Summary
Cell–cell interactions define a quintessential aspect of multicellular development. Metazoan morphogenesis depends on a handful of fundamental, conserved cellular interaction mechanisms, one of which is defined by the Notch signaling pathway. Signals transmitted through the Notch surface receptor have a unique developmental role: Notch signaling links the fate of one cell with that of a cellular neighbor through physical interactions between the Notch receptor and the membrane-bound ligands that are expressed in an apposing cell. The developmental outcome of Notch signals is strictly dependent on the cellular context and can influence differentiation, proliferation and apoptotic cell fates. The Notch pathway is conserved across species (Artavanis-Tsakonas et al., 1999; Bray, 2006; Kopan and Ilagan, 2009). In humans, Notch malfunction has been associated with a diverse range of diseases linked to changes in cell fate and cell proliferation including cancer (Louvi and Artavanis-Tsakonas, 2012). In this Cell Science at a Glance article and the accompanying poster we summarize the molecular biology of Notch signaling, its role in development and its relevance to disease.
Introduction
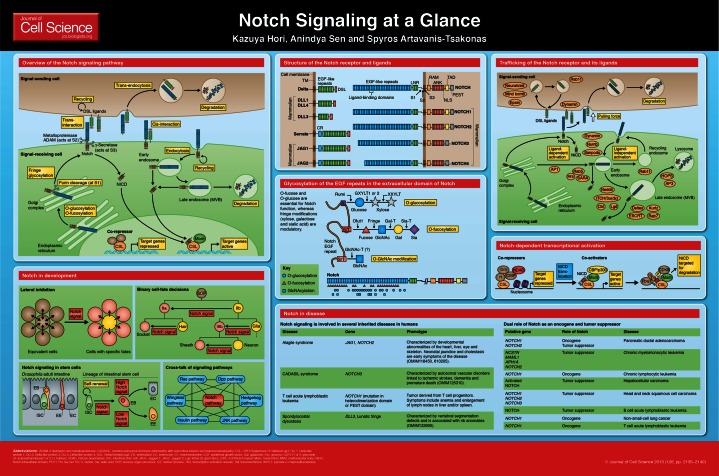
The functional Notch receptor is expressed on the cell surface as a processed heterodimer resulting from a Furin-dependent cleavage (S1 cleavage) in the Notch extracellular domain (NECD), which occurs during trafficking through the Golgi complex (Logeat et al., 1998) (see poster). The NECD undergoes O-linked glycosylation during Notch synthesis and secretion, which is crucial for proper folding of the Notch receptor and the interaction with its ligand DSL (Delta, Serrate, Lag-2) (Rana and Haltiwanger, 2011). The Notch receptor on the signal-receiving cell binds directly to ligands located on the apposing signal-sending cell (Bray, 2006; Kopan and Ilagan, 2009). Receptor–ligand engagement triggers a second NECD cleavage (S2 cleavage) by a metalloproteinase ADAM (known as Kuzbanian in Drosophila melanogaster), which in turn facilitates a further crucial signaling cleavage within the Notch transmembrane domain by a γ-secretase complex that contains Presenilin (S3 cleavage) (Struhl and Greenwald, 1999; Brou et al., 2000); this then releases the Notch intracellular domain (NICD) from the membrane. The NECD is trans-endocytosed into the signal-sending cell together with its ligand (Gordon et al., 2008). DSL ligands can also be cleaved by a metalloproteinase ADAM, which in turn downregulates ligand activity (Zolkiewska, 2008).
Trans-interactions between ligand and receptor that are expressed in apposing cells define activating events, whereas cis-interactions – between the receptor and a ligand that is expressed in the same cell – are inhibitory in nature (Sprinzak et al., 2010; del Álamo et al., 2011). The molecular biology of cis-interactions is still being elucidated, but the interplay between cis- and trans-interactions is a crucial factor that distinguishes the signal-receiving cell from the signal-sending cell, an important decision for development and disease. In addition to cis–trans interactions, this crucial fate-determination step is also regulated by the ratio of ligand-competent receptors to receptor-competent ligands on the cell surface.
The released NICD translocates directly to the nucleus, where it forms a transcriptional complex with the DNA-binding protein CSL (CBF1, Suppressor of Hairless, Lag1), Mastermind (Mam) and transcriptional co-activators to drive the expression of Notch target genes (Bray, 2006; Kopan and Ilagan, 2009). In the absence of NICD, CSL forms complexes with a variety of co-repressors to suppress the transcription of Notch target genes (Bray, 2006; Kopan and Ilagan, 2009) (see poster).
Endocytosis and endosomal processing at different stages of Notch trafficking have been shown to have important and complex roles in regulating the activity of Notch signaling (Fortini, 2009; Yamamoto et al., 2010). Notably, endocytic trafficking has not only been implicated in ligand-dependent signaling (Coumailleau et al., 2009), but it has also been shown that within endocytic compartments the receptor can, under certain circumstances, be activated in a ligand-independent fashion. The physiological significance of such phenomena remains to be elucidated (Vaccari et al., 2008; Wilkin et al., 2008; Vaccari et al., 2009; Hori et al., 2011).
Structures of Notch and its ligands
Notch structure
Notch receptors are multidomain proteins, and have been conserved from invertebrates to humans. The poster panel ‘Structure of the Notch receptor and ligands’ shows representations of the Drosophila Notch receptor and the four mammalian Notch receptors (NOTCH 1, 2, 3 and 4). The NECD consists of 29 to 36 epidermal growth factor (EGF)-like repeats, which are post-translationally modified by a variety of glycans and have been implicated in Notch function (Rana and Haltiwanger, 2011); most notably, the EGF-like repeats 11–12 have been shown to be necessary and sufficient for receptor-ligand interactions (Rebay et al., 1991). The NECD is followed by the negative regulatory region (NRR), which is composed of the three cysteine-rich LNR Notch repeats and the heterodimerization domain. The NRR has been reported to prevent the access of metalloproteinases to the S2 cleavage site of Notch in the absence of ligand (Bray, 2006; Kopan and Ilagan, 2009). The NICD consist of a RAM domain, ankyrin (ANK) repeats flanked by two nuclear localization signals (NLS), a transcriptional activation domain (TAD) and a C-terminal Pro Glu Ser Thr (PEST) domain. The RAM and ANK domains are essential for interacting with CSL in the nucleus.
Notch ligand structure
The DSL ligands of the Notch receptors have been also conserved throughout evolution (D’Souza et al., 2008). Drosophila Notch has two DSL ligands, Delta and Serrate, whereas there are five mammalian ligands, three of which belong to the Delta-like family (DLL1, DLL3 and DLL4) and two belong to the Jagged family of Serrate homologs, Jagged 1 and 2 [also known as JAG1 and JAG2, respectively (see poster)]. DSL ligands are transmembrane proteins with an extracellular domain that contains a characteristic number of EGF-like repeats and a cysteine-rich N-terminal DSL domain. The DSL domain is a conserved motif that is found in all DSL ligands and is required for their interaction with Notch. Serrate, Jagged 1 and Jagged 2 contain an additional cysteine-rich domain. In contrast to the canonical DSL ligands, non-canonical ligands lack the DSL domain and comprise a group of structurally diverse proteins, which includes integral and glycosylphosphatidylinositol (GPI)-linked membrane proteins, and are presumed to modulate Notch receptor activity (D’Souza et al., 2010).
Glycosylation of Notch
The EGF repeats of Notch are subjected to three types of O-linked modification: O-glucosylation, O-fucosylation and O-GlcNAc addition (Rana and Haltiwanger, 2011). These post-translational modifications regulate Notch activity during its synthesis and secretion (Kopan and Ilagan, 2009). An O-fucosyltransferase, which is encoded by O-fut1 (POFUT1 in mammal), adds O-fucose to several Notch EGF-like repeats that harbor the consensus C2-x-x-x-x-(S/T)-C3 motif (Okajima and Irvine, 2002). It has also been shown that O-FUT1 functions as a chaperone to promote the folding and/or export of Notch to the plasma membrane (Okajima et al., 2005; Sasamura et al., 2007). The β1,3-N-acetylglucosaminyltransferase Fringe (the three mammalian paralogs are known as Lunatic, Radical and Manic fringe) catalyzes the addition of N-acetylglucosamine (GlcNAc) to the primary O-fucose (Rana and Haltiwanger, 2011). These modifications by Fringe alter the responsiveness of the Notch receptor to DSL ligands in a context-specific manner (Panin et al., 1997). Further elongation of the sugar modification with a galactose and sialic acid has been documented for mammalian Notch, but not in Drosophila (Rana and Haltiwanger, 2011). Rumi, an endoplasmatic reticulum protein, adds O-glucose to serine at the consensus sequence C1-x-S-x-P-C2 (Acar et al., 2008; Rana et al., 2011), and the O-glucose can be elongated by the addition of xylose (Rana and Haltiwanger, 2011).
Regulation of Notch signaling by membrane trafficking
Notch trafficking
Membrane trafficking has been shown to have an important and indeed complex function in the activation and regulation of Notch signaling (Yamamoto et al., 2010; Baron, 2012). The first evidence came from genetic studies on Drosophila shibire, which encodes the GTPase Dynamin, a key regulator of endocytosis (Seugnet et al., 1997). Classical genetic analysis demonstrated a requirement of Dynamin in both the signal-sending and the signal-receiving cell (Seugnet et al., 1997). Further studies, mostly in flies, identified several factors that modulate the membrane trafficking of Notch and its ligands. Notably, such membrane trafficking events have not only been associated with ligand-dependent activation of Notch but also with its ligand-independent activation, a phenomenon that is still not well understood. In either case, the activation is dependent on the S3 cleavage by γ-secretase (Struhl and Greenwald, 1999; Wilkin et al., 2008; Coumailleau et al., 2009).
Several proteins have been involved in the early steps of endocytosis – including Dynamin (as mentioned above), Numb, a cytoplasmic protein and Sanpodo, a multipass transmembrane protein that is localized to the cell membrane – and are required in the signal-receiving cell for Notch activation (Fortini, 2009). In asymmetric cell divisions, which are crucial for the differentiation of sensory organ precursors (SOPs), the interplay between Numb and Sandopo guides the asymmetric endocytic trafficking of the Notch receptor, which determines cell fate (Berdnik et al., 2002; Hutterer and Knoblich, 2005; Babaoglan et al., 2009). In this process, elegant work demonstrated that the receptor is activated in a subpopulation of Rab5-positive early endosomes, which are marked by SARA, an adaptor protein (Coumailleau et al., 2009). The SARA-containing endosomes comprise Notch and Delta, and also possess γ-secretase activity that can generate the NICD and hence activate the receptor (Coumailleau et al., 2009) (see poster).
Upon endocytosis, internalized Notch can recycle back to the plasma membrane or can be sorted to multivesicular bodies (MVBs) or late endosomes (Yamamoto et al., 2010; Baron, 2012). Sorting of Notch into MVBs or late endosomes is regulated by the ESCRT (endosomal sorting complex required for transport) complexes, which consist of ESCRT-0 and Hrs, ESCRT-I, -II, -III, and Vps4 (Henne et al., 2011). Loss of ESCRT function can result in the ectopic activation of Notch signaling in a ligand-independent manner (Vaccari et al., 2008; Vaccari et al., 2009). Lethal (2) giant discs (lgd), which encodes a C2-containing phospholipid-binding protein, interacts with the ESCRT-III complex and regulates ligand-independent activation of Notch (Childress et al., 2006; Gallagher and Knoblich, 2006; Jaekel and Klein, 2006; Troost et al., 2012).
Ubiquitylation of Notch also has an important function in the regulation of Notch trafficking. A number of E3 ubiquitin ligases, including Suppressor of deltex [Su(dx)] (ITCH in mammals), and Nedd4, Cbl, have been shown to target Notch for degradation (Sakata et al., 2004; Wilkin et al., 2004; Jehn et al., 2002). Deltex (Dx) is a RING-finger E3 ubiquitin ligase that regulates the late-endosomal activation of Notch in a ligand-independent manner (Hori et al., 2004; Hori et al., 2005). However, when Dx forms a complex with Kurtz (Krz), a non-visual β-arrestin, it promotes the degradation of Notch (Mukherjee et al., 2005). This degradation of Notch by Dx and Krz is suppressed in the mutant form of Vps32, a component of the ESCRT-III complex (Hori et al., 2011). Thus, Dx can regulate Notch signaling in either a positive or a negative manner, depending on its interactions with other regulatory factors. Other factors that are involved in trafficking or fusion between late endosomes and lysosomes, such as the HOPS and AP3 complexes, are also required for Dx-dependent activation of Notch (Wilkin et al., 2008). Finally, the clathrin adaptor AP1 complex, which localizes to the trans-Golgi network and to recycling endosomes, has also been shown to regulate the trafficking of Notch during SOP (Benhra et al., 2011) and eye (Kametaka et al., 2012) development in Drosophila.
It has become clear that the mechanisms underlying Notch trafficking through the endosomal pathway are complex. This is reflected by the fact that genetic analysis of the various trafficking elements reveals several distinct phenotypic characteristics that influence Notch signaling in a strictly context-dependent manner (Hori et al., 2011).
DSL ligand trafficking
Endocytosis and endosomal trafficking of DSL ligands within the signal-sending cell are also essential requirements for activation of the Notch receptor (Yamamoto et al., 2010; Baron, 2012). Two RING-type E3 ligases, Neuralized (Neur) and Mindbomb (Mib), promote ligand endocytosis by ubiquitylation (Le Borgne et al., 2005; Wang and Struhl, 2005). Epsin, a ubiquitin-binding protein that interacts with phosphoinositol lipids and endocytic proteins, such as clathrin and AP-2, has been demonstrated to regulate ligand endocytosis (Wang and Struhl, 2004). Rab11-positive recycling endosomes have been shown to be required for Delta activity during SOP development in Drosophila (Emery et al., 2005). Recent studies show that ligand recycling does not change the bond strength of the ligand receptor, but appears to regulate the accumulation of ligands at the cell surface (Shergill et al., 2012). Following interactions with Notch on the surface of adjacent cells, a second endocytic event has been proposed that generates a ‘pulling force’ to activate Notch proteolysis, subsequently triggering trans-endocytosis of the NECD into the signal-sending cell, thereby releasing NICD in the signal-receiving cell (Tien et al., 2009; Musse et al., 2012).
Nuclear events in Notch signaling
The core components of the nuclear Notch complex include NICD, CSL and Mam (Bray, 2006; Kovall and Blacklow, 2010). In the absence of NICD, CSL is associated with co-repressors, such as CtBP, Hairless, Groucho, SMRT, SHRP (also known as Sav), MINT (also known as X11L) and SPEN (Schweisguth and Posakony, 1994; Zhou and Hayward, 2001; Barolo et al., 2002; Oswald et al., 2005). These co-repressors recruit histone deacetylases (HDACs) and other cofactors, thereby repressing the activation of Notch target genes until the NICD is presented in the nucleus (Hsieh et al., 1999; Zhou et al., 2000; Nagel et al., 2005). Upon activation of the Notch receptor, the released NICD moves from the cytoplasm to the nucleus. Here, the RAM domain of NICD allows its interaction with CSL, which facilitates the binding of MAM at the interface between the ANK domain of NICD and CSL (Choi et al., 2012). This complex further recruits coactivators, such as histone acetyltransferases (CBP/p300) and chromatin remodeling complexes, which mediate the transcription of Notch target genes (Wallberg et al., 2002; Kadam and Emerson, 2003; Gause et al., 2006). Kinases, such as glycogen synthase kinase 3β (GSK3β), granulocyte colony stimulating factor (G-CSF) and cyclin C/cyclin-dependent kinase-8 (CDK8), have been shown to phosphorylate NICD, which is thought to be important for the stability of NICD and hence its activity (Inglés-Esteve et al., 2001; Espinosa et al., 2003; Fryer et al., 2004). The E3 ubiquitin ligase SEL10 (also known as FBXW7) modifies NICD and targets it for proteasomal degradation (Gupta-Rossi et al., 2001; Tsunematsu et al., 2004).
The role of Notch in development
Notch function during development
Notch signaling is remarkably pleiotropic and there is hardly a tissue that is not affected by cell fate choices that are regulated by Notch signaling. The developmental consequence of either down- or upregulating of Notch signaling is strictly context-specific, and the same Notch signal might, for instance, in one context promote proliferation, whereas in another result in apoptosis. Hence, the way in which Notch signaling is integrated with other signaling pathways in the context of a particular cellular physiology, dictates how Notch activity affects cell fate. In spite of this spatial and temporal complexity, lateral specification events are the hallmark of Notch-regulated cell fate (Greenwald, 1998). Lateral specification (a more general and accurate term than ‘lateral inhibition’, which is often used when referring to Notch) describes the mechanism by which cells that are in the process of adopting one particular cell fate influence the fate of a cellular neighbor (see poster). Nevertheless, many lineages rely on Notch-mediated lateral inhibition, as is the case of neuroblast differentiation within the neurogenic region of the Drosophila embryo, the classic example of the developmental action of Notch. In this case, a cell that is about to become a neuroblast inhibits its immediate neighbors from adopting the same fate. In terms of morphogenesis, this has two important implications: Notch helps in the segregation of specific lineages from a field of developmentally equivalent cells, and Notch signaling is also one of several crucial mechanisms that specifies borders between cellular fields (Bray, 2006).
An analogous function of Notch has been documented during the differentiation of the peripheral nervous system in Drosophila, where the analysis of the SOP cell lineage has provided a useful model for understanding the role of Notch signaling in binary cell-fate decisions (see poster). After SOP cell division, one of the daughter cells becomes the signal-sending cell, whereas the other cell becomes the signal-receiving cell, leading to asymmetric activation of Notch signaling. The progeny again uses the Notch pathway in a second round of cell division to establish different cell fates (socket, hair, sheath and neuron) (Bray, 2006). As alluded to above, Notch signaling can also contribute to the establishment of boundaries between distinct cell populations to segregate the two groups of cells (see poster).
Notch in stem cells
In general, activation of the Notch signaling pathway is associated with early cell lineages in development, making Notch a very good marker for lineage tracing (Fre et al., 2011). Notch signaling has been implicated in the regulation of an increasing number of stem cells in many different tissues, leading to it being characterized as a ‘stem cell pathway’ (Liu et al., 2010). It is not possible to review the spectrum of stem cell systems, in which Notch has been implicated, in the context of this article. However, we need to emphasize that Notch activity is pervasive and important for the differentiation, maintenance and proliferation of stem cells in almost every system examined, including stem cell lineages, intestines, haematopoietic system, germline, various epithelia, muscle, mesenchyme, nervous system, and others (Liu et al., 2010; Perdigoto and Bardin, 2013; Kageyama et al., 2010; Conboy and Rando, 2002; Conboy et al., 2003; Carlson et al., 2008).
As an example, the adult Drosophila midgut contains intestinal stem cells (ISCs), which divide to self-renew and to produce committed progenitor cells called enteroblasts (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006) (see poster). The enteroblasts further differentiate terminally into absorptive enteroctye cells and secretory enteroendocrine cells (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Notch signaling has been shown to mediate asymmetric cell divisions of ISCs in the adult midgut in Drosophila. Delta is highly expressed in ISCs (Ohlstein and Spradling, 2007) and induces enteroblasts to differentiate to the enteroctye cell fate, whereas its presence at only low levels has been proposed to promote the secretory enteroendocrine cell fate (Ohlstein and Spradling, 2007). The fucosylation of Notch has been implicated in the ISC commitment (Perdigoto et al., 2011).
Proliferation and apoptosis
Notch signaling has been shown to direct cells into proliferative or apoptotic states in a context-specific manner (Pallavi et al., 2012). Interestingly, Notch has both cell-autonomous and non-cell-autonomous effects on mitotic activity, which it can either promote or suppress depending on the cellular context. Although many aspects of Notch signaling in proliferation and apoptosis remain poorly understood, its potential to link these events to differentiation might be of particular relevance to dysproliferative states, including cancer (Bray, 2006; Fiúza and Arias, 2007; Bray and Bernard, 2010).
The developmental context of a cell appears to dictate how activation of Notch affects the cell cycle. The underlying basis appears complex, as, for instance, activation of Notch signaling can have either oncogenic or tumor-suppressive effects in tumors of the same type. As is the case in stem cell differentiation, this apparent complexity is likely to be the result of crosstalk between Notch signaling and other signaling pathways (Hurlbut et al., 2007; Koch and Radtke, 2010; Louvi and Artavanis-Tsakonas, 2012; Colombo et al., 2013).
The role of Notch signaling in disease
Given the profound and widespread roles of Notch signaling across a range of tissues, it is perhaps no surprise that abnormal Notch signaling is involved in several inherited diseases in humans, notably cancer. The link between Notch signaling and disease has been reviewed recently (Gridley, 2003; Fiúza and Arias, 2007; Louvi and Artavanis-Tsakonas, 2012; Penton et al., 2012); the tables shown on the poster summarize the inherited syndromes that are associated with abnormal Notch signaling and emphasize the highly pleiotropic nature of Notch, as these syndromes present a broad range of clinical symptoms. Notch has been increasingly appreciated to have a major direct and indirect role in cancer. Both solid tumors and leukemias have been associated with Notch where it has been shown, depending on the context, to act both as an oncogene and a tumor suppressor.
Perspectives
Since the discovery of the Notch locus in Drosophila almost 100 years ago, great progress has been made in elucidating the core mechanism of the canonical Notch signaling pathway. Recent genome-scale studies reveal an extraordinarily complex network of genes that can affect Notch activity in Drosophila (Guruharsha et al., 2012), and this cohort of genes is complemented and extended by studies in other organisms. This highly interconnected network contrasts with any conventional view of the Notch signaling pathway as a simple linear sequence of events. Although we now have an unprecedented insight into the way in which such a fundamental signaling mechanism is controlled by the genome, we are faced with serious challenges in analyzing the underlying molecular mechanisms of Notch signal control. However, systems-level approaches should shed light on the complex molecular circuitry that governs this pathway and provide insights into the mechanisms that are relevant to Notch-related pathologies.
Acknowledgments
A short, schematic summary as this one, involving a pathway with so many different developmental actions is inevitably incomplete and hence also somewhat biased, in spite of our best efforts. We apologize to all those colleagues whose important studies could not be covered due to space limitations. We would like to thank Robert A. Obar for critically reading the manuscript.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant numbers NS26084 and CA98402 to S.A.-T.]; a JSPS Postdoctoral Fellowship for Research Abroad (to K.H.); and a Postdoctoral Fellowship from the FSMA (to A.S.). Deposited in PMC for release after 12 months.
A high-resolution version of the poster is available for downloading in the online version of this article at jcs.biologists.org. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.127308/-/DC1
References
- Acar M., Jafar-Nejad H., Takeuchi H., Rajan A., Ibrani D., Rana N. A., Pan H., Haltiwanger R. S., Bellen H. J. (2008). Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132, 247–258 10.1016/j.cell.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 10.1126/science.284.5415.770 [DOI] [PubMed] [Google Scholar]
- Babaoglan A. B., O’Connor-Giles K. M., Mistry H., Schickedanz A., Wilson B. A., Skeath J. B. (2009). Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development 136, 4089–4098 10.1242/dev.040386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S., Stone T., Bang A. G., Posakony J. W. (2002). Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 16, 1964–1976 10.1101/gad.987402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M. (2012). Endocytic routes to Notch activation. Semin. Cell Dev. Biol. 23, 437–442 10.1016/j.semcdb.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Benhra N., Lallet S., Cotton M., Le Bras S., Dussert A., Le Borgne R. (2011). AP-1 controls the trafficking of Notch and Sanpodo toward E-cadherin junctions in sensory organ precursors. Curr. Biol. 21, 87–95 10.1016/j.cub.2010.12.010 [DOI] [PubMed] [Google Scholar]
- Berdnik D., Török T., González-Gaitán M., Knoblich J. A. (2002). The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3, 221–231 10.1016/S1534-5807(02)00215-0 [DOI] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Bray S., Bernard F. (2010). Notch targets and their regulation. Curr. Top. Dev. Biol. 92, 253–275 10.1016/S0070-2153(10)92008-5 [DOI] [PubMed] [Google Scholar]
- Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000). A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 10.1016/S1097-2765(00)80417-7 [DOI] [PubMed] [Google Scholar]
- Carlson M. E., Hsu M., Conboy I. M. (2008). Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature 454, 528–532 10.1038/nature07034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress J. L., Acar M., Tao C., Halder G. (2006). Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr. Biol. 16, 2228–2233 10.1016/j.cub.2006.09.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. H., Wales T. E., Nam Y., O’Donovan D. J., Sliz P., Engen J. R., Blacklow S. C. (2012). Conformational locking upon cooperative assembly of notch transcription complexes. Structure 20, 340–349 10.1016/j.str.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Mirandola L., Platonova N., Apicella L., Basile A., Figueroa A. J., Cobos E., Chiriva-Internati M., Chiaramonte R. (2013). Notch-directed microenvironment reprogramming in myeloma: a single path to multiple outcomes. Leukemia [Epub ahead of print] doi: 10.1038/leu.2013.6 10.1038/leu.2013.6 [DOI] [PubMed] [Google Scholar]
- Conboy I. M., Rando T. A. (2002). The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 3, 397–409 10.1016/S1534-5807(02)00254-X [DOI] [PubMed] [Google Scholar]
- Conboy I. M., Conboy M. J., Smythe G. M., Rando T. A. (2003). Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577. 10.1126/science.1087573 [DOI] [PubMed] [Google Scholar]
- Coumailleau F., Fürthauer M., Knoblich J. A., González-Gaitán M. (2009). Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature 458, 1051–1055 10.1038/nature07854 [DOI] [PubMed] [Google Scholar]
- D’Souza B., Miyamoto A., Weinmaster G. (2008). The many facets of Notch ligands. Oncogene 27, 5148–5167 10.1038/onc.2008.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza B., Meloty-Kapella L., Weinmaster G. (2010). Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol. 92, 73–129 10.1016/S0070-2153(10)92003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Álamo D., Rouault H., Schweisguth F. (2011). Mechanism and significance of cis-inhibition in Notch signalling. Curr. Biol. 21, R40–R47 10.1016/j.cub.2010.10.034 [DOI] [PubMed] [Google Scholar]
- Emery G., Hutterer A., Berdnik D., Mayer B., Wirtz-Peitz F., Gaitan M. G., Knoblich J. A. (2005). Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 122, 763–773 10.1016/j.cell.2005.08.017 [DOI] [PubMed] [Google Scholar]
- Espinosa L., Inglés-Esteve J., Aguilera C., Bigas A. (2003). Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278, 32227–32235 10.1074/jbc.M304001200 [DOI] [PubMed] [Google Scholar]
- Fiúza U. M., Arias A. M. (2007). Cell and molecular biology of Notch. J. Endocrinol. 194, 459–474 10.1677/JOE-07-0242 [DOI] [PubMed] [Google Scholar]
- Fortini M. E. (2009). Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633–647 10.1016/j.devcel.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Fre S., Bardin A., Robine S., Louvard D. (2011). Notch signaling in intestinal homeostasis across species: the cases of Drosophila, Zebrafish and the mouse. Exp. Cell Res. 317, 2740–2747 10.1016/j.yexcr.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Fryer C. J., White J. B., Jones K. A. (2004). Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509–520 10.1016/j.molcel.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Gallagher C. M., Knoblich J. A. (2006). The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev. Cell 11, 641–653 10.1016/j.devcel.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Gause M., Eissenberg J. C., Macrae A. F., Dorsett M., Misulovin Z., Dorsett D. (2006). Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development. Mol. Cell. Biol. 26, 2347–2359 10.1128/MCB.26.6.2347-2359.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon W. R., Arnett K. L., Blacklow S. C. (2008). The molecular logic of Notch signalling – a structural and biochemical perspective. J. Cell Sci. 121, 3109–3119 10.1242/jcs.035683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. (1998). LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 12, 1751–1762 10.1101/gad.12.12.1751 [DOI] [PubMed] [Google Scholar]
- Gridley T. (2003). Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 12 Spec. No 1, R9–R13 10.1093/hmg/ddg052 [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. (2001). Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276, 34371–34378 10.1074/jbc.M101343200 [DOI] [PubMed] [Google Scholar]
- Guruharsha K. G., Kankel M. W., Artavanis-Tsakonas S. (2012). The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 13, 654–666 10.1038/nrg3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W. M., Buchkovich N. J., Emr S. D. (2011). The ESCRT pathway. Dev. Cell 21, 77–91 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Hori K., Fostier M., Ito M., Fuwa T. J., Go M. J., Okano H., Baron M., Matsuno K. (2004). Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 131, 5527–5537 10.1242/dev.01448 [DOI] [PubMed] [Google Scholar]
- Hori K., Fuwa T. J., Seki T., Matsuno K. (2005). Genetic regions that interact with loss- and gain-of-function phenotypes of deltex implicate novel genes in Drosophila Notch signaling. Mol. Genet. Genomics 272, 627–638 10.1007/s00438-004-1098-1 [DOI] [PubMed] [Google Scholar]
- Hori K., Sen A., Kirchhausen T., Artavanis-Tsakonas S. (2011). Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J. Cell Biol. 195, 1005–1015 10.1083/jcb.201104146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. J., Zhou S., Chen L., Young D. B., Hayward S. D. (1999). CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96, 23–28 10.1073/pnas.96.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut G. D., Kankel M. W., Lake R. J., Artavanis-Tsakonas S. (2007). Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 19, 166–175 10.1016/j.ceb.2007.02.012 [DOI] [PubMed] [Google Scholar]
- Hutterer A., Knoblich J. A. (2005). Numb and alpha-Adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 6, 836–842 10.1038/sj.embor.7400500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglés-Esteve J., Espinosa L., Milner L. A., Caelles C., Bigas A. (2001). Phosphorylation of Ser2078 modulates the Notch2 function in 32D cell differentiation. J. Biol. Chem. 276, 44873–44880 10.1074/jbc.M104703200 [DOI] [PubMed] [Google Scholar]
- Jaekel R., Klein T. (2006). The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev. Cell 11, 655–669 10.1016/j.devcel.2006.09.019 [DOI] [PubMed] [Google Scholar]
- Jehn B. M., Dittert I., Beyer S., von der Mark K., Bielke W. (2002). c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J. Biol. Chem. 277, 8033–8040 10.1074/jbc.M108552200 [DOI] [PubMed] [Google Scholar]
- Kadam S., Emerson B. M. (2003). Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11, 377–389 10.1016/S1097-2765(03)00034-0 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Niwa Y., Shimojo H., Kobayashi T., Ohtsuka T. (2010). Ultradian oscillations in Notch signaling regulate dynamic biological events. Curr. Top Dev. Biol. 92, 311–331 10.1016/S0070-2153(10)92010-3 [DOI] [PubMed] [Google Scholar]
- Kametaka S., Kametaka A., Yonekura S., Haruta M., Takenoshita S., Goto S., Waguri S. (2012). AP-1 clathrin adaptor and CG8538/Aftiphilin are involved in Notch signaling during eye development in Drosophila melanogaster. J. Cell Sci. 125, 634–648 10.1242/jcs.090167 [DOI] [PubMed] [Google Scholar]
- Koch U., Radtke F. (2010). Notch signaling in solid tumors. Curr. Top. Dev. Biol. 92, 411–455 10.1016/S0070-2153(10)92013-9 [DOI] [PubMed] [Google Scholar]
- Kopan R., Ilagan M. X. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R. A., Blacklow S. C. (2010). Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 92, 31–71 10.1016/S0070-2153(10)92002-4 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Remaud S., Hamel S., Schweisguth F. (2005). Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 3, e96 10.1371/journal.pbio.0030096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sato C., Cerletti M., Wagers A. (2010). Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 92, 367–409 10.1016/S0070-2153(10)92012-7 [DOI] [PubMed] [Google Scholar]
- Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., Seidah N. G., Israël A. (1998). The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95, 8108–8112 10.1073/pnas.95.14.8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. (2012). Notch and disease: a growing field. Semin. Cell Dev. Biol. 23, 473–480 10.1016/j.semcdb.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C. A., Perrimon N. (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Veraksa A., Bauer A., Rosse C., Camonis J., Artavanis-Tsakonas S. (2005). Regulation of Notch signalling by non-visual beta-arrestin. Nat. Cell Biol. 7, 1191–1201 10.1038/ncb1327 [DOI] [PubMed] [Google Scholar]
- Musse A. A., Meloty-Kapella L., Weinmaster G. (2012). Notch ligand endocytosis: mechanistic basis of signaling activity. Semin. Cell Dev. Biol. 23, 429–436 10.1016/j.semcdb.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel A. C., Krejci A., Tenin G., Bravo-Patiño A., Bray S., Maier D., Preiss A. (2005). Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 25, 10433–10441 10.1128/MCB.25.23.10433-10441.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988–992 10.1126/science.1136606 [DOI] [PubMed] [Google Scholar]
- Okajima T., Irvine K. D. (2002). Regulation of notch signaling by o-linked fucose. Cell 111, 893–904 10.1016/S0092-8674(02)01114-5 [DOI] [PubMed] [Google Scholar]
- Okajima T., Xu A., Lei L., Irvine K. D. (2005). Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science 307, 1599–1603 10.1126/science.1108995 [DOI] [PubMed] [Google Scholar]
- Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knöchel W., Borggrefe T. (2005). RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 25, 10379–10390 10.1128/MCB.25.23.10379-10390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallavi S. K., Ho D. M., Hicks C., Miele L., Artavanis-Tsakonas S. (2012). Notch and Mef2 synergize to promote proliferation and metastasis through JNK signal activation in Drosophila. EMBO J. 31, 2895–2907 10.1038/emboj.2012.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin V. M., Papayannopoulos V., Wilson R., Irvine K. D. (1997). Fringe modulates Notch-ligand interactions. Nature 387, 908–912 10.1038/43191 [DOI] [PubMed] [Google Scholar]
- Penton A. L., Leonard L. D., Spinner N. B. (2012). Notch signaling in human development and disease. Semin. Cell Dev. Biol. 23, 450–457 10.1016/j.semcdb.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto C. N., Bardin A. J. (2013). Sending the right signal: Notch and stem cells. Biochim. Biophys. Acta 1830, 2307–2322 10.1016/j.bbagen.2012.08.009 [DOI] [PubMed] [Google Scholar]
- Perdigoto C. N., Schweisguth F., Bardin A. J. (2011). Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development 138, 4585–4595 10.1242/dev.065292 [DOI] [PubMed] [Google Scholar]
- Rana N. A., Haltiwanger R. S. (2011). Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 21, 583–589 10.1016/j.sbi.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana N. A., Nita-Lazar A., Takeuchi H., Kakuda S., Luther K. B., Haltiwanger R. S. (2011). O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J. Biol. Chem. 286, 31623–31637 10.1074/jbc.M111.268243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I., Fleming R. J., Fehon R. G., Cherbas L., Cherbas P., Artavanis-Tsakonas S. (1991). Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67, 687–699 10.1016/0092-8674(91)90064-6 [DOI] [PubMed] [Google Scholar]
- Sakata T., Sakaguchi H., Tsuda L., Higashitani A., Aigaki T., Matsuno K., Hayashi S. (2004). Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr. Biol. 14, 2228–2236 10.1016/j.cub.2004.12.028 [DOI] [PubMed] [Google Scholar]
- Sasamura T., Ishikawa H. O., Sasaki N., Higashi S., Kanai M., Nakao S., Ayukawa T., Aigaki T., Noda K., Miyoshi E. et al. (2007). The O-fucosyltransferase O-fut1 is an extracellular component that is essential for the constitutive endocytic trafficking of Notch in Drosophila. Development 134, 1347–1356 10.1242/dev.02811 [DOI] [PubMed] [Google Scholar]
- Schweisguth F., Posakony J. W. (1994). Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development 120, 1433–1441 [DOI] [PubMed] [Google Scholar]
- Seugnet L., Simpson P., Haenlin M. (1997). Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 192, 585–598 10.1006/dbio.1997.8723 [DOI] [PubMed] [Google Scholar]
- Shergill B., Meloty-Kapella L., Musse A. A., Weinmaster G., Botvinick E. (2012). Optical tweezers studies on Notch: single-molecule interaction strength is independent of ligand endocytosis. Dev. Cell 22, 1313–1320 10.1016/j.devcel.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D., Lakhanpal A., Lebon L., Santat L. A., Fontes M. E., Anderson G. A., Garcia-Ojalvo J., Elowitz M. B. (2010). Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86–90 10.1038/nature08959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Greenwald I. (1999). Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398, 522–525 10.1038/19091 [DOI] [PubMed] [Google Scholar]
- Tien A. C., Rajan A., Bellen H. J. (2009). A Notch updated. J. Cell Biol. 184, 621–629 10.1083/jcb.200811141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troost T., Jaeckel S., Ohlenhard N., Klein T. (2012). The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J. Cell Sci. 125, 763–776 10.1242/jcs.097261 [DOI] [PubMed] [Google Scholar]
- Tsunematsu R., Nakayama K., Oike Y., Nishiyama M., Ishida N., Hatakeyama S., Bessho Y., Kageyama R., Suda T., Nakayama K. I. (2004). Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279, 9417–9423 10.1074/jbc.M312337200 [DOI] [PubMed] [Google Scholar]
- Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008). Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180, 755–762 10.1083/jcb.200708127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Rusten T. E., Menut L., Nezis I. P., Brech A., Stenmark H., Bilder D. (2009). Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J. Cell Sci. 122, 2413–2423 10.1242/jcs.046391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A. E., Pedersen K., Lendahl U., Roeder R. G. (2002). p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 22, 7812–7819 10.1128/MCB.22.22.7812-7819.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Struhl G. (2004). Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development 131, 5367–5380 10.1242/dev.01413 [DOI] [PubMed] [Google Scholar]
- Wang W., Struhl G. (2005). Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132, 2883–2894 10.1242/dev.01860 [DOI] [PubMed] [Google Scholar]
- Wilkin M. B., Carbery A. M., Fostier M., Aslam H., Mazaleyrat S. L., Higgs J., Myat A., Evans D. A., Cornell M., Baron M. (2004). Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 14, 2237–2244 10.1016/j.cub.2004.11.030 [DOI] [PubMed] [Google Scholar]
- Wilkin M., Tongngok P., Gensch N., Clemence S., Motoki M., Yamada K., Hori K., Taniguchi-Kanai M., Franklin E., Matsuno K. et al. (2008). Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev. Cell 15, 762–772 10.1016/j.devcel.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Charng W. L., Bellen H. J. (2010). Endocytosis and intracellular trafficking of Notch and its ligands. Curr. Top. Dev. Biol. 92, 165–200 10.1016/S0070-2153(10)92005-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Hayward S. D. (2001). Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol. Cell. Biol. 21, 6222–6232 10.1128/MCB.21.18.6222-6232.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Fujimuro M., Hsieh J. J., Chen L., Miyamoto A., Weinmaster G., Hayward S. D. (2000). SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol. Cell. Biol. 20, 2400–2410 10.1128/MCB.20.7.2400-2410.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolkiewska A. (2008). ADAM proteases: ligand processing and modulation of the Notch pathway. Cell. Mol. Life Sci. 65, 2056–2068 10.1007/s00018-008-7586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]