). Although infected erythrocytes lack MHC molecules with which to present parasite-derived peptides, it is thought that effector CD4+ T cells can enhance clearance of opsonized parasitized red blood cells (pRBCs) by macrophages in the spleen; orchestrate the induction of parasiticidal pro-inflammatory serum cytokine responses; and/or provide polarizing help for B cells leading to the induction of protective cytophilic IgG subclasses that may better interact with innate cellular effectors such as monocytes or neutrophils (17, 18). CD8+ T cells have been shown to be protective in the P. yoelii mouse model at both the liver-stage (19, 20) and blood-stage of malaria infection (21). CD8+ T cells specific for blood-stage antigens potentially target merozoite derived antigens during the late stages of pre-erythrocytic parasite development within infected hepatocytes (20).
We have thus aimed to develop clinically-relevant subunit vaccine delivery platforms that are capable of inducing antibody responses against the transgene of interest in conjunction with strong cellular immunity (22, 23), and in a recent series of Phase I/IIa clinical trials in Oxford, UK we have shown that viral vectored delivery of the MSP1 and AMA1 blood-stage malaria antigens can achieve this goal (24, 25). In these trials, the two antigens were separately delivered utilizing a heterologous prime-boost immunization regime consisting of a priming vaccination with a recombinant replication-deficient chimpanzee adenovirus serotype 63 (ChAd63) vector, followed eight weeks later by a boosting vaccination with an attenuated modified vaccinia virus Ankara (MVA) vector recombinant for the same antigen (Figure S1). This regime was shown to be safe and immunogenic for both antibody and T cell responses in healthy adult human volunteers and, when the vectors for both antigens were co-administered, sterilizing efficacy was observed in 1/9 individuals against controlled human malaria infection (CHMI) with vaccine homologous P. falciparum 3D7 strain sporozoites (26).
Importantly, antibodies against MSP1 and AMA1 have been shown to elicit vaccine-strain specific efficacy in non-human primate studies (12, 14) as well as most recently humans, in the case of a monovalent 3D7 strain AMA1 protein/adjuvant vaccine tested in Malian children (27). Attempts to address this issue of antigenic polymorphism have involved the development of multivalent vaccine formulations containing multiple allelic variants of the MSP1 or AMA1 target antigen (28-30) or artificial diversity covering consensus sequences (31). Similarly, both of the viral vectored vaccine transgene inserts had been previously designed to address the issues surrounding target antigen polymorphism by encoding bi-allelic vaccine inserts for AMA1 (32, 33) and MSP1 (34, 35). Although these strategies aim to confront the difficulties surrounding the induction of cross-strain humoral immunity, other reports have raised important concerns about this approach in the context of T cell immunity (36). Such studies utilizing malaria antigens have concluded that inclusion of multiple allelic variants in a vaccine may be detrimental to both the priming and in vivo re-stimulation of antigen-experienced T cells (37, 38). These immunological studies arose from questions relating to the population dynamics of natural P. falciparum malaria infection, host-parasite co-evolution and how allelic dimorphisms are maintained.
Although the population of P. falciparum parasite strains circulating in an endemic population will be influenced by host human leukocyte antigen (HLA) type and other genetic factors, studies of parasite populations have also led to questions as to whether parasite strain co-habitation within hosts can affect T cell responses. A previous study has described how IFN-γ responses to allele-specific CD4+ T cell epitopes in MSP1 failed to correlate with differential antigenic exposure in The Gambia (39). A follow-up study demonstrated that allelic altered peptide ligand (APL) T cell epitopes of MSP1 mutually inhibited IFN-γ secretion of CD4+ T cells in naturally-exposed Gambian volunteers, and that the same variant epitopes were also able to impair priming of T cells from malaria naïve individuals (38). This observed effect of priming bias termed ‘immune interference’ (37) is dependent on the presentation of APLs on the same antigen presenting cell (APC). This effect is distinguished from effector level ‘immune antagonism’ which describes the phenomenon whereby the simultaneous presence of an APL epitope pair causes a significantly reduced T cell recall response compared to that observed in the presence of one epitope alone, and is not dependant on presentation of APLs on the same APC (36). Together these mechanisms could provide a significant in vivo immune evasion mechanism and thus facilitate survival of co-habiting parasites that bear such antagonistic allelic epitope regions. Gilbert et al. (36) proposed a mathematical model for co-habitation by such mechanisms and in a follow up study (37) showed evidence for immune interference in the dimorphic CD8+ cytotoxic T lymphocyte (CTL) epitopes, cp26 and cp29, from the liver-stage circumsporozoite protein (CSP). Moreover, they also described ‘immune diversion’ a mechanism whereby after priming CTLs to one epitope, exposure to the second APL variant elicited a narrow response to the original epitope and reduced cross-reactivity (37). This phenomenon is also observed in human immunodeficiency virus (HIV) (40) and hepatitis C virus (HCV) infection (41). Consequently, the identification of such immune evasionary mechanisms led to the concern that attempts to address the well-recognized problem of antigenic polymorphism, by including bi-allelic sequences of P. falciparum antigens in the same vaccine, may be more detrimental than beneficial for both the in vivo priming and boosting of broad and effective T cell responses.
Until now, there has been no bi-valent blood-stage malaria vaccine tested in clinical trials that was designed to induce strong T cell responses. Here we analyze the vaccine-induced CD8+ and CD4+ T cell responses to both alleles of MSP1 and AMA1 delivered by the viral vectored vaccines in malaria-naïve adult volunteers. We show that whilst there is a significant bias towards the 3D7/MAD20 allele (over the Wellcome allele) in terms of T cell responses to the dimorphic 33kDa region of MSP1 (MSP133), this is not observed for the 19kDa region of MSP1 (MSP119) or for the AMA1 antigen. Although the bias to a stronger 3D7 allele MSP133-specific response could be due to ‘immune interference’, we offer an alternative explanation that limited recognition of class I HLA epitopes in the Wellcome allele may result in similar observations. We also confirm that ‘immune antagonism’ does occur at the minimal epitope level in vitro, as previously shown for the MSP1 antigen (38, 39), but we conclude these effects were too small to influence the overall antigen-specific responses as measured by ex-vivo IFN-γ ELISPOT assay. Similarly, exposure of volunteers to infectious challenge with 3D7 strain P. falciparum parasites does not appear to ‘immune divert’ cellular responses towards the Wellcome allele, although interestingly we also observed no apparent boosting of responses post-exposure. Overall, the data suggest host HLA type may play a prominent role in determining the magnitude of measured T cell responses to different alleles within these bi-valent blood-stage malaria vaccine candidates and should be taken into consideration for immuno-monitoring in future studies.
MATERIALS & METHODS
Immunization Groups and Peripheral Blood Mononuclear Cells (PBMCs)
Frozen PBMC samples were used throughout this study and were obtained from Phase Ia safety and immunogenicity clinical trials for the MSP1 (24) and AMA1 (25) candidate vaccines, as well as a Phase IIa efficacy study where immunized volunteers underwent CHMI with vaccine homologous P. falciparum 3D7 strain sporozoites delivered by mosquito bite (Figure S1)(26). In all cases, the two antigens were separately delivered by a heterologous prime-boost immunization regime consisting of a priming intramuscular (i.m.) vaccination with a recombinant replication-deficient ChAd63 vector (doses: 5 × 109 – 5 × 1010 viral particles (vp)), followed eight weeks later by a boosting vaccination i.m. with MVA vector (doses: 1.25 – 5 × 108 plaque forming units (pfu)) recombinant for the same antigen. All necessary regulatory and ethical approvals were granted as previously described (24-26), and the trials were registered with ClinicalTrials.gov. All volunteers gave written informed consent prior to participation, and the studies were conducted according to the principles of the Declaration of Helsinki and in accordance with Good Clinical Practice (GCP). All volunteers participating in these clinical trials gave permission for samples to be used for exploratory immunology analysis and HLA typing (Transplant, Immunology & Immunogenetics Department, Churchill Hospital, Oxford NHS, UK). PBMC samples from the trials were all prepared and frozen as previously described (24).
Antigens and Peptides
The composition of the bi-allelic vaccine inserts for MSP1 (34, 35) and AMA1 (32, 33) used in both the ChAd63 and MVA vaccine vectors have been previously described. In the case of AMA1, a bivalent transgene was optimized to consist of the 3D7 and FVO strain alleles fused in tandem; whilst for MSP1 an insert was designed comprising both the 3D7/MAD20 and Wellcome alleles of the dimorphic 42kDa C-terminal region (MSP142 / sequence blocks 16+17) fused in tandem and preceded by the naturally conserved regions of MSP1 sequence (blocks 1, 3, 5 and 12)(35). The MSP142 region is composed of an N-terminal 33kDa region (MSP133, block 16) followed by a C-terminal 19kDa region (MSP119, block 17). Peptides were designed as 20mers overlapping by 10 amino acids (αα) to cover the entire sequence of each transgene insert, and were pooled according to sequence block for the MSP1 insert, or by whether they were 3D7 allele-specific, FVO allele-specific or common to both in the case of AMA1, all as previously described (24, 25). All new peptides used for the studies here are listed in Table I. The epitope prediction software ‘SBS EpiToolKit’ (http://www.epitoolkit.org/epipred) was used to select the minimal epitopes described in Table II. All new peptides were purchased from Peptide Protein Research Ltd., UK, at a purity of >75%. Peptides were reconstituted in 100% DMSO at 100-200mg/mL and then used to create working peptide pool stocks of 10μg/mL for ELISPOT assay (2x final concentration).
Table I. Amino acid sequences and allelic identity of peptides used to study in vitro antagonism.
For the ELISPOT assays, two MSP1 derived peptides, M7* (3D7) and M8* (Wellcome) were synthesized. These peptides are identical to those used by Lee et al. (M7, M8) and were described as non-conserved sequenced paired APL antagonists (38). These epitopes were present in two of the 20mer peptides previously used in the MSP1 vaccine clinical trials (24): M92 (containing M7*) and M53 (containing M8* with one αα change, Y-V indicated in bold). Additionally two more peptides from this trial M85 and M48 were used. These were similarly dimorphic and contained African epitopes shown to be immunogenic (39) but have not previously been tested for antagonism. For both pairs, the 3D7 or Wellcome peptides were made up individually and in a combined pool, with each peptide at a concentration of 10μg/mL. Final concentration in the ELISPOT assay was 5μg/mL.
| Peptide Name | Strain | Sequence |
|---|---|---|
| M7* | 3D7 | D Y L I N L K A K I N D C |
| M8* | Wellcome | L F V I H L E A K V L N V |
| M92 | 3D7 | L V N K I D D Y L I N L K A K I N D C N |
| M53 | Wellcome | N D K I D L F V I H L E A K V L N Y T Y |
| M85 | 3D7 | F A Q E G I S Y Y E K V L A K Y K D D L |
| M48 | Wellcome | A N D V L G Y Y K I L S E K Y K S D L D |
Table II. Amino acid sequences of predicted CD8+ T cell minimal epitopes.
Twelve peptides representing minimal epitopes from the Wellcome MSP133 sequence were designed based on the epitope prediction software ‘SBS EpiToolKit’. These minimal epitopes are unique to the Wellcome strain and are not present in the 3D7 sequence. These were predicted to bind different HLA-B alleles possessed by the trial volunteers, with strong responses predicted for minimal epitopes predicted to bind to HLA-B*1801. All epitopes are also found in 20mer trial peptides (located in the trial peptide pool 33a previously described (24)). The amino acid sequence of the Wellcome MSP133 fragment is also shown in Figure 7C.
| Peptide Name | Predicted HLA-B association |
8-mer Sequence | 20-mer trial peptide containing 8-mer |
Trial peptide pool |
|---|---|---|---|---|
| Well33a1 | 0702 | T P S V I D N I L | M36 | 33a |
| Well33a2 | 1801 | I E N E Y E V L | M37 | 33a |
| Well33a3 | 1801 | N E Y E V L Y L | M37 | 33a |
| Well33a4 | 1801 | Y E V L Y L K P | M37 & M38 | 33a |
| Well33a5 | 1801 | L E N N V M T F | M39& M40 | 33a |
| Well33a6 | 1801 | R E N F K N V L | M42 | 33a |
| Well33a7 | 1801 | L E S D L I P Y | M42 & M43 | 33a |
| Well33a8 | 5101 | E A V T P S V I | M36 | 33a |
| Well33a9 | 5101 | T P S V I D N I | M36 | 33a |
| Well33a10 | 5101 | L A G V Y R S L | M38 & M39 | 33a |
| Well33a11 | 0801 | Y R S L K K Q L | M39 | 33a |
| Well33a12 | 0801 | N V N V K D I L | M40 | 33a |
Ex-vivo IFN-γ ELISPOT Assay
The ex-vivo IFN-γ ELISPOT was performed as previously described (24, 25). However instead of using freshly isolated cells, frozen PBMC were thawed in a water bath at 37°C and immediately transferred into warm medium. Cells were washed twice in medium, before incubation in medium containing 1μL/mL benzonase (25units/μL) (Sigma-Aldrich) for a minimum of 1h, then counted and resuspended at 5×106/mL. 50μL of cells were added to the relevant wells of the ELISPOT plate to give 250,000 cells per well. Cells were re-stimulated in duplicate with 50μL of the relevant peptide(s) (final concentration 5μg/mL of each peptide), with a media/DMSO unstimulated negative control, and a PHA/SEB positive control before incubation and development, all as previously described (24, 25). Results are expressed as IFN-γ spot-forming units (SFU) per million PBMC. Background responses in unstimulated control wells were almost always less than 20 spots, and were subtracted from those measured in peptide-stimulated wells.
CD4+ and CD8+ T cell Depletion ELISPOT Assay
CD4+ and CD8+ T cell depletions were performed according to the manufacturer’s instructions for the MACS magnetic column system, with human CD4 and CD8 microbeads (Miltenyi Biotec, UK). Typically 10 million PBMC were thawed as previously described and divided equally to generate three samples (~3 million cells/sample after losses): i) Undepleted; ii) CD4+ cells (CD8+ cell depleted) and iii) CD8+ cells (CD4+ cell depleted). Microbeads were added to specifically bind to the fraction intended for depletion and then incubated before application to the magnetic column. Labeled (unwanted) cells are attracted to the magnetic field trapping them within the column whilst the remaining cells pass through. The flow-through fraction was then resuspended to the original volume and a standard ex-vivo IFN-γ ELISPOT assay performed, whilst the depleted cells were discarded. A subset of samples was also analyzed by flow cytometry to assess the efficiency of depletion. Cells were extracellularly stained using anti-human CD3 FITC (clone UCHT1), anti-human CD4 PerCP-Cy5 (clone OKT4) and anti-human CD8α APC (clone RPA-T8) antibodies (eBioscience) before analysis using a LSRII Flow Cytometer (BD Biosciences, Franklin Lakes, NJ). Cells were gated by Lymphocytes/Singlets/CD3+ before gating on CD4+ and CD8+. Depletion efficiency was >98% for CD4+ and >99% for CD8+ (Figure S2).
Statistical Analysis
Data were analyzed using GraphPad Prism version 5.03 for Windows (GraphPad Software Inc., California, USA). Wilcoxon matched-pairs signed rank tests were carried out to compare responses to pairs of alleles. A P value >0.05 was considered significant in all cases.
RESULTS
Human vaccination with bi-allelic AMA1 and MSP1 vaccines induces IFN-γ T cell responses to both alleles
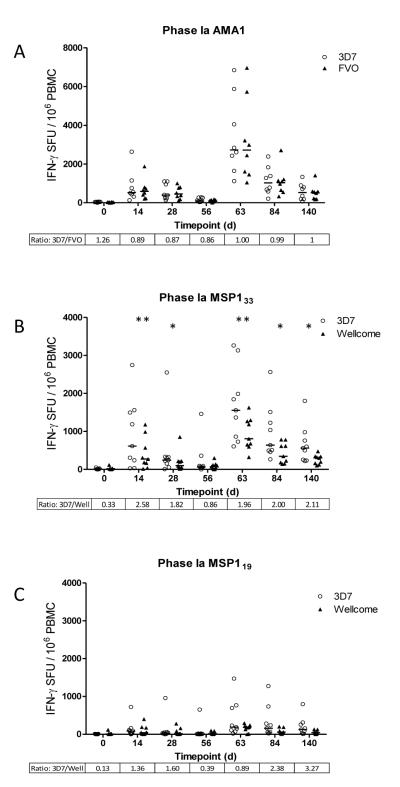
Previous data have suggested that co-habiting P. falciparum parasites could interfere with the priming of T cell responses by the phenomenon of ‘immune interference’ as well as re-stimulation of memory responses by ‘immune antagonism’, whereby APL epitopes present in dimorphic antigens antagonize responses to each other (38). This led to concerns about the ability to prime and boost cellular immune responses with vaccines encoding more than one allele of an antigen. In this study, healthy malaria-naïve adult volunteers were immunized in a heterologous prime-boost immunization regime with viral vectored vaccines encoding bi-allelic inserts for the MSP142 or AMA1 blood-stage malaria antigens (Figure S1). Initially total IFN-γ producing T cell responses were assessed to each allele by ex-vivo IFN-γ ELISPOT assay. AMA1-specific responses were calculated by summing the response to pools containing peptides unique to each individual allele (3D7 or FVO) with the response to pools containing peptides that were conserved between the two alleles. We have previously reported that the responses at the peak time-points (day (d) 14 after the ChAd63 prime and d63 after the MVA boost) are broad and spread over each of the ten peptide pools assayed (25), however the C-terminal domain (also included in the vaccine construct and unique to the FVO sequence (32, 33) was excluded from the analysis reported here. Figure 1A shows the allele-specific responses for all time-points assayed in the Phase Ia study. There were no significant differences between overall responses to the 3D7 or FVO alleles of AMA1 by a Wilcoxon matched-pairs signed rank test at all time-points measured, including those measured after the ChAd63 prime (d0 – d56) and after the MVA boost (d63 – d140).
Figure 1.
Comparison of allele specific IFN-γ T cell responses to 3D7 (open circles) and FVO/Wellcome (closed triangles) AMA1 and MSP1 in volunteers at key time-points during two independent Phase Ia clinical trials of bi-allelic AMA1 and MSP1 vaccines. Graphs represent the individual response and group median for T cell responses, expressed as IFN-γ SFU/106 PBMC summed from pools containing overlapping peptides from the vaccine insert. (A) AMA1 allele specific responses calculated by adding the allele specific response (3D7 or FVO) to the common pool response for each individual. MSP133 (B) and MSP119 (C) fragment specific responses for each allele (3D7 or Wellcome) calculated by adding responses from relevant peptide pools. Day 0 calculations are excluded from statistical analysis. Ratios for the two median responses are also included (>1 = stronger 3D7 response). *P = <0.05, **P = <0.01.
MSP1 allele-specific IFN-γ T cell responses were also calculated by summing the response to pools containing peptides specific to each individual allele (3D7 or Wellcome) of MSP133 and MSP119 contained within the vaccine antigen. Similar to AMA1, we have also previously reported that the responses at the peak time-points (d14 after the ChAd63 prime and d63 after the MVA boost) are broad and spread over each of the 12 peptide pools assayed (24). Here we show the allele-specific responses for all time-points assayed in the Phase Ia study for both MSP133 (Figure 1B) and MSP119 (Figure 1C). In the case of the MSP133 region, significant differences were observed between the responses to the 3D7 and Wellcome alleles in a Wilcoxon matched-pairs signed rank test for all time points after the priming and boosting vaccinations except d0 and d56, with 3D7 being the dominant allele as reflected in the 3D7:Wellcome ratio. At the peak response post-prime (d14) and post-boost (d63) the differences were most significant. Responses to MSP119 were, however, comparable between the two alleles, with no significant differences observed. It should be noted that the MSP119 3D7 pool contains three more peptides than its Wellcome equivalent (24). These three peptide sequences are duplicated in the Wellcome sequence and therefore were excluded in the second pool to avoid duplication of the response. It is highly unlikely that these peptides contain strong epitopes given the lack of significant differences between the responses to the two pools. These responses to MSP119 were also of much weaker magnitude in comparison to MSP133 (in agreement with the smaller size of this domain) but were not absent, as might be expected from preclinical data suggesting this conformational region of MSP1 (34) is refractory to antigen processing and presentation to T cells (42, 43). Overall, these data confirm that it is possible to both prime and boost T cell responses to each component allele encoded in the bi-allelic vaccines, and only in the case of MSP133 was a dominant allele noted.
Infectious challenge with 3D7 clone parasites does not divert T cell responses
Previous data from studies of malaria, HCV and HIV have also described the phenomenon of ‘immune diversion’ – a mechanism whereby after priming to one epitope, exposure to a second APL variant elicited a narrow response to the original epitope and reduced cross-reactivity. We explored this effect in the context of a CHMI study, whereby immunized volunteers were experimentally exposed to five mosquito bites harboring infectious 3D7 clone P. falciparum sporozoites (26, 44). All the volunteers immunized with the MSP1 or AMA1 vaccines alone developed blood-stage malaria infection, and were treated at the time of patency as diagnosed by thin blood film microscopy (26). Volunteers had thus been immunized with bi-allelic T cell-inducing vaccines followed by subsequent exposure to a single malaria parasite clone (3D7 in this case). Total IFN-γ producing T cell responses were again assessed to each allele by ex-vivo ELISPOT assay before and after malaria exposure.
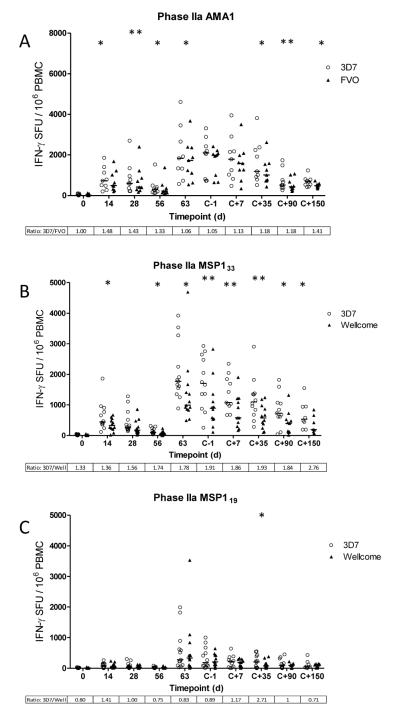
As before, the allele-specific responses are shown for all time-points assayed in the Phase IIa efficacy study (Figure 2). In contrast to the Phase Ia trial (Figure 1A), small but significant differences were observed between the 3D7 and FVO AMA1-specific responses by Wilcoxon matched-pairs signed rank test after the ChAd63 prime (d0 – d56), although this difference was less pronounced following MVA boost and prior to malaria infection (d63 – dC-1) (Figure 2A). Following malaria sporozoite infection, blood-stage merozoites are predicted to rupture out of the liver and enter the blood on days 6.5-7 post-challenge (dC+6.5 and dC+7). Median time to patent parasitemia was 10.8 days, for both MSP1 and AMA1 vaccinated volunteers compared to 9.7 for the controls (26). While PBMC samples assayed up to 150 days post-challenge (dC+150) showed some small but significant differences between the 3D7 and FVO allele-specific responses, there was no significant boosting of 3D7 AMA1-specific T cell responses. Overall the median ratio of responses between 3D7 and FVO AMA1 remained close to 1, and the significant differences observed were primarily due to the paired analysis and slightly stronger 3D7 responses in the case of most individuals.
Figure 2.
Comparison of allele specific IFN-γ T cell responses to 3D7 (open circles) and FVO/Wellcome (closed triangles) AMA1 and MSP1 in volunteers at key time points during a Phase IIa clinical trial of bi-allelic AMA1 and MSP1 vaccines. Volunteers underwent CHMI ~16 days post boosting vaccination. Graphs represent the individual response and group median for T cell responses, expressed as IFN-γ SFU/106 PBMC summed from pools containing overlapping peptides from the vaccine insert. (A) AMA1 allele specific responses calculated by adding the allele specific response (3D7 or FVO) to the common pool response for each individual. MSP133 (B) and MSP119 (C) fragment specific responses for each allele (3D7 or Wellcome) calculated by adding responses from representative pools. Day 0 calculations are excluded from statistical analysis. Ratios for the two median response are also included (>1 = stronger 3D7 response). *P = <0.05, **P = <0.01.
In agreement with AMA1, very similar results were observed for the dimorphic MSP133 and MSP119 responses (Figure 2B, C). Comparable results were observed to the Phase Ia clinical trial data prior to malaria infection, and following challenge, the ratio of 3D7 and Wellcome responses for MSP133 were not affected. There was no significant boosting of 3D7 allele responses, and no apparent diversion towards Wellcome-specific responses. Although the dominance of the 3D7 MSP133 allele does become significantly greater at the dC+7 and dC+35 time-points, this significance level is also observed at dC-1 prior to infection, and is less pronounced at dC+90 and dC+150. For MSP119 there was also no change to the 3D7:Wellcome ratios towards a 3D7 bias after challenge except for dC+35 where there was a small but significant enhancement of the 3D7-specific response. Overall, these data suggest that controlled 3D7 clone blood-stage parasite exposure does not boost the IFN-γ T cell response induced by the vaccine, nor is there significant diversion of responses towards the non-infecting heterologous allele.
Priming of T cell responses in unimmunized control volunteers is predominantly strain-specific
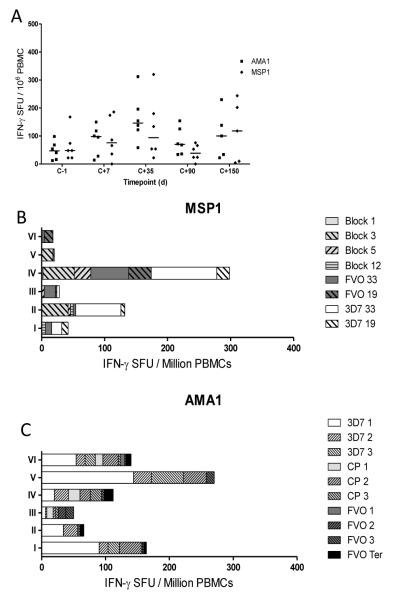
In the Phase IIa efficacy study, six malaria-naïve infectivity control volunteers were also infected, and responses against MSP1 and AMA1 were assessed by ex-vivo IFN-γ ELISPOT post-infection (Figure 3). An acute blood-stage exposure induced low level responses against both antigens, peaking at dC+35 (Figure 3A). Analysis of the breakdown of the responses to the MSP1 antigen (dC+35 after subtracting background at dC-1) showed that infection induced T cells reactive to the conserved blocks of MSP1 sequence, in particular Block 3 in 3/6 volunteers, as well as responses to the 3D7 MSP142 region in 3/6 volunteers (Figure 3B). Four out of six volunteers also showed detectable responses to the Wellcome strain peptide pools, most notably for the MSP133 region. In contrast, for the AMA1 antigen, the vast majority of the T cell response was to unique 3D7-specific peptide pools along with some to the conserved peptides (Figure 3C), whilst those to the FVO specific-peptide pools were negative or extremely low. The potential for the induction of cross-reactive T cell responses following 3D7 clone parasite infection thus appears to be more common for MSP133 than AMA1. Given that MSP133 3D7 and Wellcome only share 51% similarity, in comparison to 95% similarity between 3D7 and FVO in AMA1, it appears that natural exposure may favor the induction of AMA1 strain-specific T cell responses. This is in strong contrast to vaccination with the bi-allelic AMA1 vaccine, where comparable T cell responses were observed to both the strain-specific and common peptides (25).
Figure 3.
CHMI induces allele-specific responses in unvaccinated control volunteers in a Phase IIa efficacy study. (A) The total antigen specific IFN-γ T cell response peaks 35 days post CHMI (dC+35). Peak response breakdown to individual pools for MSP1 (B) and AMA1 (C) following 3D7 challenge, calculated by subtracting baseline response before CHMI (dC-1) from response 35 days after CHMI (dC+35). Pools can be identified as containing conserved sequences (light grey), 3D7-specific sequences (white), Wellcome/FVO-specific sequences (dark grey), or FVO AMA1 C-terminal region sequences (black).
Immune antagonism does occur at the minimal epitope level but does not impact overall allele-specific responses
The previous analyses assessed overall IFN-γ T cell responses by ELISPOT assay, and indicated that following secondary immunization there was no apparent immune antagonism that prevented boosting of T cell responses to both alleles of the vaccine antigen. However, it remained possible that more subtle effects would occur at the epitope-specific level or within the CD4+ or CD8+ T cell subsets. Lee et al., have previously reported HLA class II APL antagonistic epitope pairs within dimorphic regions of MSP1 (38, 39), and one pair of epitopes (M7/8) reported by Lee et al. were also contained within the MSP133 region of the vaccine construct (Table I).
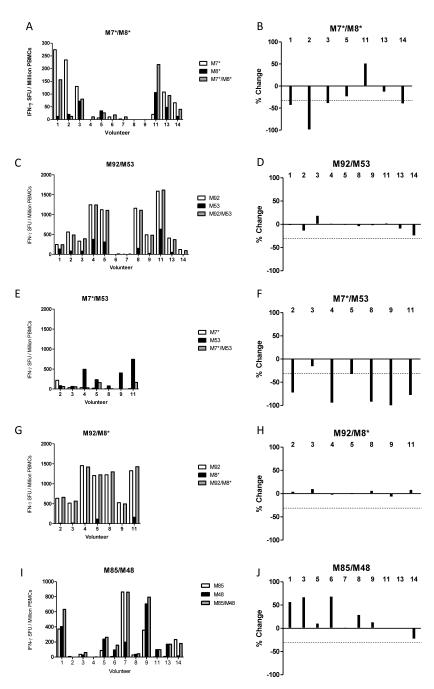
Immune antagonism was assessed in vitro in the same manner as Lee et al., by ex-vivo IFN-γ ELISPOT assay using peptides either alone or combined to re-stimulate PBMC from the d63 time-point (peak response). Using this pair of minimal epitopes, referred to here as M7* and M8*, we identified seven volunteers who responded significantly to at least one of the epitopes (Figure 4A), indicating that responses had been induced in vivo by vaccination to these epitopes and not completely prevented due to immune interference or antagonism. In their studies, Lee et al. described antagonism as an arbitrary ≥30% reduction in the response following re-stimulation with both peptides (38). To increase stringency we increased the cut-off to a ≥50% reduction.
Figure 4.
IFN-γ T cell responses in MSP1 vaccinated volunteers at d63 to single peptides from allelic pairs both individually and combined in a single pool; (A) M7*/M8* 8-mers, (C) M92/M53 20-mers, (E) M7*/M53, (G) M92/M8* and (I) M85/M48 20mers. In the right hand graphs, bars shows % change from the strongest single allele peptide response, to the response to the pool containing both peptides; (B) M7*/M8* 8-mers, (D) M92/M53 20-mers, (F) M7*/M53, (H) M92/M8* and (I) M85/M48 20mers. Antagonism is defined as a ≥50% reduction (signified by dotted line) from the IFN-γ response measured to a single peptide compared to when that peptide is combined with its opposing allele. Two volunteers (#10 and #12) are not included in this analysis due to limited d63 PBMC availability, but are included in later analyses. Only 7/12 volunteers were tested for E-H due to limited availability of d63 PBMC samples. Responses are measured in duplicate, averaged and background subtracted.
Of the individuals tested, only one volunteer (#2) showed almost complete antagonism (98% reduction) whilst three volunteers fell just short of the 50% cut-off (Figure 4B). It should be noted that a single amino acid difference does exist between the sequence for M8* and that found in the vaccine antigen sequence (V-Y substitution at the final position of M8*), although vaccinated volunteers still responded to the M8* peptide re-stimulation, indicating the final amino acid may not be critical for HLA class II molecule binding.
When we performed the same experiment using 20mer peptides (routinely used in the clinical assays) that contain the M7/8 minimal epitopes (referred to here as M92 and M53, Table I) (24) we observed a different result. Ten out of 12 volunteers responded to the extended peptides (Figure 4C), possibly indicating the presence of other epitopes within the extended peptides (e.g. volunteers #4, 5, 8, 9 and 11 responded more strongly in the assay than when using the minimal peptides). However, none of these samples, including volunteer #2, showed antagonism in vitro, and in most cases the combined M92/M53 pool roughly equaled the response seen with the M92 pool alone (Figure 4D). These data suggest that the 20mer peptides may be detecting responses to alternative epitopes that are not antagonized, or that the 20mer is not sufficiently processed into the minimal antagonistic APL.
To address this, we next tested each minimal epitope with its heterologous 20mer. Volunteer #2 who had the notable M7* response, was again antagonized in the presence of M53, in agreement with the observation in Figure 4A, and suggesting that the 20mer peptide M53 can be processed into an antagonistic APL (Figure 4E,F). We also observed volunteers (#4, 5, 8, 9, and 11) who responded better to M53 (Figure 4C,E) than to M8* (Figure 4A), suggesting either the presence of another epitope, or that the optimal HLA binding sequence in fact contains flanking αα only present in the 20mer peptide. In agreement with the latter, these responses were all highly antagonized in the presence of M7* peptide (Figure 4F) greatly exceeding the ≥50% cut-off. The opposite experiment, testing peptide M92 with M8*, confirmed the absence of responses to M8* but improved responsiveness to the M92 20mer in comparison to M7* (Figure 4G), suggesting the presence of another epitope. In agreement with this, these responses were not antagonized in the presence of M8* peptide (Figure 4H).
We also tested a combination of two 20mer peptides, M85 and M48, which contained the dimorphic minimal epitopes M30 and M31 shown by Lee et al. to be immunogenic (39) but not previously tested for antagonism. In this case, 10/12 volunteers responded to these peptides (Figure 4I) with none displaying antagonism (Figure 4J). In fact, for 3/12 volunteers, re-stimulation with both peptides gave an additive effect (>50% increase) in comparison to the single peptide responses. Overall these data confirm that it is possible to observe immune antagonism in vitro between APL variants, however, these differences can be subtle and only observed when using specific peptide sequences.
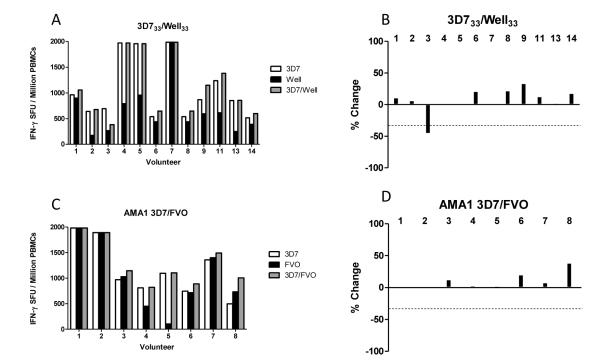
We further speculated that the previous observations of antagonism could be specific to the M7/8 epitope pair alone. Therefore in order to confirm whether an effect of immune antagonism in vitro could be detected to the vaccine antigen overall, we performed similar experiments using pools of 20mer peptides spanning the whole MSP133 regions. As before, pools representing the 3D7 and Wellcome alleles were tested alone or in combination using PBMC from the peak time-point following the MVA booster immunization (day 63). All 12 volunteers responded (Figure 5A) and only one (volunteer #3) showed moderate antagonism of 45% that approached the arbitrary cut-off (Figure 5B). AMA1 was also tested in the same format using pools of peptides unique for the 3D7 and FVO alleles and PBMC from AMA1 vaccinated volunteers. No antagonism was observed, and for 5/8 volunteers the ratios of the 3D7:FVO responses were roughly even and only one volunteer (#5) displayed a bias towards the 3D7 allele (Figure 5C,D).
Figure 5.
IFN-γ T cell responses were measured against allele-specific pools both individually and combined for MSP133 3D7/Well (A) and AMA1 3D7/FVO (C). In the right hand graphs, bars shows % change from the strongest single allele peptide pool response, to the response to the combined pool containing allelic peptides; MSP133 3D7/Well (B) and AMA1 3D7/FVO (D). Antagonism is defined as a ≥50% reduction (signified by dotted line) from the IFN-γ gamma response measured to a single peptide compared to when that peptide is combined with its opposing allele. Two volunteers (#10 and #12) are not included in this analysis due to limited d63 PBMC availability, but are included in later analyses. Responses are measured in duplicate, averaged and background subtracted.
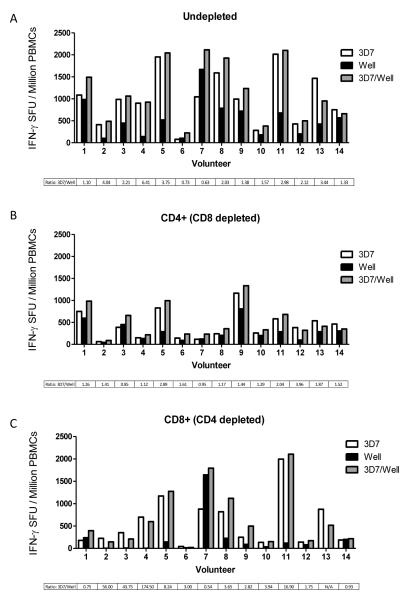
Depletions show a lack of a Wellcome allele-specific CD8+ T cell response to MSP133
We had so far only observed immune antagonism in vitro when testing a previously described minimal HLA class II epitope from MSP133 (38). Immune antagonism has been reported for both HLA class I and class II malaria epitopes (37, 38), and we therefore also assessed whether we could observe this effect in either the CD4+ or CD8+ T cell subsets. Frozen PBMCs from 14 volunteers who had received the MSP1 immunization regime were used for CD4+ and CD8+ T cell depletion studies. PBMC samples showing strong immunogenicity at various time-points, and for which frozen cells were available (Figure S3), were selected and depleted of the relevant T cell subset, before performing the ELISPOT assays as before using the same MSP133 peptide pools. T cell depletion efficiency was confirmed by flow cytometric analysis (as described in Methods) in four of the volunteers and was shown to be >98% efficient (Figure S2). Undepleted cells (Figure 6A) showed comparable responses to those seen previously (Figure 5A). Volunteer #3, who previously displayed some evidence of antagonism at the d63 time-point, no longer showed this effect when using PBMC from d84. Only 1/14 (volunteer #7) showed a stronger response to the Wellcome allele, with the remaining 13 showing a stronger 3D7 response in agreement with the previous result (Figure 1B, 2B). When the same assay was performed with CD4+ (CD8+ depleted) T cells, the results showed roughly even 3D7/Wellcome responses for 7/14 of the volunteers, with the remainder still having stronger 3D7 allele-specific responses (Figure 6B). Moreover, no evidence of in vitro antagonism was observed. The results for the CD8+ (CD4+ depleted) T cells showed that most volunteers had high 3D7 responses and low to negligible Wellcome allele responses (Figure 6C). Two volunteers (#1 and #14) displayed low responses but a roughly equal 3D7/Wellcome ratio, while a third (#7) showed a strong, dominant Wellcome response. There was also no in vitro antagonism observed for these CD8+ T cell responses. Taken together, these data indicated that the previously observed dominance of the 3D7 MSP133 response over the Wellcome MSP133 response is possibly due to an apparently low CD8+ T cell response against the Wellcome allele sequence.
Figure 6.
IFN-γ T cell subset responses in MSP1 vaccinated volunteers against peptide pools containing either MSP133 3D7 peptides, MSP133 Wellcome peptides or a combined pool of both allelic peptides. To compare IFN-γ responses from different T cell subsets ‘Undepleted’ PBMC (A), CD4+ (CD8+ depleted) PBMC (B) and CD8+ (CD4+ depleted) PBMC (C) were prepared using a MACS magnetic column depletion kit and responses to peptide pools measured. Graphs represent the individual responses expressed as IFN-γ SFU/106 PBMC from various time-points selected for high responses and PBMC availability (#10, 12 = d14; #1, 9, 14 = d63; #2, 3, 4, 8, 11 = d84; #5 = dC-1; #13 = dC+35; #7 = dC+90; #6 = d140; see Figure S3). Ratios for the 3D7/Well median response are included (>1 = stronger 3D7 response).
Volunteers with HLA-B*1801 mount a CD8+ T cell response to Wellcome MSP133
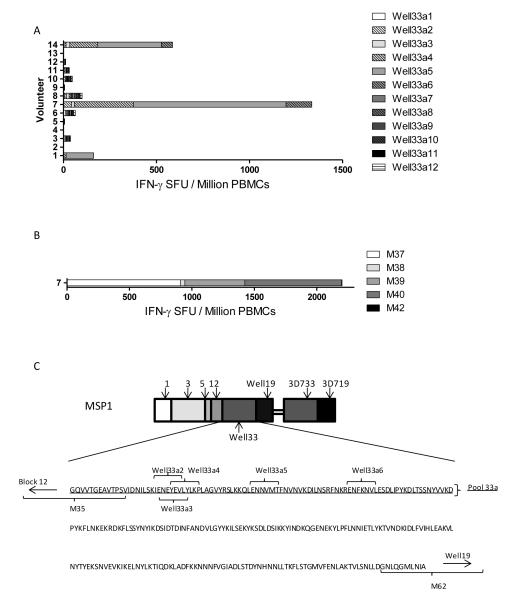
We decided to further investigate why only a small subset of volunteers produce a CD8+ T cell response to the Wellcome allele of MSP133. Volunteers are HLA typed routinely as part of the clinical trial program and review of these data identified that volunteers #1 and #14, both of which showed 3D7/Wellcome MSP133 ratios below 1 (indicating a stronger Wellcome response) were heterozygous for HLA-B*1801. Moreover, volunteer #7 who showed a much stronger Wellcome allele-specific response (as compared to 3D7) was found to be homozygous for this same HLA type. To test for HLA-B*1801-specific CD8+ T cell minimal epitopes, we ran the Wellcome MSP133 vaccine sequence through the epitope prediction software ‘SBS EpiToolKit’ for HLA-B*1801 as well as other common HLA-B types displayed in the volunteer population. Initially we analysed the routine trial ELISPOT assay readout for volunteers, where the total responses to Wellcome MSP133 is measured using three peptide pools (24). A breakdown of responses in these volunteers revealed a dominance of responses in the “33a pool”, which contains the 10 N-terminal peptides. This was most noticeable in volunteer #7 who had 92% of their Wellcome MSP133-specific response in pool 33a. A summary of individual responses to the Well33a pool over time is shown in Figure S3. In agreement with this, the prediction software for HLA-B*1801 predicted a large number of epitopes in this pool which were not present in the 3D7 sequence. Several epitopes for other common HLA-B types were also predicted for sequences in the 33a pool, many overlapping with the sequences predicted for HLA-B*1801. Based on this observation twelve minimal epitopes were selected based on strongly predicted HLA association, broad coverage of HLA types and no homology with the 3D7 sequence (Table II). These were then tested in a CD8+ (CD4+ depleted) T cell ELISPOT assay. Of the 12 epitopes, three were found to give strong positive responses in volunteers #1, #7 and #14, with volunteer #7 displaying the strongest response as would be predicted for this HLA-B*1801 homozygote individual (Figure 7A). All other volunteers gave essentially negative responses with no individual peptide response >15 SFU/million PBMC. To confirm this observation, we tested the CD8+ T cell response in volunteer #7 using the equivalent 20mers peptides (used in the routine trial assays) containing the predicted epitopes Well33a2-6 (Figure 7B). Results indicated a strong response to both 20mers (M39 and M40) containing epitope Well33a5 and the 20mer (M37) containing epitopes Well33a2, 3 and 4 (see Table II). Overall these results suggest that the strong response to Wellcome MSP133 in HLA-B*1801 volunteers is primarily due to a number of HLA class I epitopes, situated towards the N-terminus of the MSP133 sequence (Figure 7C), and this may account for weaker Wellcome MSP133 responses in volunteers lacking this HLA-B class I allele.
Figure 7.
(A) IFN-γ CD8+ T cell responses to predicted minimal epitopes from the MSP133 Wellcome allele. Volunteers and time-points tested are equivalent to those in Figure 6. HLA-B*1801 is heterozygous in volunteers, #1 and #14 and homozygous in volunteer #7. Complete epitope sequences, predicted HLA associations, and equivalent trial peptide location for all minimal epitopes can be found in Table II. (B) IFN-γ CD8+ T cell responses from volunteer #7 to 20mer peptides containing minimal epitopes recognized previously. (C) Location of CD8+ T cell minimal epitopes recognized by volunteers with HLA-B*1801. Epitopes are located towards the N-terminus of the Wellcome MSP133 fragment.
DISCUSSION
We have developed viral vectored blood-malaria vaccine candidates that encode bi-allelic transgene inserts for the MSP1 and AMA1 antigens. These vaccines were developed with the aim of providing a broad immunological response that can protect against multiple strains of P. falciparum. The use of a ChAd63 vector to prime responses followed by a boost with MVA eight weeks later has been shown to induce high levels of T cells and substantial antibody responses in both animal models and Phase I/IIa clinical trials in humans for both blood-stage vaccine candidate antigens (8, 24, 25). However, previous studies have raised concerns that inclusion of multiple allelic variants in a vaccine may be detrimental to both the priming and in vivo re-stimulation of antigen-experienced memory T cells owing to the immune evasion effects of antagonistic APLs present in polymorphic antigens. Here we explored the concepts of immune interference, antagonism and diversion of IFN-γ T cell responses in the context of human vaccination with viral vectored vaccines and subsequent malaria exposure following a Phase IIa CHMI study.
Previous studies have suggested that dimorphic malaria antigen variants arose and were maintained in parasite populations due to a survival advantage in the context of parasite co-habitation within the human host. MSP1 allelic T cell responses failed to correlate with differential antigen exposure in The Gambia (39), and associated with this observation was the inference from in vitro studies that co-habiting P. falciparum parasites could interfere with the priming of T cell responses by the phenomenon of ‘immune interference’, and that the same would be true for re-stimulation of memory responses by ‘immune antagonism’ (38). Such immune evasion mechanisms could be achieved by APL epitopes from both HLA class I and class II molecules present in dimorphic antigens leading to antagonism of responses (36-38). Ultimately, concerns were raised about the ability to prime and boost cellular immune responses with vaccines encoding more than one allele of an antigen. Until now, these concerns could not be fully explored in the context of human vaccination due to the lack of bi-allelic malaria vaccine candidates that are capable of inducing strong cellular immune responses in human volunteers. Here we investigated these effects following human vaccination with the ChAd63-MVA viral vectored vaccine platform – one that has been optimized for the induction of both humoral and cellular immunity (22, 23).
Initially we investigated the priming and boosting of T cell responses using the ex-vivo IFN-γ ELISPOT assay. T cell responses, as measured against the 3D7 and FVO alleles of AMA1, appeared to be primed and boosted consistently, and the ratio of the responses was roughly equal and also stable throughout the monitoring period. Similar results were obtained for the MSP119 region of MSP1. In agreement with this, the two alleles of AMA1 (as encoded in the vaccine) differ by 24 αα and the ectodomains share 95% sequence identity, and similarly, MSP119 shows 96% αα conservation between the 3D7 and Wellcome alleles. In contrast, the MSP133 fragment only shows 51% αα conservation between the two and in this case, responses towards the 3D7 allele were dominant over the Wellcome allele in most volunteers. This was apparent after priming and was maintained after boosting and throughout the memory phase. In the absence of a control GMP grade vaccine encoding a single allele that could be used for human vaccination, it remains impossible to establish whether responses against APL variants could display interference/antagonism at the priming and/or boosting phase, thus leading to reduced responses in the presence of both variants compared to the presence of one. However, the data clearly establish that strong and detectable IFN-γ T cell responses can be primed and boosted by immunization against both component alleles, and this was independently observed in two clinical trials of the same vaccines. The observation of strong cellular responses against the MSP133 region (in comparison to MSP119) is in good agreement with many previous studies that have assessed T cell recognition within MSP142 in both animal models (20, 45) and humans (46-49). It should also be noted that future studies could further assess such phenomena in the context of other T cell phenotype readouts, including different cytokine production.
We next assessed the potential effects of single strain blood-stage malaria parasite exposure on vaccine-induced T cells responses. In this case, 3D7 clone parasites were used to assess vaccine efficacy, following immunization with the bi-allelic vaccines (26). In this experimental context, re-stimulation of heterologous epitope-specific T cell responses (Wellcome allele in the case of MSP142 or FVO allele in the case of AMA1) by antagonistic APLs from the 3D7 clone sequence could occur in conjunction with re-stimulation of the homologous 3D7 responses. No significant boosting was observed to either allele of either antigen during natural in vivo infection by 3D7 clone blood-stage malaria parasites. In all cases, IFN-γ T cell responses post-malaria challenge continued to contract into the memory phase with similar kinetics to those observed in the Phase Ia trials where volunteers were not experimentally infected. It remains possible that the lack of boosting is due to a form of immune diversion, however in the absence of a single allele comparator clinical-grade vaccine, we cannot confirm responses were curtailed in such a manner. Interestingly, despite any apparent boosting of vaccine-induced T cell responses, de novo T cell responses were observed against both MSP1 and AMA1 in control volunteers who had not received any vaccine. In the case of AMA1, responses were almost exclusively detected with peptides specific for the 3D7 allele, rather than those in common with the FVO allele. This is in contrast to vaccination where the common peptides are better recognized (25), and may mean that immunization with two alleles can better focus T cell responses on conserved epitopes. In the case of MSP133, responses that were cross-reactive with the Wellcome allele were noted in some volunteers. This is potentially due to conserved sequences between the two dimorphic MSP133 regions, or potentially represents T cells primed by 3D7 parasite exposure that cross-react with Wellcome allele sequence variants.
Given we had seen no apparent effects in terms of the overall IFN-γ T cell response following vaccination with bi-allelic antigens, we sought to confirm whether the APL antagonism previously reported by Lee et al. (38) could also be observed here in vitro. One pair of previously reported peptides (M7/8) represented minimal class II epitopes that showed APL antagonism. These sequences were contained within our vaccine, and responses were detectable to either one epitope or both in about half of the volunteers tested. Interestingly, and in agreement with the previous data from naturally-exposed individuals, this epitope was shown to be convincingly antagonistic in 1/7 volunteers who possessed detectable IFN-γ responses. However, when 20mer peptides were used that contained the minimal epitopes, this antagonistic effect was no longer observed. Furthermore when minimal epitopes were combined with heterologous 20mers, antagonism was observed between M7* and M53 but not M8* and M92. The data suggested that the 20mer peptides could be processed into antagonistic APLs, but also that more optimal HLA binding sequences and/or other epitopes may be present within in the 20mers in comparison to the minimal peptides. Further testing with larger pools of 20mer peptides representing both alleles of MSP133 and AMA1 confirmed that there was no overall or obvious effect from antagonism, as measured using this ELISPOT assay readout for either the CD4+ or CD8+ T cell subsets. Neither antigen produced any antagonism in vitro and responses to the combined allele peptide pools roughly equaled or were slightly greater than those seen to the strongest single allele. This observation was the same for volunteers who had a reduced or equivalent Wellcome MSP133 response as compared to 3D7, indicating that APL antagonism at the time of booster vaccination is unlikely to be contributing to the observed dominance of 3D7 responses.
Whilst performing the CD8+ T cell depletion assays, it became apparent that the 3D7 MSP133-specific CD8+ T cell responses were dominant over the Wellcome allele in most volunteers, except those possessing HLA-B*1801. For two volunteers (#1, #14), who were heterozygous for HLA-B*1801, the 3D7:Wellcome ratio of response was roughly equal, whilst a third volunteer (#7) showed a dominant Wellcome response and was the only person homozygous for this HLA type. This observation led us to hypothesize that it was possibly a lack of HLA class I-specific epitopes in the Wellcome MSP133 sequence that may be responsible for the observed difference in immunogenicity between the alleles. We subsequently predicted twelve HLA-B*1801 epitopes, and of these, one showed dominant responses in 3 of the 14 volunteers screened, with four other epitopes also showing responses across the same volunteers. These were the three volunteers who possessed HLA-B*1801, and the broadest observed response was seen in the homozygous volunteer. These epitopes are all located close together toward the N-terminus of the Wellcome MSP133 fragment and are not present in the 3D7 sequence. Two previous studies (47, 49) have reported that T cell responses to MSP133 are directed to conserved epitopes, however, our data indicate this may not always be the case. Obvious differences exist between the studies, and most notably those related to study of class I versus class II epitopes, as well as host population genetics. Moreover, in the absence of single allele comparator clinical-grade vaccines, we cannot formally exclude the possibility that the observed differences between the 3D7 and Wellcome MSP133 responses are due to immune interference during T cell priming. It remains possible that the presence of the 3D7 MSP133 allele interferes with priming to the Wellcome allele in individuals, except in the case of certain HLA types such as HLA-B*1801.
It also remains of interest to assess such vaccine-induced responses in a malaria-endemic target population. A number of studies have characterized HLA types within and between West African, South African (50) and East African populations (51). Focusing on the HLA B locus, a number of typically African alleles such as HLA-B53 are seen across the populations at high frequencies, while other alleles vary in frequency depending on location. HLA-B*1801 was observed as the fifth highest allele frequency (6.21%) in a study in Kenya (51), while lower frequencies have been described in West African populations such as The Gambia (5%), and Mali (2.4%), with Europeans around 10% (50). Given that 3D7:Wellcome parasite ratios vary in different locations (52-54), a lack of recognition by local HLA types as well as immune interference during co-infection would be of potential advantage to parasites. Similarly, lack of vaccine antigen recognition by local HLA types following vaccination may lead to different levels of vaccine efficacy, highlighting the importance of considering factors such as HLA types when undertaking vaccine field trials.
In summary, previous mechanistic studies have concluded that inclusion of multiple alleles of an antigen in a vaccine may be detrimental to both the priming and boosting of T cells. Our data, from human volunteers vaccinated with T cell-inducing viral vectored vaccines against two different but polymorphic blood-stage malaria antigens, suggest that T cell responses to bi-allelic antigens can be primed and boosted through vaccination. Following controlled infection with a single P. falciparum strain, vaccine-induced responses did not boost but neither were they apparently immune diverted to the heterologous strain, whilst strain-specific de novo responses could be measured in unvaccinated controls. Responses against the more conserved sequences of MSP1 and AMA1 are evenly distributed across the two alleles; however for the more divergent regions such as the MSP133 fragment, there does appear to be bias towards the 3D7 allele, most notably for CD8+ T cells. We cannot exclude the possibility that immune interference due to invariant APLs occurred at the time of priming, but we do report a paucity of HLA class I epitopes in the Wellcome allele sequence recognized by UK volunteers. We could also observe APL antagonism to minimal epitopes in vitro, however this effect did not appear to have a significant impact on the overall antigen-specific IFN-γ T cell response. Although the fine epitope specificity of responses is likely to be important in the context of vaccine-induced immunity, these data suggest host HLA-type may more significantly affect the magnitude or breadth of T cell responses. It will therefore remain of interest to further assess in target endemic populations the potential impact of circulating parasite strains, HLA type distributions and prior malaria exposure on vaccine immunogenicity.
Supplementary Material
ACKNOWLEDGEMENTS
For clinical and logistical support we thank C. Bateman, M. Smith, J. Meyer, R. Lopez-Ramon, P. Lillie, N. Anagnostou, R. Antrobus, I. Poulton, A. Lawrie, L. Dinsmore, K. Gantlett, R. Sinden, and S. Gilbert (Oxford University, UK); T. Havelock, C. Grocott, F. Martins, and S. Faust (Wellcome Trust Clinical Research Facility, Southampton, UK); T. Mahungu, R. Singzon, J. Ryu, and T. Doherty (University College London Clinical Research Facility, London, UK). We also thank J. Furze, A. Spencer, and D. Worth for laboratory assistance; the Jenner Institute Flow Cytometry Core Facility for technical assistance; S. Colloca, R. Cortese, and A. Nicosia (Okairòs, Italy) for provision of the ChAd63 vector; and all the study volunteers.
FUNDING STATEMENT
This work was supported by the UK Medical Research Council (MRC) [grant number G0700735]; the EMVDA (European Malaria Vaccine Development Association, a European Commission FP6-funded consortium [LSHP-CT-2007-037506]; the UK National Institute of Health Research through the Oxford Biomedical Research Centre; the Wellcome Trust [084113/Z/07/Z]; and by EVIMalaR, a European Community FP7-funded programme [Grant agreement No. 242095]. AVSH and SJD are Jenner Investigators; and SJD is a UK MRC Career Development Fellow [G1000527].
Glossary
- AMA1
Apical membrane antigen 1
- APL
Altered peptide ligand
- ChAd63
Chimpanzee adenovirus serotype 63
- CHMI
Controlled human malaria infection
- MSP1
Merozoite surface protein 1
- MVA
Modified vaccinia virus Ankara
Footnotes
COMPETING INTEREST STATEMENT AVSH and SJD are named inventors on patent applications covering malaria vectored vaccines and immunization regimes.
REFERENCES
- 1.Hill AV. Vaccines against malaria. Philos Trans R Soc Lond B Biol Sci. 2011;366:2806–2814. doi: 10.1098/rstb.2011.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman AL, Draper SJ. Blood-stage malaria vaccines - recent progress and future challenges. Ann Trop Med Parasitol. 2010;104:189–211. doi: 10.1179/136485910X12647085215534. [DOI] [PubMed] [Google Scholar]
- 3.Holder AA. The carboxy-terminus of merozoite surface protein 1: structure, specific antibodies and immunity to malaria. Parasitology. 2009;136:1445–1456. doi: 10.1017/S0031182009990515. [DOI] [PubMed] [Google Scholar]
- 4.Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24:74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Riley EM, Allen SJ, Wheeler JG, Blackman MJ, Bennett S, Takacs B, Schonfeld HJ, Holder AA, Greenwood BM. Naturally acquired cellular and humoral immune responses to the major merozoite surface antigen (PfMSP1) of Plasmodium falciparum are associated with reduced malaria morbidity. Parasite Immunol. 1992;14:321–337. doi: 10.1111/j.1365-3024.1992.tb00471.x. [DOI] [PubMed] [Google Scholar]
- 6.Polley SD, Mwangi T, Kocken CH, Thomas AW, Dutta S, Lanar DE, Remarque E, Ross A, Williams TN, Mwambingu G, Lowe B, Conway DJ, Marsh K. Human antibodies to recombinant protein constructs of Plasmodium falciparum Apical Membrane Antigen 1 (AMA1) and their associations with protection from malaria. Vaccine. 2004;23:718–728. doi: 10.1016/j.vaccine.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Fowkes FJ, Richards JS, Simpson JA, Beeson JG. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: A systematic review and meta-analysis. PLoS Med. 2010;7:e1000218. doi: 10.1371/journal.pmed.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draper SJ, Moore AC, Goodman AL, Long CA, Holder AA, Gilbert SC, Hill F, Hill AV. Effective induction of high-titer antibodies by viral vector vaccines. Nat Med. 2008;14:819–821. doi: 10.1038/nm.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Hodder AN, Yan H, Crewther PE, Anders RF, Good MF. CD4+ T cells acting independently of antibody contribute to protective immunity to Plasmodium chabaudi infection after apical membrane antigen 1 immunization. J Immunol. 2000;165:389–396. doi: 10.4049/jimmunol.165.1.389. [DOI] [PubMed] [Google Scholar]
- 10.Biswas S, Spencer AJ, Forbes EK, Gilbert SC, Holder AA, Hill AV, Draper SJ. Recombinant Viral-Vectored Vaccines Expressing Plasmodium chabaudi AS Apical Membrane Antigen 1: Mechanisms of Vaccine-Induced Blood-Stage Protection. J Immunol. 2012;188:5041–5053. doi: 10.4049/jimmunol.1101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darko CA, Angov E, Collins WE, Bergmann-Leitner ES, Girouard AS, Hitt SL, McBride JS, Diggs CL, Holder AA, Long CA, Barnwell JW, Lyon JA. The clinical-grade 42-kilodalton fragment of merozoite surface protein 1 of Plasmodium falciparum strain FVO expressed in Escherichia coli protects Aotus nancymai against challenge with homologous erythrocytic-stage parasites. Infect Immun. 2005;73:287–297. doi: 10.1128/IAI.73.1.287-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyon JA, Angov E, Fay MP, Sullivan JS, Girourd AS, Robinson SJ, Bergmann-Leitner ES, Duncan EH, Darko CA, Collins WE, Long CA, Barnwell JW. Protection induced by Plasmodium falciparum MSP1(42) is strain-specific, antigen and adjuvant dependent, and correlates with antibody responses. PLoS ONE. 2008;3:e2830. doi: 10.1371/journal.pone.0002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahdi Abdel Hamid M, Remarque EJ, van Duivenvoorde LM, van der Werff N, Walraven V, Faber BW, Kocken CH, Thomas AW. Vaccination with Plasmodium knowlesi AMA1 formulated in the novel adjuvant co-vaccine HT protects against blood-stage challenge in rhesus macaques. PLoS One. 2011;6:e20547. doi: 10.1371/journal.pone.0020547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta S, Sullivan JS, Grady KK, Haynes JD, Komisar J, Batchelor AH, Soisson L, Diggs CL, Heppner DG, Lanar DE, Collins WE, Barnwell JW. High antibody titer against apical membrane antigen-1 is required to protect against malaria in the Aotus model. PLoS One. 2009;4:e8138. doi: 10.1371/journal.pone.0008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good MF, Engwerda C. Defying malaria: Arming T cells to halt malaria. Nat Med. 2011;17:49–51. doi: 10.1038/nm0111-49. [DOI] [PubMed] [Google Scholar]
- 16.Good MF, Xu H, Wykes M, Engwerda CR. Development and regulation of cell-mediated immune responses to the blood stages of malaria: implications for vaccine research. Annu Rev Immunol. 2005;23:69–99. doi: 10.1146/annurev.immunol.23.021704.115638. [DOI] [PubMed] [Google Scholar]
- 17.Bouharoun-Tayoun H, Attanath P, Sabchareon A, Chongsuphajaisiddhi T, Druilhe P. Antibodies that protect humans against Plasmodium falciparum blood stages do not on their own inhibit parasite growth and invasion in vitro, but act in cooperation with monocytes. J Exp Med. 1990;172:1633–1641. doi: 10.1084/jem.172.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joos C, Marrama L, Polson HE, Corre S, Diatta AM, Diouf B, Trape JF, Tall A, Longacre S, Perraut R. Clinical protection from falciparum malaria correlates with neutrophil respiratory bursts induced by merozoites opsonized with human serum antibodies. PLoS One. 2010;5:e9871. doi: 10.1371/journal.pone.0009871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belnoue E, Voza T, Costa FT, Gruner AC, Mauduit M, Rosa DS, Depinay N, Kayibanda M, Vigario AM, Mazier D, Snounou G, Sinnis P, Renia L. Vaccination with live Plasmodium yoelii blood stage parasites under chloroquine cover induces cross-stage immunity against malaria liver stage. J Immunol. 2008;181:8552–8558. doi: 10.4049/jimmunol.181.12.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draper SJ, Goodman AL, Biswas S, Forbes EK, Moore AC, Gilbert SC, Hill AV. Recombinant viral vaccines expressing merozoite surface protein-1 induce antibody- and T cell-mediated multistage protection against malaria. Cell Host Microbe. 2009;5:95–105. doi: 10.1016/j.chom.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai T, Shen J, Chou B, Duan X, Tu L, Tetsutani K, Moriya C, Ishida H, Hamano S, Shimokawa C, Hisaeda H, Himeno K. Involvement of CD8+ T cells in protective immunity against murine blood-stage infection with Plasmodium yoelii 17XL strain. Eur J Immunol. 2010;40:1053–1061. doi: 10.1002/eji.200939525. [DOI] [PubMed] [Google Scholar]
- 22.Hill AVS, Reyes-Sandoval A, O’Hara G, Ewer K, Lawrie AM, Goodman AL, Nicosia A, Folgori A, Colloca S, Cortese R, Gilbert SC, Draper SJ. Prime-boost vectored malaria vaccines: progress and prospects. Hum Vaccin. 2010;6:78–83. doi: 10.4161/hv.6.1.10116. [DOI] [PubMed] [Google Scholar]
- 23.Draper SJ, Heeney JL. Viruses as vaccine vectors for infectious diseases and cancer. Nat Rev Microbiol. 2010;8:62–73. doi: 10.1038/nrmicro2240. [DOI] [PubMed] [Google Scholar]
- 24.Sheehy SH, Duncan CJ, Elias SC, Collins KA, Ewer KJ, Spencer AJ, Williams AR, Halstead FD, Moretz SE, Miura K, Epp C, Dicks M, Poulton ID, Lawrie AM, Berrie E, Moyle S, Long CA, Colloca S, Cortese R, Gilbert SC, Nicosia A, Hill AV, Draper SJ. Phase Ia Clinical Evaluation of the Plasmodium falciparum Blood-stage Antigen MSP1 in ChAd63 and MVA Vaccine Vectors. Mol Ther. 2011;19:2269–2276. doi: 10.1038/mt.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehy SH, Duncan CJ, Elias SC, Biswas S, Collins KA, O’Hara GA, Halstead FD, Ewer KJ, Mahungu T, Spencer AJ, Miura K, Poulton ID, Dicks MD, Edwards NJ, Berrie E, Moyle S, Colloca S, Cortese R, Gantlett K, Long CA, Lawrie AM, Gilbert SC, Doherty T, Nicosia A, Hill AV, Draper SJ. Phase Ia Clinical Evaluation of the Safety and Immunogenicity of the Plasmodium falciparum Blood-Stage Antigen AMA1 in ChAd63 and MVA Vaccine Vectors. PLoS One. 2012;7:e31208. doi: 10.1371/journal.pone.0031208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheehy SH, Duncan CJ, Elias SC, Choudhary P, Biswas S, Halstead FD, Collins KA, Edwards NJ, Douglas AD, Anagnostou NA, Ewer KJ, Havelock T, Mahungu T, Bliss CM, Miura K, Poulton ID, Lillie PJ, Antrobus RD, Berrie E, Moyle S, Gantlett K, Colloca S, Cortese R, Long CA, Sinden RE, Gilbert SC, Lawrie AM, Doherty T, Faust SN, Nicosia A, Hill AVS, Draper SJ. ChAd63-MVA–vectored Blood-stage Malaria Vaccines Targeting MSP1 and AMA1: Assessment of Efficacy Against Mosquito Bite Challenge in Humans. Mol Ther. 2012 doi: 10.1038/mt.2012.223. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, Guindo AB, Traore K, Traore I, Kouriba B, Diallo DA, Diarra I, Daou M, Dolo A, Tolo Y, Sissoko MS, Niangaly A, Sissoko M, Takala-Harrison S, Lyke KE, Wu Y, Blackwelder WC, Godeaux O, Vekemans J, Dubois MC, Ballou WR, Cohen J, Thompson D, Dube T, Soisson L, Diggs CL, House B, Lanar DE, Dutta S, Heppner DG, Jr., Plowe CV. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365:1004–1013. doi: 10.1056/NEJMoa1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis RD, Martin LB, Shaffer D, Long CA, Miura K, Fay MP, Narum DL, Zhu D, Mullen GE, Mahanty S, Miller LH, Durbin AP. Phase 1 trial of the Plasmodium falciparum blood stage vaccine MSP1(42)-C1/Alhydrogel with and without CPG 7909 in malaria naive adults. PLoS One. 2010;5:e8787. doi: 10.1371/journal.pone.0008787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duncan CJ, Sheehy SH, Ewer KJ, Douglas AD, Collins KA, Halstead FD, Elias SC, Lillie PJ, Rausch K, Aebig J, Miura K, Edwards NJ, Poulton ID, Hunt-Cooke A, Porter DW, Thompson FM, Rowland R, Draper SJ, Gilbert SC, Fay MP, Long CA, Zhu D, Wu Y, Martin LB, Anderson CF, Lawrie AM, Hill AV, Ellis RD. Impact on Malaria Parasite Multiplication Rates in Infected Volunteers of the Protein-in-Adjuvant Vaccine AMA1-C1/Alhydrogel+CPG 7909. PLoS One. 2011;6:e22271. doi: 10.1371/journal.pone.0022271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusi KA, Faber BW, Thomas AW, Remarque EJ. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One. 2009;4:e8110. doi: 10.1371/journal.pone.0008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remarque EJ, Faber BW, Kocken CH, Thomas AW. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect Immun. 2008;76:2660–2670. doi: 10.1128/IAI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biswas S, Dicks MD, Long CA, Remarque EJ, Siani L, Colloca S, Cottingham MG, Holder AA, Gilbert SC, Hill AV, Draper SJ. Transgene Optimization, Immunogenicity and In Vitro Efficacy of Viral Vectored Vaccines Expressing Two Alleles of Plasmodium falciparum AMA1. PLoS One. 2011;6:e20977. doi: 10.1371/journal.pone.0020977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Draper SJ, Biswas S, Spencer AJ, Remarque EJ, Capone S, Naddeo M, Dicks MDJ, Faber BW, de Cassan SC, Folgori A, Nicosia A, Gilbert SC, Hill AVS. Enhancing blood-stage malaria subunit vaccine immunogenicity in rhesus macaques by combining adenovirus, poxvirus, and protein-in-adjuvant vaccines. J Immunol. 2010;185:7583–7595. doi: 10.4049/jimmunol.1001760. [DOI] [PubMed] [Google Scholar]
- 34.Goodman AL, Epp C, Moss D, Holder AA, Wilson JM, Gao GP, Long CA, Remarque EJ, Thomas AW, Ammendola V, Colloca S, Dicks MD, Biswas S, Seibel D, van Duivenvoorde LM, Gilbert SC, Hill AV, Draper SJ. New candidate vaccines against blood-stage Plasmodium falciparum malaria: prime-boost immunization regimens incorporating human and simian adenoviral vectors and poxviral vectors expressing an optimized antigen based on merozoite surface protein 1. Infect Immun. 2010;78:4601–4612. doi: 10.1128/IAI.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanabe K, Mackay M, Goman M, Scaife JG. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert SC, Plebanski M, Gupta S, Morris J, Cox M, Aidoo M, Kwiatkowski D, Greenwood BM, Whittle HC, Hill AV. Association of malaria parasite population structure, HLA, and immunological antagonism. Science. 1998;279:1173–1177. doi: 10.1126/science.279.5354.1173. [DOI] [PubMed] [Google Scholar]
- 37.Plebanski M, Lee EA, Hannan CM, Flanagan KL, Gilbert SC, Gravenor MB, Hill AV. Altered peptide ligands narrow the repertoire of cellular immune responses by interfering with T-cell priming. Nat Med. 1999;5:565–571. doi: 10.1038/8444. [DOI] [PubMed] [Google Scholar]
- 38.Lee EA, Flanagan KL, Minigo G, Reece WH, Bailey R, Pinder M, Hill AV, Plebanski M. Dimorphic Plasmodium falciparum merozoite surface protein-1 epitopes turn off memory T cells and interfere with T cell priming. Eur J Immunol. 2006;36:1168–1178. doi: 10.1002/eji.200526010. [DOI] [PubMed] [Google Scholar]
- 39.Lee EA, Flanagan KL, Odhiambo K, Reece WH, Potter C, Bailey R, Marsh K, Pinder M, Hill AV, Plebanski M. Identification of frequently recognized dimorphic T-cell epitopes in plasmodium falciparum merozoite surface protein-1 in West and East Africans: lack of correlation of immune recognition and allelic prevalence. Am J Trop Med Hyg. 2001;64:194–203. doi: 10.4269/ajtmh.2001.64.194. [DOI] [PubMed] [Google Scholar]
- 40.Klenerman P, Meier UC, Phillips RE, McMichael AJ. The effects of natural altered peptide ligands on the whole blood cytotoxic T lymphocyte response to human immunodeficiency virus. Eur J Immunol. 1995;25:1927–1931. doi: 10.1002/eji.1830250720. [DOI] [PubMed] [Google Scholar]
- 41.Kaneko T, Moriyama T, Udaka K, Hiroishi K, Kita H, Okamoto H, Yagita H, Okumura K, Imawari M. Impaired induction of cytotoxic T lymphocytes by antagonism of a weak agonist borne by a variant hepatitis C virus epitope. Eur J Immunol. 1997;27:1782–1787. doi: 10.1002/eji.1830270728. [DOI] [PubMed] [Google Scholar]
- 42.Quin SJ, Seixas EM, Cross CA, Berg M, Lindo V, Stockinger B, Langhorne J. Low CD4(+) T cell responses to the C-terminal region of the malaria merozoite surface protein-1 may be attributed to processing within distinct MHC class II pathways. Eur J Immunol. 2001;31:72–81. doi: 10.1002/1521-4141(200101)31:1<72::aid-immu72>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 43.Stanisic DI, Martin LB, Liu XQ, Jackson D, Cooper J, Good MF. Analysis of immunological nonresponsiveness to the 19-kilodalton fragment of merozoite surface Protein 1 of Plasmodium yoelii: rescue by chemical conjugation to diphtheria toxoid (DT) and enhancement of immunogenicity by prior DT vaccination. Infect Immun. 2003;71:5700–5713. doi: 10.1128/IAI.71.10.5700-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauerwein RW, Roestenberg M, Moorthy VS. Experimental human challenge infections can accelerate clinical malaria vaccine development. Nat Rev Immunol. 2011;11:57–64. doi: 10.1038/nri2902. [DOI] [PubMed] [Google Scholar]
- 45.Hensmann M, Li C, Moss C, Lindo V, Greer F, Watts C, Ogun SA, Holder AA, Langhorne J. Disulfide bonds in merozoite surface protein 1 of the malaria parasite impede efficient antigen processing and affect the in vivo antibody response. Eur J Immunol. 2004;34:639–648. doi: 10.1002/eji.200490004. [DOI] [PubMed] [Google Scholar]
- 46.Huaman MC, Martin LB, Malkin E, Narum DL, Miller LH, Mahanty S, Long CA. Ex vivo cytokine and memory T cell responses to the 42-kDa fragment of Plasmodium falciparum merozoite surface protein-1 in vaccinated volunteers. J Immunol. 2008;180:1451–1461. doi: 10.4049/jimmunol.180.3.1451. [DOI] [PubMed] [Google Scholar]
- 47.Egan A, Waterfall M, Pinder M, Holder A, Riley E. Characterization of human T- and B-cell epitopes in the C terminus of Plasmodium falciparum merozoite surface protein 1: evidence for poor T-cell recognition of polypeptides with numerous disulfide bonds. Infect Immun. 1997;65:3024–3031. doi: 10.1128/iai.65.8.3024-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crisanti A, Fruh K, Muller HM, Bujard H. The T cell reactivity against the major merozoite protein of Plasmodium falciparum. Immunol Lett. 1990;25:143–148. doi: 10.1016/0165-2478(90)90106-z. [DOI] [PubMed] [Google Scholar]
- 49.Udhayakumar V, Anyona D, Kariuki S, Shi YP, Bloland PB, Branch OH, Weiss W, Nahlen BL, Kaslow DC, Lal AA. Identification of T and B cell epitopes recognized by humans in the C-terminal 42-kDa domain of the Plasmodium falciparum merozoite surface protein (MSP)-1. J Immunol. 1995;154:6022–6030. [PubMed] [Google Scholar]
- 50.Allsopp CE, Harding RM, Taylor C, Bunce M, Kwiatkowski D, Anstey N, Brewster D, McMichael AJ, Greenwood BM, Hill AV. Interethnic genetic differentiation in Africa: HLA class I antigens in The Gambia. Am J Hum Genet. 1992;50:411–421. [PMC free article] [PubMed] [Google Scholar]
- 51.Luo M, Embree J, Ramdahin S, Ndinya-Achola J, Njenga S, Bwayo JB, Pan S, Mao X, Cheang M, Stuart T, Brunham RC, Plummer FA. HLA-A and HLA-B in Kenya, Africa: allele frequencies and identification of HLA-B*1567 and HLA-B*4426. Tissue Antigens. 2002;59:370–380. doi: 10.1034/j.1399-0039.2002.590503.x. [DOI] [PubMed] [Google Scholar]
- 52.Conway DJ, Greenwood BM, McBride JS. Longitudinal study of Plasmodium falciparum polymorphic antigens in a malaria-endemic population. Infect Immun. 1992;60:1122–1127. doi: 10.1128/iai.60.3.1122-1127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, Kone AK, Traore K, Ouattara A, Djimde AA, Sehdev PS, Lyke KE, Diallo DA, Doumbo OK, Plowe CV. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babiker HA, Creasey AM, Fenton B, Bayoumi RA, Arnot DE, Walliker D. Genetic diversity of Plasmodium falciparum in a village in eastern Sudan. 1. Diversity of enzymes, 2D-PAGE proteins and antigens. Trans R Soc Trop Med Hyg. 1991;85:572–577. doi: 10.1016/0035-9203(91)90347-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.