Abstract
Voltage-gated sodium (Nav) channels and their Na+/K+ selectivity are of great importance in the mammalian neuronal signaling. According to mutational analysis, the Na+/K+ selectivity in mammalian Nav channels is mainly determined by the Lys and Asp/Glu residues located at the constriction site within the selectivity filter. Despite successful molecular dynamics simulations conducted on the prokaryotic Nav channels, the lack of Lys at the constriction site of prokaryotic Nav channels limits how much can be learned about the Na+/K+ selectivity in mammalian Nav channels. In this work, we modeled the mammalian Nav channel by mutating the key residues at the constriction site in a prokaryotic Nav channel (NavRh) to its mammalian counterpart. By simulating the mutant structure, we found that the Na+ preference in mammalian Nav channels is collaboratively achieved by the deselection from Lys and the selection from Asp/Glu within the constriction site.
Introduction
Voltage-gated sodium (Nav) channels are responsible for the rising phase of the action potential by mediating the permeation of Na+ ions across the membrane of most electrically excitable cells (1,2). The dysfunction of Nav channels frequently causes severe diseases such as epileptic seizures, migraine, and cardiac arrhythmias (3). In contrast to the extensive structural and mechanistic studies on the ion selection in voltage-gated potassium (Kv) channels (4–11), the corresponding research studies for Nav channels have been lagging behind for many years, due to the lack of atomic-resolution structures. The crystal structures of prokaryotic Nav channels, NavAb (12,13) and NavRh (14), were successively solved recently. Prokaryotic Nav channels are composed of four identical subunits, each of which contains six transmembrane helices (12,14–17). Helices S1–S4 of each protomer form a voltage-sensing domain located at periphery, which regulates the channel opening upon membrane depolarization (15–17). Helices S5–S6 as well as the intervening half helices and P-loops from the four protomers are clustered at the center to constitute the pore domain, which allows the transmembrane permeation of cations (15–17,19). Within the pore domain, each P-loop provides five (12,13,20) or six (14,21) residues to constitute a narrow vestibule that determines the ion selectivity and is therefore named as the selectivity filter (SF). (Fig. 1 A).
Figure 1.
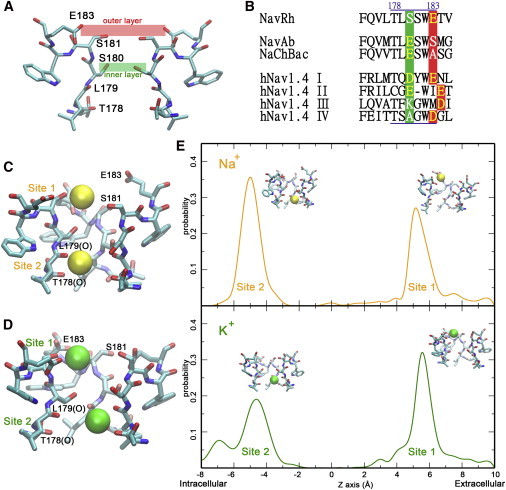
The SF of NavRh is less discriminative between Na+ and K+. (A) The SF of NavRh contains two layers of residues that correspond to the conserved residues in mammalian Nav channels. Only the two diagonal protomers of NavRh are shown for clarity. (B) Sequence alignment of the SF regions of several Nav channels. (C and D) In simulations, two binding sites are identified in the SF of WT-NavRh for both Na+ (C) and K+ (D). (E) The calculated residence probability along the permeation pathway for Na+ (upper panel) and K+ (lower panel).
Unlike the architecture of homotetramer in prokaryotic Nav channels, the corresponding structural units in mammalian Nav channels (repeats I to IV in Fig. S1 in the Supporting Material) are linearly connected in a single polypeptide chain (16), which allows the asymmetrical distribution of amino acid residues lining the SF. Notably, the SF of mammalian Nav channels contains two layers of highly conserved residues (16), as shown on the NavRh structure (14) in Fig. 1 A based on sequence alignment. The outer layer, which is composed of three or four acidic residues (shaded in red in Fig. 1 B and Fig. S1), is associated with the efficiency of ion permeation (22). The inner layer, which is invariantly composed of Asp, Glu, Lys, and Ala (or DEKA as shaded in green in Fig. 1 B and Fig. S1), forms the geometrically narrowest region, which is called the constriction site. The inner layer has been identified as the major determinant on ion selectivity in mammalian Nav channels by mutation experiments (23–25). Among the four key residues (DEKA), Lys is absolutely indispensable in maintaining ion selection, because its mutation to Arg, Ala, or Gln either disrupts or cripples the ion selectivity (23–25). Besides Lys, the presence of at least one acidic residue (Asp or Glu) is required for proper Na+/K+ selection (24).
The Na+/K+ selectivity of mammalian Nav channels was analyzed theoretically before the crystallographic studies. According to the static hypothesis proposed by Favre et al. (24), the salt bridge between Lys and Asp/Glu at the constriction site constrained the pore size to better accommodate Na+ rather than K+. Lipkind and Fozzard (26,27) later proposed a dynamic hypothesis, stating that as a stronger Lewis acid, Na+ was more likely to displace the positive side chain of Lys, which blocked the SF by forming salt bridge with the opposing Asp/Glu. Dudev and Lim (28–30) mimicked the constriction site of the SF using model compounds and systematically studied the factors affecting the Na+/K+ selectivity in Nav channels by quantum mechanics calculation (28).
Since the determination of the first crystal structure of prokaryotic Nav channels, molecular dynamics (MD) simulations have been engaged to explore the ion selectivity by several independent groups (31–38). The simulations on NavAb showed a marked difference in the free energy profiles of Na+ and K+ ions (32,33). Size restraint and hydration number have been assumed to be responsible for the selectivity (31–34). In our simulation on NavRh, we found that Ca2+ permeation was disfavored due to the presence of a huge energy barrier at the constriction site (35).
Despite the success of simulation studies on the prokaryotic Nav channels, insightful hints on the mechanism of ion selectivity in mammalian Nav channels are missing due to the absence of Lys and Asp/Glu in prokaryotic Nav channels. Although some mechanistic models have been proposed to illustrate the roles of these key residues, the validity of these hypotheses is impaired by the lack of atomistic details. In this work, we first investigated the Na+/K+ selectivity in the wild-type NavRh (WT-NavRh) channel. After that, residues (Ser-180) at the constriction site of NavRh were mutated to DEKA to mimic the SF of mammalian Nav channels. By simulating the mutant structure, we finally proposed a model to explain the Na+/K+ selectivity in mammalian Nav channels: Lys screens most weakly Na+-selective locations within the constriction site through electrostatic repulsion, and repels the cation to pass through a highly Na+-selective location formed by the clustered carboxylate groups of Asp and Glu.
Results
WT-NavRh SF has weak Na+/K+ selectivity
Similar to our previous research (35), the pore domain of the NavRh crystal structure was simulated in 70 mM NaCl and KCl, respectively (simulation 1 and 2 in Table S1). In both simulations, extracellular cations initially lingered around the binding site 1, which is composed of the side groups of Ser-181 and Glu-183, and then quickly squeezed into the SF and stably resided at binding site 2, a site composed of eight carbonyl oxygens from Thr-178 and Leu-179 (Fig. 1 and Fig. S2; Movie S1 and Movie S2). The two binding sites described here are consistent with the ones observed in our previous research (35).
The Na+/K+ selectivity was then evaluated at both binding sites using the free energy perturbation (FEP) (39,40), by calculating the relative binding affinity ΔΔG(Na+→K+) (see Fig. S3), which is positive for a Na+-selective site and negative for a K+-selective site. As shown in Table 1 and Table S2, both sites are weakly Na+-selective (ΔΔG(Na+→K+) < 1.5 kcal/mol), and the ion bound at the inner site does not raise the ion selectivity at the outer site. The weak Na+/K+ selectivity is also indirectly implicated from the lack of a significant difference between the binding times of Na+ and K+ (Table S3 and Fig. S4) when equal amounts of these ions compete for site 1 in the presence of a cation bound at site 2 (simulations 3 and 4 in Table S1).
Table 1.
FEP calculations to evaluate ion selectivity
| ID | ΔΔG(Na+→K+) in WT-NavRh (kcal/mol) |
||
|---|---|---|---|
| Site 1 | Site 2 | ||
| 1 | 0.70 (0.08) | Ø | |
| 2 | Ø | 0.63 (0.56) | |
| 3 | 0.74 (0.85) | Na+ | |
| 4 | 0.04 (0.09) |
K+ |
|
| ΔΔG(Na+→K+) in the DEKA mutant (kcal/mol) |
|||
| Site 1 | Site HFS | Site 2 | |
| 5 | 0.74 (0.55) | Ø | Ø |
| 6 | Ø | 1.25 (1.65) | Ø |
| 7 | Ø | Ø | 0.68 (0.16) |
| 8 | 0.33 (0.91) | Ø | Na+ |
| 9 | Ø | 4.57 (0.03) | Na+ |
| 10 | 0.17 (0.39) | Ø | K+ |
| 11 | Ø | 2.38 (0.44) | K+ |
| 12 | 1.06 (1.12) | Na+ | Na+ |
| 13 | Na+ | 3.39 (1.42) | Na+ |
| 14 | Na+ | Na+ | 1.85 (1.21) |
Ø indicates unoccupied site; Na+ and K+ indicate the presence of these ions at the site; detailed information for each calculation is listed in Table S2, labeled by the calculation ID. The uncertainties, which are roughly estimated from the difference between the forward and backward FEP calculations, are listed in parenthesis. Results obtained from independently repeated FEP calculations are listed in Table S8 to show the good reproducibility of the FEP calculations.
Notably, the lack of strong Na+/K+ selectivity will not impair the survivability of the host (Rickettsiales sp. HIMB114), a species living in the deep ocean where the K+:Na+ ratio is greatly lower than that in the mammalian extracellular environment (41,42). Furthermore, the high ion selectivity, which is critical for the fast and accurate neuronal signaling in the mammalian nerve system, is possibly nonessential for a marine bacterium channel whose function may be related with cell motility and chemotaxis (43,44).
NavRh was mutated to model the mammalian Nav channels
In the highly Na+-selective mammalian Nav channels, the constriction site is composed of four nonidentical amino acid residues (DEKA), which are equivalent to the Ser-180 in NavRh. (Fig. 1 B and Fig. S1). To explore the roles of these residues in the ion selectivity of mammalian Nav channels, we mutated Ser-180 in the four protomers of NavRh crystal structure to DEKA, respectively (Fig. S5 A). The mutation did not disrupt the structure of the channel, as shown by the small structural deviation (<2 Å) in the backbones of both the SF and the pore domain after mutation. (Fig. S5 B and Fig. S6).
The permeability of DEKA-mutant SF depends on Lys-180 position
The DEKA mutant was simulated in 70 mM NaCl and KCl solutions, respectively. In both cases, the SF of the mutant can either allow or reject ion permeation, depending on the status of Lys-180 side chain. In a couple of simulations (simulations 6 and 8 in Table S1), the side chain of Lys-180 (chain C) stretches into the center of SF and interacts with the opposing carboxylate groups of Asp-180 and Glu-180, a posture that blocks the permeation pathway (Fig. S5 C, upper panel; Movie S3). Only when the side chain of Lys-180 moves upward to interact with the carboxylate group of Glu-183 in the neighboring chain (chain D), could the ion permeate across the constriction site (Fig. S5 C, lower panel; Movie S4). This can be clearly seen from the marked difference in the atomic distances between the side-chain nitrogen of Lys-180 (chain C) and the carboxylate oxygens of Glu-183 (chain D) measured from trajectories of successful and failed ion permeations, respectively (Fig. S7). In the permeable SF, the distance is mainly distributed at <3.5 Å, a condition defined as the on-state of Lys-180 side chain in our later simulations. In contrast, the distance exceeds 4 Å in the impermeable SF, and the Lys-180 side chain in those simulations is defined as the off-state. The dependence of the permeability on the status of Lys-180 side chain was further validated by the free energy profiles estimated using the adaptive biasing force method (45–48), when the Lys-180 side chain was constrained to the on and off states, respectively. As shown in Fig. S8, both Na+ and K+ can permeate through the SF free of barrier when Lys-180 side chain is at the on-state. In contrast, high free energy barriers prevent ion passage when Lys-180 side chain is at the off-state.
The previous observation conspicuously denies the static hypothesis (24) because neither cation can penetrate when Lys-180 interacts with Asp-180 or Glu-180. Although Na+ and K+ apparently show similar propensities to displace Lys-180 and to bind with Asp-180 and Glu-180, as also suggested by Dudev and Lim (28), the limited number of equilibrium simulations conducted here cannot provide sufficient evidence to exclude the dynamic hypothesis (26,27). More systematic equilibrium simulations should be conducted in the future work to further test the correctness of this hypothesis. To further explore the mechanism of ion selection, the Lys-180 side chain is restrained to the on-state in the following simulations to guarantee the opening of the SF. Notably, the switch between the on/off states of Lys-180 may be affected by the change of the cross-membrane potential during neuronal signaling. For instance, depolarization will favor the upward movement of the positively charged Lys-180 side chain and promote the opening of the SF. Correspondingly in the rising phase of an action potential, the cross-membrane potential grows slowly in the early stage but rises much more rapidly once it reaches a threshold, partially because of the improved conductance associated with the elevated probability of Lys to switch to its on-state at the depolarizing potential. However, because the conductance of Nav channels is determined by factors other than the SF region, the previous speculation still awaits experimental validation.
Na+ and K+ permeate across the DEKA-mutant SF in distinct manners
Once Lys-180 is constricted to the on-state, both Na+ and K+ can permeate across the SF. In Na+ permeation (simulation 5 in Table S1), the first Na+ ion quickly approached the carboxylate groups of Glu-183. After transiently lingering around the carboxylate groups of Asp-180 and Glu-180, the ion finally reached the site formed by the carbonyl groups of Thr-178 and Leu-179 (Movie S5). The outer and inner sites are identical to those in WT-NavRh, and are therefore still named as site 1 and site 2, respectively. Between them is one additional site, site HFS (high field strength) formed by the carboxylate groups of Asp-180 and Glu-180 (Fig. 2 A and the upper panel of Fig. 2 C; Fig. S9, upper panel; Fig. S10). After the first ion occupied site 2, two more extracellular Na+ ions approached site HFS and site 1 successively. Ultimately, three Na+ ions were bound in the SF simultaneously (Movie S5). Notably, the single-ion free energy profile only indicates one energetically favored binding site, which corresponds to site 2 (Fig. S8). This can explain the natural downhill permeation of the first ion from the extracellular solution to the inner site. After the occupancy of site 2, the free energy profile changes drastically, and two more energetically favorable binding sites show up (Fig. S11), which agree well with the site HFS and site 1 identified in the equilibrium simulations (Fig. 2 C).
Figure 2.
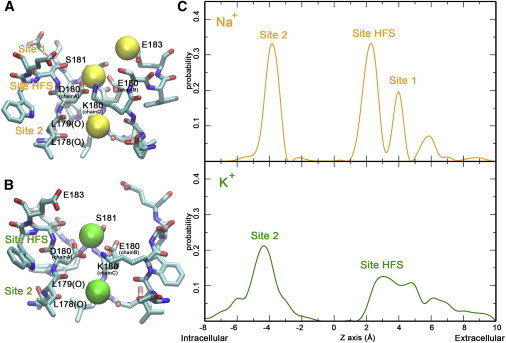
The SF of the DEKA mutant of NavRh exhibits distinct binding sites for Na+ and K+, respectively. (A and B) Three and two binding sites are identified in the SF of the DEKA mutant of NavRh for Na+ (A) and K+ (B), respectively. (C) The calculated residence probability along the permeation pathway for Na+ (upper panel) and K+ (lower panel).
The profile is different in K+ permeation. Once the first K+ ion reaches the inner site (site 2), only one additional binding site (site HFS) emerges (Fig. 2 B and the lower panel of Fig. 2 C; Fig. S9, lower panel). In addition, the site HFS of K+ is slightly shifted toward the extracellular solution (Fig. 2 C and Fig. S11), and therefore interferes with the ion binding at site 1 (Fig. 2 C, lower panel). Consequently, only two K+ ions were observed to bind in the SF in the equilibrium simulation (simulation 7 in Table S1, Movie S6).
Strong Na+/K+ selectivity at site HFS after conformational change
As shown in Table 1 and Table S2, all three sites of the DEKA mutant are weakly Na+-selective (ΔΔG(Na+→K+) < 1.5 kcal/mol). However, once site 2 is preoccupied by Na+ or K+, the ΔΔG(Na+→K+) at site HFS grows to 4.57 and 2.38 kcal/mol, respectively, which roughly correspond to 1000-fold and 50-fold selectivity at the body temperature according to the Boltzmann factor exp(−ΔΔG/RT). This can also be verified from the Na+/K+ competition at site HFS in the presence of an ion bound at site 2 (simulations 9 and 10 in Table S1). In both simulations, a Na+ ion won the competition after several rounds of turnovers and resided stably at the site HFS until the end of the trajectory (Fig. S4). The previous high difference (50-fold and 1000-fold) in the binding affinity of Na+ and K+ at site HFS apparently contradict with the 10- to 30-fold difference in the permeability of Na+ and K+ measured in the electrophysiological experiment (23). This contradiction can be reconciled by considering that the permeability is determined kinetically by the free energy profile in the overall permeation pathway rather than by the binding affinity at one specific energy minimum in the pathway. Notably, the presence of Na+ ions at the two extracellular sites (site 1 and site HFS) also improves the Na+ preference at the inner site (site 2) by >1 kcal/mol (Table 1 and Table S2), which may reduce the interference by intracellular K+ during the consecutive cross-membrane conduction of Na+ in mammalian Nav channels.
The previous significant improvement in Na+/K+ selectivity at site HFS is associated with the conformational change of the channel after site 2 is occupied. When the first ion passes site HFS, the carboxylate groups from Asp-180 and Glu-180 are spatially separated. In this situation, only one carboxylate group can favorably interact with the permeating ion without backbone movement (Fig. S12, left panel). Once the inner site is occupied, the preclusion of the forward permeation leads to the long residence of the incoming cation at site HFS, which allows the backbone movement of both chain A and chain B. The face-to-face backbone movement of both chains then geometrically permits the approximation of the side-chain carboxylate groups of Asp-180 and Glu-180 on them (Fig. S12, right panel). Finally, the cation is sandwiched between both carboxylate groups, a conformation that sufficiently stabilizes the cation bound at site HFS (Fig. S11). The new conformation is highly Na+ selective, as indicated by the large ΔΔG(Na+→K+) of 4.79 kcal/mol in the control experiment in which the ion located at site 2 was removed to exclude its effect and the protein backbone conformation was maintained by strong positional restraints (Table S2).
The role of Lys in Na+/K+ selectivity
Although the significant improvement in ion selectivity by the DEKA mutant indicates the crucial roles of these four residues, they do not have equal contribution to Na+/K+ selectivity. In fact, Lys was reported to be the most important residue, because a single K to A mutation completely disrupted the high Na+/K+ selectivity in mammalian Nav channels (24). To better understand the role of Lys in Na+/K+ selectivity, a DEAA mutant of NavRh was generated and was simulated as a control (simulations 11 and 12 in Table S1). Due to the lack of the Lys side chain at the constriction site, this mutant is always permeable for both Na+ and K+. To analyze the ion selectivity at site HFS, the SF structures were extracted from the highly Na+-selective conformation of the DEKA mutant and from one conformation of the DEAA mutant (after sufficient equilibration in 70 mM NaCl), respectively, and all atoms outside the SF were neglected to accelerate the calculation. The SF regions were blocked at the chain termini and were immersed in water boxes with strong positional constraints to maintain their backbone conformations (Fig. S13 A and Fig. S13 B). Later, a cation (Na+ or K+) was allowed to sample in the plane of the constriction site (Fig. S13 C) to derive the two-dimensional free energy profiles using the adaptive biasing force method (Fig. 3 A and B).
Figure 3.
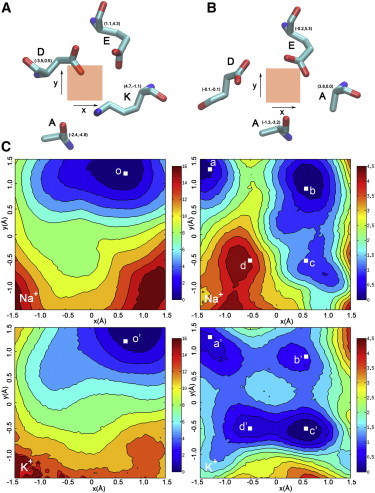
The two-dimensional free-energy profiles at the constriction site suggest the role of Lys in Na+/K+ selection. (A and B) A cation is forced to sample in the plane of the constriction site of the DEKA (A) and DEAA (B) mutants, where the shaded squares at the center indicate the allowed sampling space. The key residues at the constriction site taken from one snapshot of the according simulations are shown around the squares. The relative coordinate of the α-carbon is shown for each key residue. (C) The two-dimensional free-energy maps of the DEKA (left column) and DEAA (right column) mutants were calculated for Na+ (upper row) and K+ (lower row), respectively. A representative cation coordinate (labeled by white squares) was selected from each local minimum, and FEP calculation was engaged to evaluate the ion selectivity at the location (Table S4). The representative structures for the previous selected locations are shown in Fig. S14 with the same letter labeling.
In the DEKA mutant, only one energetically favorable location is present for both Na+ and K+ (o and o′ in Fig. 3 C, left column), where the cation is sandwiched by the carboxylate groups of Asp-180 and Glu-180, a conformation indicative of high Na+/K+ selectivity (Fig. S14). Accordingly, ΔΔG(Na+→K+) at this location is 2.05 kcal/mol (Table S4). In contrast, the free-energy maps of both Na+ and K+ in the DEAA mutant are much less restrictive (Fig. 3 C, right column). In particular, K+ has four energetically favorable locations, three of which are also local minima for Na+. The upper left minimum (a and a′) is highly Na+ selective (ΔΔG(Na+→K+) = 2.05 kcal/mol in Table S4), because the cation is sandwiched by the carboxylate groups of Asp-180 and Glu-180 (Fig. S14). At the upper right (b and b′) and lower right minima (c and c′), Na+ is less preferred (ΔΔG(Na+→K+) = 1.16 and 1.64 kcal/mol, respectively, in Table S4), because the cation is only coordinated by the carboxylate group of Glu-180 (Fig. S14). The local minimum in the lower left energy well (d and d′) on the K+ free-energy map is disfavored for Na+, as indicated by the ΔΔG(Na+→K+) of −1.09 kcal/mol (Table S4). At this location, the cation interacts more intimately with the carbonyl oxygen of Ala-180 than with the carboxylate groups (Fig. S14).
In summary, in the absence of Lys side chain, the cation can stay at several locations within the constriction site, most of which are weakly Na+ selective or even K+ selective. The presence of Lys side chain, however, precludes all less Na+-selective locations by electrostatic and geometrical repulsion. Consequently, the cation has to permeate through the only available location (between Asp and Glu), which is highly Na+ selective. The free energy analysis suggests that Lys at the constriction site renders Na+/K+ selectivity through a negative design (or deselection).
The role of Asp/Glu in Na+/K+ selectivity
The previous analysis leads to another question: Why is Na+ highly preferred when sandwiched by two carboxylate groups? To address this question, we first evaluated the Na+/K+ selectivity of a carboxylate group (in acetate) and a carbonyl group (in N-methylacetylamide or NMA) (Fig. S15). As shown in Table S5, the carboxylate group slightly prefers Na+, whereas the carbonyl group weakly favors K+. This difference can partially rationalize the distinct structural design in the SF of Nav and Kv channels: Kv channels expose backbone carbonyls to favor the K+ coordination (5–7), whereas Nav channels use side-chain carboxylate groups to prioritize the passage of Na+ ions (12–14,20). This agrees with multiple previous research studies, where both the field strength and the number of the coordinating groups have been proposed to be consequential in ion selection by solvating the target ion in a symmetrical environment composed of homogenous ion-coordinating groups to mimic the binding site in potassium channels (50–55). Although the cation is highly hydrated and frequently coordinates asymmetrically to only one or two protein groups in sodium channels, the field strength of the coordinating group can still weakly determine the ion selectivity.
In the previous calculation, the cations are restrained within 3 Å to the center of the carboxylate oxygens, which slightly allows monodentate interaction between the cation and the carboxylate group. When cations are restricted in a bidentate binding state (within 2.6 Å to both carboxylate oxygens), Na+ preference rises to the level of the site HFS in the DEKA mutant (ΔΔG(Na+→K+) = 2.62 kcal/mol in Table S5). On the other hand, in a control simulation where asymmetrical binding is allowed to a larger extent (by restraining the cation within 5 Å to the center of the carboxylate oxygens), Na+ is more likely to stay in a bidentate state than K+, as clearly suggested from their distinct distributions of the difference in ion-oxygen distances (Fig. S16).
In the following simulations, the cations were forced to locate at equal distances to both carboxylate oxygens of acetate to explore why Na+ is highly preferred at the bidentate binding state (Fig. S17 A). The coordination numbers of Na+ and K+ were then counted and statistically fitted with Gaussian distribution, respectively (Fig. S17 B). When the cation is well chelated by the carboxylate group (both ion-oxygen distances ≤2.4 Å), as in the site HFS (Fig. S16 D), the coordination number of K+ ions drops greatly from the ideal value (sevenfold coordination in water) (Fig. S17 C, lower panel). Therefore, K+ has to sacrifice the favorable electrostatic interaction in the bidentate binding state and choose a monodentate binding manner to preserve its ideal coordination state. Na+, on the contrary, can optimize the favorable electrostatic interaction by bidentate binding but with only a tiny loss in its ideal coordination number (sixfold coordination in water) (Fig. S17 C, upper panel). This difference leads to the high Na+/K+ selectivity at site HFS in the mammalian Nav channels, where the electrostatic bias for the bidentate binding is amplified by the closely clustered carboxylate groups of Asp and Glu (Fig. S16 D). Accordingly, K+ has to pay more energetic cost to maintain the monodentate interactions with both carboxylate groups. These observations agree with the research of Bostick et al. (50), where the ion coordination was systematically analyzed by simulations using polarizable force field. In this insightful work, K+ was proposed to be highly favored when constrained in a state of eightfold coordination by the carbonyl oxygens in the SF of potassium channels. Similarly, according to their free energy analysis (50), Na+ should be favored when constrained in a state of sixfold coordination, the situation when the cation is well chelated by the carboxylate group. Similarly, Corry and coworkers (51–53) proved that larger ions such as K+ are effectively favored in over coordinating environment in a series of studies. In addition, through quantum mechanics calculations, Dudev and Lim also proposed that disruption on the coordination state of ions exerted by the protein coordinating groups favors the cation with a smaller coordination number (28).
In summary, the high Na+/K+ selectivity at site HFS is generated by the chemical property of the carboxylate group, which is naturally designed to prefer Na+ to K+ in the chelated state. Consistent with mutational analysis (24), at least one acidic residue is required for Na+ selection and the presence of both Asp and Glu improves the Na+/K+ selectivity by enforcing the bidentate binding at site HFS. In contrast to Lys, Asp/Glu at the constriction site renders Na+/K+ selectivity through a positive design (or direct selection). Notably, the role of Asp/Glu was analyzed principally on model compounds solvated in a water box and therefore still awaits the validation by more stringent computations.
Discussion
Implications in the mammalian Nav channels
According to the sequence similarity (Fig. 1 B and Fig. S1), the SF of mammalian Nav channels is proposed to adopt a similar structural framework to that of NavRh. Therefore, the mammalian Nav channels may follow a mechanism of ion permeation and ion selection similar to the DEKA mutant of NavRh (Fig. 4).
Figure 4.
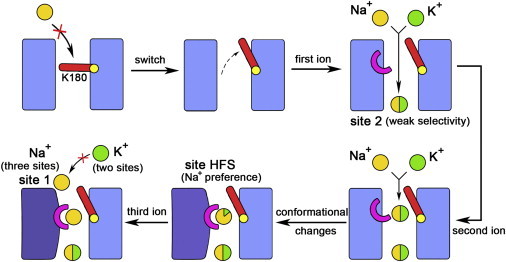
Schematic representation for the mechanism of the mammalian Nav channels. Na+ and K+ ions are represented by yellow and green spheres, respectively, and the ion selectivity is reflected by the ratio of yellow and green colors in one sphere. Lys and Asp/Glu are represented by the red bar and the pink half ring, respectively. The conformational changes are indicated by changes in both the color and the shape of the rectangles, which represent the pore domain.
In the mammalian Nav channels, the side chain of Lys at the constriction site may control the opening and closing of the SF (Fig. 4). Once the Lys side chain moves upward to allow ion permeation, an acidic residue in the outer layer may hold the Lys side chain at the on-state to sustain the opening of the SF. Otherwise, the occasional downward movement of Lys side chain will occlude the SF and impair the efficiency of ion permeation. This important acidic residue, which is equivalent to the Glu-183 (chain D) in the DEKA mutant of NavRh, is invariably Asp in all mammalian Nav channels (shaded red in domain IV in Fig. S1). Despite its shorter side chain, Asp can make favorable interaction with the Lys side chain, as suggested by our simulation (simulation 13 in Table S1) in which Glu-183 in chain D of the DEKA mutant was further mutated to Asp (Fig. S18).
When Lys side chain is stabilized at the on-state, both Na+ and K+ can permeate across the constriction site composed of DEKA (Fig. 4). After the first ion occupies the inner site, the arrival of the second cation at the constriction site elicits a conformational change of the SF by drawing the carboxylate groups of both Asp and Glu at the constriction site to its proximity (Fig. 4). The new conformation becomes highly Na+ selective, a property that is cooperatively contributed by the deselection from Lys and the selection from Asp/Glu. Specifically, all less Na+-selective locations within the constriction site are disallowed for permeation by the electrostatic repulsion from Lys and the ion has to pass through the naturally highly Na+-selective location sandwiched between the carboxylate groups of Asp and Glu. Therefore, even if K+ accidentally arrives at the constriction site, it will be eventually displaced by the more favorable Na+ ions, as shown in the competition simulations (simulations 9 and 10 in Table S1; Fig. S4; Movie S7). Finally, the outer binding site is also occupied by a Na+ ion, and the three simultaneously bound Na+ ions allow the continuous Na+ conduction once the intracellular activation gate is open (Fig. 4).
The limitation of our simulation
Although the model proposed in this work presents an indication on how the Na+/K+ selectivity arises in mammalian Nav channels, there are still some limitations in our simulations. First, we applied a nonpolarizable force field to accelerate computation. In this manner, the nonnegligible charge transfer from the protein to the bound ion is omitted (56). Second, the high Na+/K+ selectivity in mammalian Nav channels was modeled through point mutation on the crystal structure of the prokaryotic channel NavRh. Third, the crystal structure of NavRh has a collapsed SF. Although the SF spontaneously opens in simulation (35), its conformation may have some different properties from those of the truly open SF in an active channel. Therefore, the model proposed here still awaits the validation by electrophysiological experiments and by the crystallographic studies on eukaryotic Nav channels.
Methods
The protein structure was processed in a similar way to our previous work (35). Atomic coordinates of the pore domain of NavRh channel were taken from the Protein Data Bank (PDB code: 4DXW). After membrane insertion, the system was simulated in 70 mM XCl, where X corresponds to Na+, K+, or their combination. Mutation on the protein structure was accomplished by the Mutator plugin of VMD 1.9.1 (57). All simulations were performed using NAMD 2.8 (58) in an NPT ensemble using the CHARMM27 force field with CMAP correction (59) and NBFIX (54). More detailed instructions can be found in the Supporting Material.
Several FEP calculations have been repeated using a different force field parameter set for the ions (38), and the result has been listed in the last table in the Supporting Material to show that our FEP calculations are robust on the choice of force field parameters.
Acknowledgments
This work was supported by National Natural Science Foundation of China (#31170674 and #31021002) and the Tsinghua National Laboratory for Information Science and Technology.
Supporting Material
References
- 1.Catterall W.A. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 2.Hille B. Sinauer; Sunderland, MA: 2001. Ion Channels of Excitable Membranes. [Google Scholar]
- 3.Mantegazza M., Curia G., Avoli M. Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet Neurol. 2010;9:413–424. doi: 10.1016/S1474-4422(10)70059-4. [DOI] [PubMed] [Google Scholar]
- 4.Durell S.R., Guy H.R. Structural model of the outer vestibule and selectivity filter of the Shaker voltage-gated K+ channel. Neuropharmacology. 1996;35:761–773. doi: 10.1016/0028-3908(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y.F., Morais-Cabral J.H., MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y.X., Lee A., MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 7.Long S.B., Campbell E.B., Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 8.Treptow W., Tarek M. K+ conduction in the selectivity filter of potassium channels is monitored by the charge distribution along their sequence. Biophys. J. 2006;91:L81–L83. doi: 10.1529/biophysj.106.095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baştuğ T., Kuyucak S. Importance of the peptide backbone description in modeling the selectivity filter in potassium channels. Biophys. J. 2009;96:4006–4012. doi: 10.1016/j.bpj.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furini S., Domene C. Selectivity and permeation of alkali metal ions in K+-channels. J. Mol. Biol. 2011;409:867–878. doi: 10.1016/j.jmb.2011.04.043. [DOI] [PubMed] [Google Scholar]
- 11.Bernèche S., Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414:73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 12.Payandeh J., Scheuer T., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payandeh J., Gamal El-Din T.M., Catterall W.A. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Ren W., Yan N. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charalambous K., Wallace B.A. NaChBac: the long lost sodium channel ancestor. Biochemistry. 2011;50:6742–6752. doi: 10.1021/bi200942y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu F.H., Catterall W.A. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci. STKE. 2004;2004:1–17. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 17.Koishi R., Xu H., Clapham D.E. A superfamily of voltage-gated sodium channels in bacteria. J. Biol. Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 18.Reference deleted in proof.
- 19.Guy H.R., Seetharamulu P. Molecular model of the action potential sodium channel. Proc. Natl. Acad. Sci. USA. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCusker E.C., Bagnéris C., Wallace B.A. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue L., Navarro B., Clapham D.E. The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 2002;120:845–853. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNulty M.M., Edgerton G.B., Lipkind G.M. Charge at the lidocaine binding site residue Phe-1759 affects permeation in human cardiac voltage-gated sodium channels. J. Physiol. 2007;581:741–755. doi: 10.1113/jphysiol.2007.130161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlief T., Schönherr R., Heinemann S.H. Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur. Biophys. J. 1996;25:75–91. doi: 10.1007/s002490050020. [DOI] [PubMed] [Google Scholar]
- 24.Favre I., Moczydlowski E., Schild L. On the structural basis for ionic selectivity among Na+, K+, and Ca2+ in the voltage-gated sodium channel. Biophys. J. 1996;71:3110–3125. doi: 10.1016/S0006-3495(96)79505-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y.M., Favre I., Moczydlowski E. On the structural basis for size-selective permeation of organic cations through the voltage-gated sodium channel. Effect of alanine mutations at the DEKA locus on selectivity, inhibition by Ca2+ and H+, and molecular sieving. J. Gen. Physiol. 1997;110:693–715. doi: 10.1085/jgp.110.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipkind G.M., Fozzard H.A. KcsA crystal structure as framework for a molecular model of the Na(+) channel pore. Biochemistry. 2000;39:8161–8170. doi: 10.1021/bi000486w. [DOI] [PubMed] [Google Scholar]
- 27.Lipkind G.M., Fozzard H.A. Voltage-gated Na channel selectivity: the role of the conserved domain III lysine residue. J. Gen. Physiol. 2008;131:523–529. doi: 10.1085/jgp.200809991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudev T., Lim C. Factors governing the Na(+) vs. K(+) selectivity in sodium ion channels. J. Am. Chem. Soc. 2010;132:2321–2332. doi: 10.1021/ja909280g. [DOI] [PubMed] [Google Scholar]
- 29.Dudev T., Lim C. Why voltage-gated Ca2+ and bacterial Na+ channels with the same EEEE motif in their selectivity filters confer opposite metal selectivity. Phys. Chem. Chem. Phys. 2012;14:12451–12456. doi: 10.1039/c2cp00036a. [DOI] [PubMed] [Google Scholar]
- 30.Dudev T., Lim C. Competition among Ca2+, Mg2+, and Na+ for model ion channel selectivity filters: determinants of ion selectivity. J. Phys. Chem. B. 2012;116:10703–10714. doi: 10.1021/jp304925a. [DOI] [PubMed] [Google Scholar]
- 31.Carnevale V., Treptow W., Klein M.L. Sodium ion binding sites and hydration in the lumen of a bacterial ion channel from molecular dynamics simulations. J. Phys. Chem. Lett. 2011;2:2504–2508. [Google Scholar]
- 32.Corry B., Thomas M. Mechanism of ion permeation and selectivity in a voltage gated sodium channel. J. Am. Chem. Soc. 2012;134:1840–1846. doi: 10.1021/ja210020h. [DOI] [PubMed] [Google Scholar]
- 33.Furini S., Domene C. On conduction in a bacterial sodium channel. PLOS Comput. Biol. 2012;8:e1002476. doi: 10.1371/journal.pcbi.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu H., Shen R., Guo W. Ion solvation and structural stability in a sodium channel investigated by molecular dynamics calculations. BBA-Biomembranes. 2012;1818:2529–2535. doi: 10.1016/j.bbamem.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X., Xia M., Gong H. Analysis of the selectivity filter of the voltage-gated sodium channel Na(v)Rh. Cell Res. 2013;23:409–422. doi: 10.1038/cr.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ke S., Zangerl E.M., Stary-Weinzinger A. Distinct interactions of Na(+) and Ca(2+) ions with the selectivity filter of the bacterial sodium channel Na(V)Ab. Biochem. Biophys. Res. Commun. 2012;430:1272–1276. doi: 10.1016/j.bbrc.2012.12.055. [DOI] [PubMed] [Google Scholar]
- 37.Corry B. Na+/Ca2+selectivity in the bacterial voltage-gated sodium channel NavAb. PeerJ. 2013;1:e16. doi: 10.7717/peerj.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zwanzig R.W. High-temperature equation of state by a perturbation method. II. Polar gases. J. Chem. Phys. 1955;23:1915–1922. [Google Scholar]
- 40.Kollman P. Free energy calculations: Applications to chemical and biochemical phenomena. Chem. Rev. 1993;93:2395–2417. [Google Scholar]
- 41.Perter Castro M.H. McGraw-Hill; New York: 2009. Marine Biology. [Google Scholar]
- 42.Donald Voet J.G.V. John Wiley & Sons; New York: 2001. Biochemistry. [DOI] [PubMed] [Google Scholar]
- 43.McCarter L.L. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yorimitsu T., Homma M. Na(+)-driven flagellar motor of Vibrio. Biochim. Biophys. Acta. 2001;1505:82–93. doi: 10.1016/s0005-2728(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 45.Darve E., Pohorille A. Calculating free energies using average force. J. Chem. Phys. 2001;115:9169–9183. [Google Scholar]
- 46.Darve E., Wilson M., Pohorille A. Calculating free energies using a scaled-force molecular dynamics algorithm. Mol. Simul. 2002;28:113–144. [Google Scholar]
- 47.Rodriguez-Gomez D., Darve E., Pohorille A. Assessing the efficiency of free energy calculation methods. J. Chem. Phys. 2004;120:3563–3578. doi: 10.1063/1.1642607. [DOI] [PubMed] [Google Scholar]
- 48.Darve E., Rodríguez-Gómez D., Pohorille A. Adaptive biasing force method for scalar and vector free energy calculations. J. Chem. Phys. 2008;128:144120. doi: 10.1063/1.2829861. [DOI] [PubMed] [Google Scholar]
- 49.Reference deleted in proof.
- 50.Bostick D.L., Brooks C.L., 3rd Selectivity in K+ channels is due to topological control of the permeant ion’s coordinated state. Proc. Natl. Acad. Sci. USA. 2007;104:9260–9265. doi: 10.1073/pnas.0700554104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas M., Jayatilaka D., Corry B. The predominant role of coordination number in potassium channel selectivity. Biophys. J. 2007;93:2635–2643. doi: 10.1529/biophysj.107.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas M., Jayatilaka D., Corry B. Mapping the importance of four factors in creating monovalent ion selectivity in biological molecules. Biophys. J. 2011;100:60–69. doi: 10.1016/j.bpj.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas M., Jayatilaka D., Corry B. How does overcoordination create ion selectivity? Biophys. Chem. 2013;172:37–42. doi: 10.1016/j.bpc.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Noskov S.Y., Bernèche S., Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 55.Eisenman G., Sandblom J., Neher E. Interactions in cation permeation through the gramicidin channel. Cs, Rb, K, Na, Li, Tl, H, and effects of anion binding. Biophys. J. 1978;22:307–340. doi: 10.1016/S0006-3495(78)85491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Illingworth C.J., Domene C. Many-body effects and simulations of potassium channels. Proc. R. Soc. A. 2009;465:1701–1716. [Google Scholar]
- 57.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 58.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


